Advances in Microbiology
Vol. 2 No. 4 (2012) , Article ID: 25863 , 7 pages DOI:10.4236/aim.2012.24065
The LysR Transcription Factor, HexS, Is Required for Glucose Inhibition of Prodigiosin Production by Serratia marcescens
1Charles T. Campbell Laboratory of Ophthalmic Microbiology, Department of Ophthalmology, University of Pittsburgh, Pittsburgh, USA
2College of Medicine, University of Vermont, Burlington, USA
Email: stellan@upmc.edu, jef474@yahoo.com, rml40@pitt.edu, eric.kalivoda@uvm.edu, *shanksrm@upmc.edu
Received August 31, 2012; revised October 10, 2012; accepted October 23, 2012
Keywords: Pigment; Antibiotic; Transcription Factor; Secondary Metabolite
ABSTRACT
Generation of many useful microbe-derived secondary metabolites, including the red pigment prodigiosin of the bacterium Serratia marcescens, is inhibited by glucose. In a previous report, a genetic approach was used to determine that glucose dehydrogenase activity (GDH) is required for inhibiting prodigiosin production and transcription of the prodigiosin biosynthetic operon (pigA-N). However, the transcription factor(s) that regulate this process were not characterized. Here we tested the hypothesis that HexS, a LysR-family transcription factor similar to LrhA of Escherichia coli, is required for inhibition of prodigiosin by growth in glucose. We observed that mutation of the hexS gene in S. marcescens allowed the precocious production of prodigiosin in glucose-rich medium conditions that completely inhibited prodigiosin production by the wild type. Unlike previously described mutants able to generate prodigiosin in glucoserich medium, hexS mutants exhibited GDH activity and medium acidification similar to the wild type. Glucose inhibittion of pigA expression was shown to be dependent upon HexS, suggesting that HexS is a key transcription factor in secondary metabolite regulation in response to medium pH. These data give insight into the prodigiosin regulatory pathway and could be used to enhance the production of secondary metabolites.
1. Introduction
The production of many secondary metabolites by microorganisms, such as antibiotics, is inhibited by growth in glucose-rich medium [1,2]. This is unfortunate as glucose is an ideal carbon source for growth of many microorganisms. The Gram-negative bacterium Serratia marcescens is a model to study secondary metabolites because it generates the tractable tripyrrole pigment, prodigiosin [3]. Glucose inhibits prodigiosin production by S. marcescens [4] due to acidification of the medium [1,2], but the mechanism of this inhibition is incompletely understood.
Fender, et al., recently identified genes required for glucose dehydrogenase (GDH)-dependent inhibition of prodigiosin by medium acidification [5]. It was demonstrated that GDH activity in glucose rich medium is necessary for the rapid culture pH reduction, resulting in a complete inhibition of prodigiosin production. Expression of the prodigiosin biosynthetic operon, pigA-N [6,7] was inhibited by glucose-induced acidification, suggesting that prodigiosin inhibition is largely controlled at the transcriptional level. The transcription factors responsible for this regulation are unknown. However, a clue to the identification of the unknown factor(s) was revealed in a recent genetic screen [5]. Fender and colleagues observed that mutation in the gene that codes for the LysR-family transcription factor, HexS, allowed the bacterium to generate prodigiosin in glucose-rich medium in a crp mutant strain. However, the impact of mutation of hexS on glucose inhibition of secondary metabolites was not characterized and the phenotype was not verified in a strain with a functional CRP protein.
The goal of this study was to determine whether the transcription factor HexS is necessary for glucose-mediated inhibition of secondary metabolite production, using prodigiosin as a model secondary metabolite.
HexS was previously found in a genetic screen for genes that control production of the secondary metabolites, serratamolide and prodigiosin, and was also shown to directly regulate the prodigiosin biosynthetic operon [8]. The only other report featuring HexS indicates that it negatively regulates production of the antimicrobial biosurfactant serratamolide [9]. HexS is highly similar LrhA of Escherichia coli [10] and is well conserved among the Enterobacteriaceae. HexS is also similar to HexA proteins from Pectobacterium and Photorhabdus species, and PigU from Serratia sp. ATCC 39006, which are regulators of secreted enzymes and metabolites [11- 14]. LrhA regulates a number of processes including motility, chemotaxis, and aspects of stationary phase physiology partially through indirect control of sigma factor S [10].
This brief report provides genetic and biochemical evidence that HexS is necessary for glucose to inhibit prodigiosin production and transcription from the prodigiosin biosynthetic operon by S. marcescens.
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
All microorganisms are listed in Table 1 and were grown in lysogeny broth (LB) [15,16] with or without D-glucose (110 mM), with the exception of Saccharomyces cerevisiae which was grown with SC-uracil or YPD broth [17]. Cultures were grown in 5 mL aliquots of medium and rotated at high speed on a TC-7 tissue culture rotor (New Brunswick Scientific). Antibiotics used were tetracycline (10 µg/mL), kanamycin (100 µg/mL) and gentamicin (10 µg/mL). Conjugations were performed using Escherichia coli donor strain S17-1 as previously described [5]. Time course experiments were performed as previously described [5].
2.2. Cloning and Genetic Manipulations
The hexS open reading frame (ORF) was cloned using primers gaattgtgagcggataacaatttcacacaggaaac agctATGACAACTGCAAATCGTCCG (uppercase describes priming DNA, and lowercase represents recombination targeting sequence) and gttttatcagaccgcttctgcgttctgatttagTTATTCTTCTTCGTCCACCAGG CTGGC using yeast in vivo cloning [18] into integrative plasmid pMQ236 [19] to generate pMQ294. The hexS-∆1 allele was made by

Table 1. Strains and plasmids used in this study.
digesting pMQ294 with SacII, which cuts at a single site in the hexS ORF. The SacII digested plasmid was treated with the End-IT kit (Epicentre) to generate blunt ends and the plasmid was recircularized using T4 DNA ligase (New England Biolabs). The resulting plasmid, pMQ296, contained a hexS gene with a one base pair deletion (base pair 670 out of a total of 945 for the gene) creating a frame shift mutation and truncation.
For the insertional mutagenesis of alaT, hexS and nuoC, internal fragments were amplified using primers tgggtaacgccagggttttcccagtcacgacgttgtaCCGTTACGACATGCATTTGCACC and gataacaatttcacacaggaaacag ctatgaccatgatGGACGATCAGCTCAGAGACGC for alaT, gttgggtaacgccagggttttcccagtcacgacgttgtaCGCGCA CGTCAATGGCCAGC and taacaatttcacacaggaaacagctatgaccatgatCCTCGATCTGCTAAGAACCTTTG for hex S, and gacgttgtaaaacgacgggatctatcatcgtggatccTGGTGATAACCGATGTCCGGCAC and ctagagcggtttcccg actggaaagcgggcagtgagcgcCGAACTGCGCAACCGTTTTGG for nuoC. The amplicons were cloned into pMQ118 or pStvZ3 [19,20] and the resulting plasmids were introduced into S. marcescens by conjugation and selection for kanamycin resistance. The resulting strains have the plasmid inserted before the midpoint of each gene. Mutations were verified by PCR and mutant phenotype.
For purification of recombinant HexS, a N-terminal maltose binding protein (MBP) fusion was created in which the entire hexS open reading frame was fused to the malE gene using pMal-C2 (New England Biolabs). The open reading frame was amplified with primers that incorporated EcoR1 and PstI sites for subsequent digestion and T4-ligase based ligation into the pMal-C2 plasmid polylinker. The resulting construct was verified by sequencing across the malE-hexS junction.
Allelic replacement of hexS with deletion alleles was performed using pMQ296, respectively, with previously described protocols [19], and was verified by PCR, sequencing, and phenotypic analysis. Briefly, pMQ296 was introduced into the recipient strain by conjugation and selection for kanamycin resistance (100 µg/mL) to generate a merodiploid. The I-SceI expressing plasmid pMQ240 was introduced into the merodiploid by conjugation with selection for gentamicin (10 µg/mL). The merodiploid strain with pMQ240 was grown in LB with gentamicin and streaked to single colonies on LB agar plates. Colonies were tested individually for loss of kanamycin resistance, and kanamycin susceptible strains were assessed for deletion of pigA or hexS using PCR. The pigA::lacZ construct was generated using pMQ268 as previously described [20], where the pStvZ3 plasmid bearing an internal fragment of pigA was introduced into recipient strains by conjugation and selection for kanamycin (100 µg/mL).
2.3. Enzymatic Assays and Electrophoretic Mobility Shift Assay (EMSA)
Measurement of β-galactosidase activity was performed as previously described using 200 µg of cell lysate from stationary phase cultures that had grown for 20 hours at 30˚C using o-nitrophenyl β-D-galactopyranoside (ONPG) as a chromogenic substrate [20].
D-gluconic acid concentration was measured using an enzymatic D-gluconic acid detection kit (Megazyme, product K-GATE) as described by the manufacturer, with modifications that were previously described [5].
Glucose dehydrogenase activity (GDH) was measured using a chromogenic reaction based on the method of Matsushita, et al. [21] using 2,6-Dichlorophenol-Indophenol (Sigma, D-1878) and 13 mM phenazine methosulfate (Sigma, P-9625) as substrates as previously described [5].
A maltose binding protein (MBP) fusion of HexS and the MBP alone were purified using the manufacturers specifications (pMal Protein Fusion and Purification System, New England Biolabs). EMSA reactions were performed as previously described [20] using a comercial EMSA kit (Lightshift Chemiluminescent EMSA kit, Pierce, Rockford IL). Briefly, pigA promoter DNA was amplified using biotinylated (or non-biotinylated) oligonucleotide primer 5’-ggggtacccttcacctgcaaagtattcatc-3’ and non-biotinylated primer cggaattcagtatcgagcgcattcatgcc. The resulting amplicon was gel purified and used at 2 ng per binding reaction following the manufacturers specifications. A 10 µL aliquot of the reaction was separated in a TBE PAGE gel (Pierce), transferred to a nylon membrane, and examined through chemiluminescence according to the manufacturers specifications. MBP and MBP-HexS were used at 25 - 30 µg per reaction, and unlabelled promoter DNA was amplified with unlabelled versions of the above listed primers and the amplicon was used at 500 ng per binding reaction where specified. The EMSA reaction was performed three times with consistent results.
3. Results and Discussion
3.1. Mutation of hexS Allows Prodigiosin Production in Glucose Rich Medium
In a recently reported genetic screen, we identified mutants that produced prodigiosin on LB glucose-supplemented agar plates (LBG) unlike the parental strain that was pigmentless on LBG [5]. One mutant strain that was pigmented on LBG agar had a transposon insertion in the gene for the HexS prodigiosin-regulatory transcription factor protein. The importance of HexS in glucose inhibition of prodigiosin by glucose was not verified in that study. Furthermore the previous study was performed using a crp mutant rather than a wild-type strain. The goal of this brief study was to determine whether HexS is necessary to inhibit S. marcescens prodigiosin production in glucose rich medium.
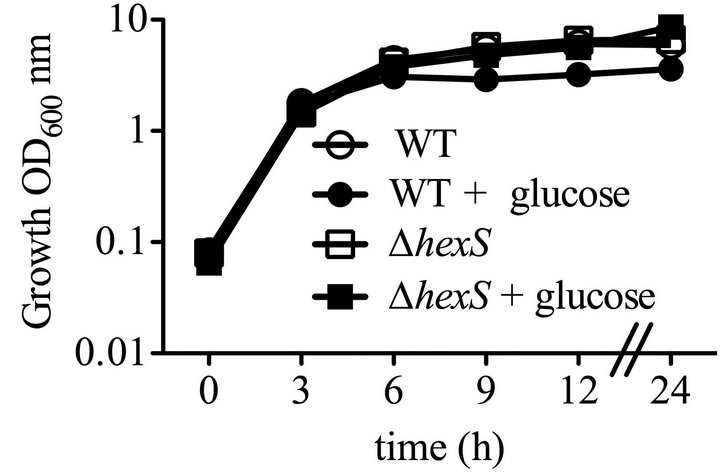
To characterize the effect of the hexS mutant phenoltype in a wild-type (WT strain CMS376) background, we generated a deletion allele hexS-∆1, a directed hexS null-mutant allele to replace the chromosomal hexS gene in the WT (CMS376) strain, generating strain CMS2210, referred to here as ∆hexS. This mutant strain exhibits all of the characteristics of a hexS null mutant strain, such as elevated prodigiosin production and serratamolide production (data not shown). The ∆hexS (CMS2210) mutant and WT (CMS376) grew at the same rate in LB medium (Figure 1(a)). Unlike its growth-inhibitory effect upon the WT (CMS376), glucose did not inhibit culture density of the ∆hexS (CMS2210) mutant (Figure 1(a)). Furthermore, the ∆hexS (CMS2210) strain produced prodigiosin when grown with glucose unlike the WT (CMS376) (Figure 1(b)).
Previously described mutants able to produce pigment in LB medium supplemented with glucose (LBG) had mutations in genes necessary for glucose dehydrogenase activity (GDH), failed to achieve WT levels of medium acidification, and medium acidification was necessary for glucose to prevent prodigiosin production [5]. To test the prediction that hexS mutants are also able to generate pigment in LBG because they fail to acidify the medium, we measured the impact of hexS mutation upon medium acidification. No difference was observed between WT (CMS376) and the ∆hexS (CMS2210) mutant with regards to pH in LB (Figure 1(c)). In LBG, the ∆hexS (CMS2210) mutant differed from the WT (CMS376). Up to hour 6, both the WT (CMS376) and hexS mutant (CMS2210) experienced a sharp drop in pH, but after 6 hours the WT (CMS376) culture continued to decrease below pH 4, whereas the ∆hexS (CMS2210) culture reproducibly stabilized close to pH 5 and then increased to neutrality by t = 24 hours (Figure 1(c)). This result is in stark contrast to all other reported pigment positive mutants that did not support a glucose-induced pH reduction [5]. Interestingly, the HexS deficient strain produced prodigiosin in cultures with very low pH. These are important observations because it shows that prodigiosin can be produced in acidic medium, indicating that the enzymatic process of generating prodigiosin is not simply unable to occur at low pH, and implying that an active inhibition of prodigiosin production takes place in glucose-rich medium. The mechanism by which HexS mediates culture pH in LBG will be the subject of subsequent research.
3.2. Quinoprotein Glucose Dehydrogenase Protein Activity Is Altered in hexS Mutants
Previous work showed that reduced quinoprotein glucose dehydrogenase (GDH) activity, which converts glucose to gluconic acid, conferred the ability to produce prodigiosin in LBG [5]. Therefore, we tested whether the hexS
 (a)
(a)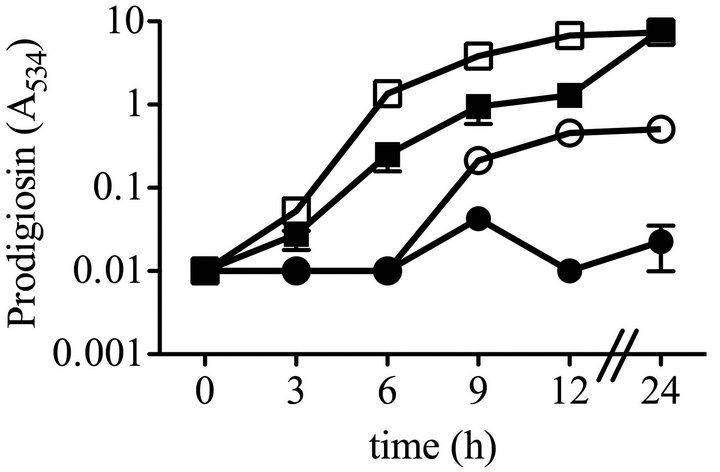 (b)
(b)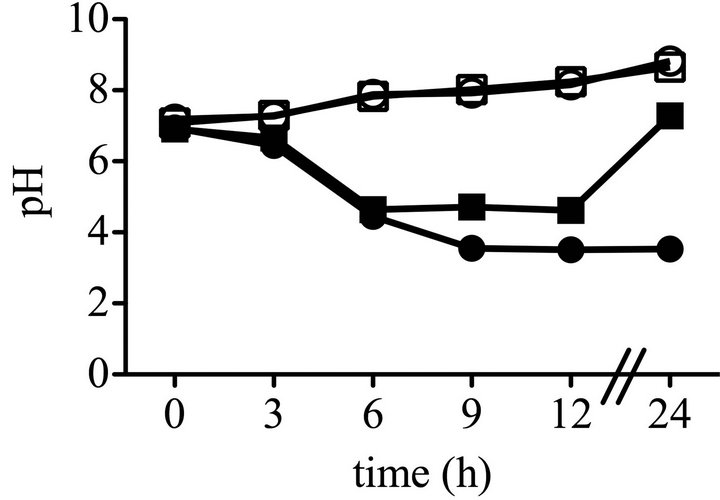 (c)
(c)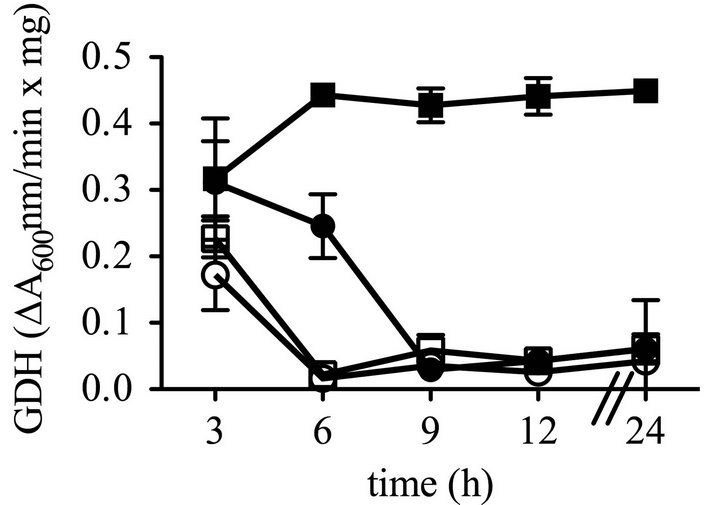 (d) time
(d) time
Figure 1. Growth, prodigiosin production, culture pH of WT and ∆hexS (CMS2210) mutant cells grown in LB and LBG. Genotype symbols are consistent for each panel. Cultures were grown in LB and in LBG (+glucose). The hexS mutant used was ∆hexS (CMS2210). (a) Culture growth as measured by turbidity (OD600 nm); (b) Extracted prodigiosin levels were measured from 1 mL of culture (A534 nm); (c) Culture pH was determined with a pH meter; (d) Glucose dehydrogenase activity measured from crude cellular extracts. Time course experiments were performed with 4 independent biological replicates per time point per condition, and performed three times with consistent results, one representative time course is shown. Error bars indicate one standard deviation.
mutant (CMS2210) would have reduced GDH activity. However, the ∆hexS (CMS2210) mutant surprisingly produced equivalent GDH activity to the WT (CMS376) when grown in LB, and higher levels than the WT (CMS376) at later time points when cultured in LBG (Figure 1(d)) suggesting the GDH activity in LBG is regulated by HexS.
As a second assay for GDH activity, we measured gluconic acid (GA) levels. GA in LBG was maximal for the wild type (CMA376) at 6 - 9 hours [5]. Unexpectedly, gluconic acid levels in the ∆hexS (CMS2210) mutant culture at six hours were significantly reduced compared to the WT (CMS376) (Student’s T-test p < 0.05). In the aforementioned time course (Figure 1) gluconic acid was measured at 12.1 ± 1.9 mM from the ∆hexS (CMS2210) strain, compared 20.2 ± 2.4 mM for the WT (CMS376) at t = 6 hours. A similar trend was observed over the time course (data not shown). These data suggest that the reduced organic acid in ∆hexS (CMS2210) mutant cultures at late time points may account of the increased culture pH at late time points.
3.3. Complementation of the ∆hexS (CMS2210) Phenotype
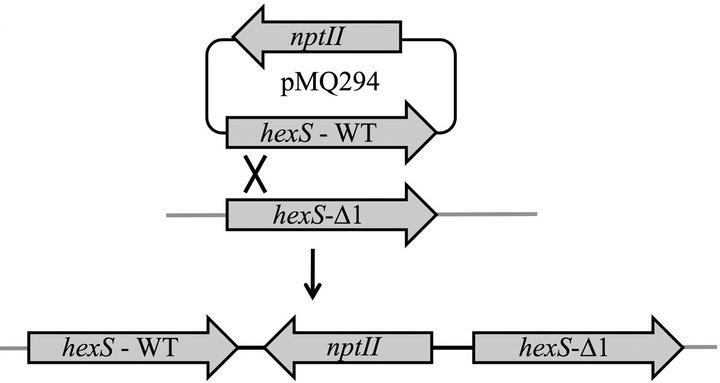
To perform complementation analysis, we introduced a single WT copy of hexS into the ∆hexS (CMS2210) chromosome with pMQ294 (diagramed in Figure 2(a)) and restored the glucose sensitivity phenotype (Figure 2(b)). We measured elevated prodigiosin levels in both the hexS deletion strain and a hexS-1 (CMS1779) insertion mutant strain compared to WT (CMS376). As an additional control, hexS was mutated by plasmid insertion. This hexS-1 (CMS1779) strain served as a control to be an independent mutation to confirm that mutation of hexS conferred the observed pigment phenotypes, and to test whether insertion of a plasmid alone into a hexS mutant (CMS2210) strain would restore sensitivity to glucose. In LBG, the WT (CMS376) and ∆hexS (CMS2210) + pMQ294 results were not significantly different in prodigiosin production, whereas the hexS mutant’s (CMS2210) pigment levels were significantly higher than either WT (CMS376) or ∆hexS (CMS2210) + pMQ294 (ANOVA, p < 0.05, Figure 2). To further ensure that the pigment phenotype of hexS mutants were not due to a polar effect on adjacent genes, mutations were introduced into the operons on either side of hexS on the chromosome. In one direction is a divergently transcribed alaT homolog greater than 650 base pairs away from the hexS gene, and on the opposite side of hexS, transcribed in the same orientation, is a predicted NADH-ubiquinone oxidoreducatase operon (nuoA-N), greater than 700 base pairs away from the hexS ORF. To further support that loss of the glucose inhibition of
 (a)
(a)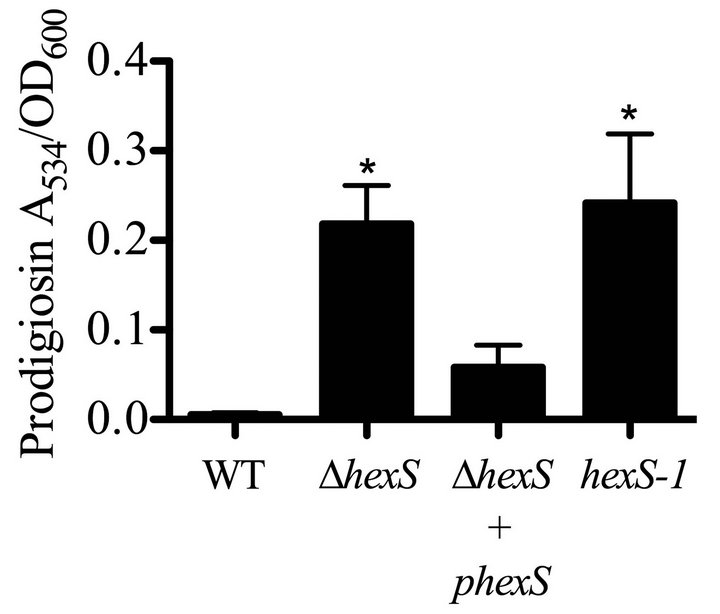 (b)
(b)
Figure 2. Complementation of the hexS mutant phenotype. (a) Complementation strategy through integrating a wildtype hexS gene on pMQ294 into a strain with a hexS deletion mutation (CMS3125). (b) Prodigiosin production in LBG shows that unlike the WT (CMS376), a hexS deletion mutant (∆hexS, CMS2210) and a hexS insertion mutant (hexS-1, CMS1779) were able to generate prodigiosin. A wild-type copy of hexS on a plasmid (pMQ294) was inserted into the chromosome of the ∆hexS (CMS2210) strain and restored glucose sensitivity. Error bars indicate one standard deviation, and asterisks indicate significant differences from the WT (CMS376) (p < 0.05, ANOVA with Tukey’s post-test).
prodigiosin phenotype of hexS mutants was not due to a polar effect on adjacent genes, alaT and nuoC were separately mutated in the WT (CMS376) strain. The resulting mutant strains did not exhibit the pigmented phenotype of the hexS insertion mutant in LBG (data not shown). Together, these data support that HexS is important for glucose inhibition of pigmentation.
3.4. HexS Is Necessary for Glucose-Inhibition of Transcription of the Prodigiosin Biosynthetic Operon
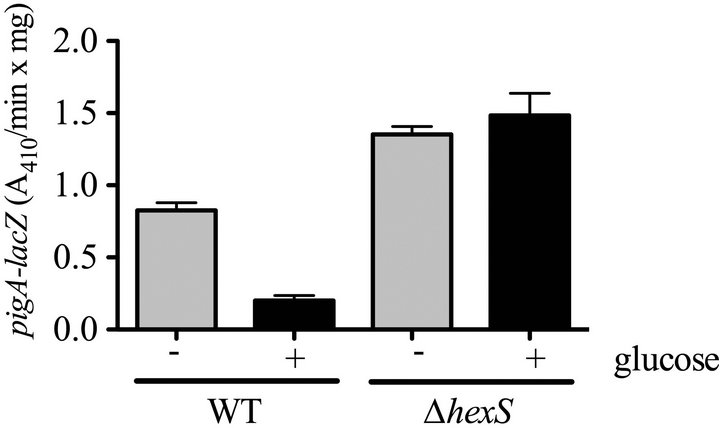
To test the hypothesis that HexS regulates the pigA promoter in LBG, we used a previously characterized chromosomal pigA-lacZ reporter [20]. Whereas pigA-lacZ associated β-galactosidase levels were severely reduced in the WT (CMS376) in LBG compared to LB medium, the ∆hexS (CMS2210) mutant exhibited similar expression levels regardless of glucose (Figure 3(a)). To ensure that this was a direct transcriptional regulation, we con
 (a)
(a)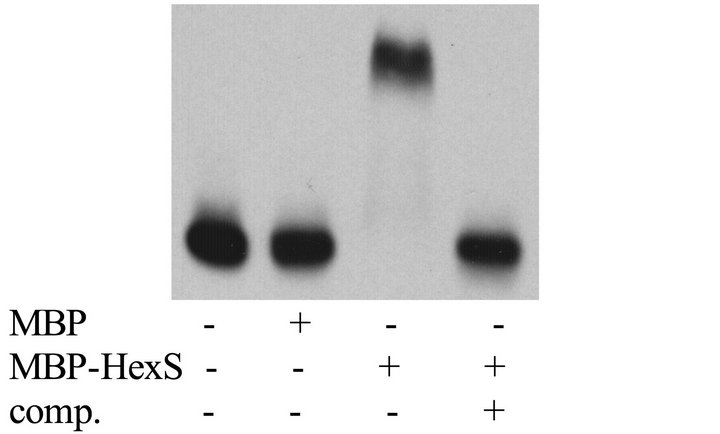 (b)
(b)
Figure 3. Glucose sensitivity of the prodigiosin biosynthetic operon is dependent upon HexS. (a) Expression from the pigA promoter was indirectly measured using a chromosomal lacZ transcriptional fusion. β-galactosidase activity was measured from cells grown to stationary phase (20 h) from culture grown in LB (−) or LBG (+) medium. Error bars indicate one standard deviation; (b) Representative EMSA showing MBP-HexS binding to the biotinylated pigA promoter. MBP = maltose binding protein, MBP-HexS = N-terminal fusion of maltose binding protein to HexS, comp. = unlabelled competitor pigA promoter DNA.
firmed a previous report that the HexS protein binds to pigA promoter DNA in vitro using a recombinant version of HexS in an EMSA assay [8], but using recombinant HexS protein that had been cloned from the CMS376 genome (Figure 3(b)).
The major goal of this study, to test whether HexS is required for inhibition of the secondary metabolite, prodigiosin, by glucose was achieved. Genetic analysis implicated the HexS transcription factor in the glucose inhibition of secondary metabolite phenotype. Interestingly, unlike the other glucose insensitive mutants isolated [5], hexS mutation neither prevented early medium acidification nor prevented glucose oxidation by glucose dehydrogenase.
Together results suggest that HexS transcriptional control of pigA-N is an important factor in inhibiting prodigiosin production in glucose-rich medium. Given that HexS negatively regulates multiple secondary metabolites [8], and the data from this study, we propose that HexS may be the major regulator involved of the inhibition of secondary metabolites by glucose. Future experimentation will be aimed at determining whether HexS activity responds directly to changes in pH. We propose that mutation of LrhA-like regulators may be useful for production of desired secondary metabolites.
4. Acknowledgements
The authors would like to thank Jean-Paul Vergnes, Kip Kinchington, Cody Bender, Aroba Sadaf, and Marissa Aston for technical help, and Pryce Haddix and Kim Brothers for critical manuscript evaluation. This study was funded by AI085570 and a Research to Prevent Blindness Career Development Award to RS. Other funding sources were NIH grant EY08098, the Eye and Ear Foundation of Pittsburgh, and unrestricted funds from Research to Prevent Blindness.
REFERENCES
- M. Sole, A. Francia, N. Rius and J. G. Loren, “The Role of pH in the ‘Glucose Effect’ on Prodigiosin Production by Non-Proliferating Cells of Serratia marcescens,” Letters in Applied Microbiology, Vol. 25, No. 1997, pp. 81- 84.
- M. Sole, N. Rius and J. G. Loren, “Rapid Extracellular Acidification Induced by Glucose Metabolism in NonProliferating Cells of Serratia marcescens,” International Microbiology, Vol. 3, No. 1, 2000, pp. 39-43.
- N. R. Williamson, P. C. Fineran, F. J. Leeper and G. P. Salmond, “The Biosynthesis and Regulation of Bacterial Prodiginines,” Nature Reviews Microbiology, Vol. 4, No. 12, 2006, pp. 887-899. doi:10.1038/nrmicro1531
- M. I. Bunting, C. F. Robinow and H. Bunting, “Factors Affecting the Elaboration of Pigment and Polysaccharide by Serratia marcescens,” Journal of Bacteriology, Vol. 58, No. 1, 1949, p. 114.
- J. E. Fender, C. M. Bender, N. A. Stella, R. M. Lahr, E. J. Kalivoda and R. M. Q. Shanks, “Serratia marcescens Quinoprotein Glucose Dehydrogenase Activity Mediates Medium Acidification and Inhibition of Prodigiosin Production by Glucose,” Applied and Environmental Microbiology, Vol. 78, No. 17, 2012, pp. 6225-6235. doi:10.1128/AEM.01778-12
- S. A. Dauenhauer, R. A. Hull and R. P. Williams, “Cloning and Expression in Escherichia coli of Serratia marcescens Genes Encoding Prodigiosin Biosynthesis,” Journal of Bacteriology, Vol. 158, No. 3, 1984, pp. 1128- 1132.
- A. K. Harris, N. R. Williamson, H. Slater, A. Cox, S. Abbasi, I. Foulds, H. T. Simonsen, F. J. Leeper and G. P. Salmond, “The Serratia Gene Cluster Encoding Biosynthesis of the Red Antibiotic, Prodigiosin, Shows Speciesand Strain-Dependent Genome Context Variation,” Microbiology, Vol. 150, No. 11, 2004, pp. 3547-3560.
- T. Tanikawa, Y. Nakagawa and T. Matsuyama, “Transcriptional Down Regulator HexS Controlling Prodigiosin and Serrawettin W1 Biosynthesis in Serratia marcescens,” Microbiology and Immunology, Vol. 50, No. 8, 2006, pp. 587-596.
- R. M. Shanks, N. A. Stella, R. M. Lahr, S. Wang, T. I. Veverka, R. P. Kowalski and X. Liu, “Serratamolide Is a Hemolytic Factor Produced by Serratia marcescens,” PLoS One, Vol. 7, No. 5, 2012, Article ID: e36398. doi:10.1371/journal.pone.0036398
- K. E. Gibson and T. J. Silhavy, “The LysR Homolog LrhA Promotes RpoS Degradation by Modulating Activity of the Response Regulator sprE,” Journal of Bacteriology, Vol. 181, No. 2, 1999, pp. 563-571.
- P. C. Fineran, H. Slater, L. Everson, K. Hughes and G. P. Salmond, “Biosynthesis of Tripyrrole and Beta-Lactam Secondary Metabolites in Serratia: Integration of Quorum Sensing with Multiple New Regulatory Components in the Control of Prodigiosin and Carbapenem Antibiotic Production,” Molecular Microbiology, Vol. 56, No. 6, 2005, pp. 1495-1517. doi:10.1111/j.1365-2958.2005.04660.x
- S. J. Harris, Y. L. Shih, S. D. Bentley and G. P. Salmond, “The hexA Gene of Erwinia carotovora Encodes a LysR Homologue and Regulates Motility and the Expression of Multiple Virulence Determinants,” Molecular Microbiology, Vol. 28, No. 4, 1998, pp. 705-717. doi:10.1046/j.1365-2958.1998.00825.x
- A. Mukherjee, Y. Cui, W. Ma, Y. Liu and A. K. Chatterjee, “hexA of Erwinia carotovora ssp. carotovora Strain Ecc71 Negatively Regulates Production of RpoS and rsmB RNA, a Global Regulator of Extracellular Proteins, Plant Virulence and the Quorum-Sensing Signal, N-(3- oxohexanoyl)-L-homoserine Lactone,” Environmental Microbiology, Vol. 2, No. 2, 2000, pp. 203-215. doi:10.1046/j.1462-2920.2000.00093.x
- S. A. Joyce and D. J. Clarke, “A hexA Homologue from Photorhabdus Regulates Pathogenicity, Symbiosis and Phenotypic Variation,” Molecular Microbiology, Vol. 47, No. 5, 2003, pp. 1445-1457. doi:10.1046/j.1365-2958.2003.03389.x
- G. Bertani, “Studies on Lysogenesis. I. The Mode of Phage Liberation by Lysogenic Escherichia coli,” Journal of Bacteriology, Vol. 62, No. 3, 1951, pp. 293-300.
- G. Bertani, “Lysogeny at Mid-Twentieth Century: P1, P2, and Other Experimental Systems,” Journal of Bacteriology, Vol. 186, No. 3, 2004, pp. 595-600. doi:10.1128/JB.186.3.595-600.2004
- D. D. Burke D. S. Dawson and T. Stearns, “Methods in Yeast Genetics, a Cold Spring Harbor Laboratory Course Manual,” Cold Harbor Laboratory Press, Plainview, 2000.
- R. M. Shanks, N. C. Caiazza, S. M. Hinsa, C. M. Toutain and G. A. O’Toole, “Saccharomyces cerevisiae-Based Molecular Tool Kit for Manipulation of Genes from Gram-Negative Bacteria,” Applied and Environmental Microbiology, Vol. 72, No. 7, 2006, pp. 5027-5036. doi:10.1128/AEM.00682-06
- R. M. Shanks, D. E. Kadouri, D. P. MacEachran and A. G. O’Toole, “New Yeast Recombineering Tools for Bacteria,” Plasmid, Vol. 62, No. 2, 2009, pp. 88-97. doi:10.1016/j.plasmid.2009.05.002
- E. J. Kalivoda, N. A. Stella, M. A. Aston, J. E. Fender, P. P. Thompson, R. P. Kowalski and R. M. Shanks, “Cyclic AMP Negatively Regulates Prodigiosin Production by Serratia marcescens,” Research in Microbiology, Vol. 161, No. 2, 2010, pp. 158-167. doi:10.1016/j.resmic.2009.12.004
- K. Matsushita, E. Shinagawa, O. Adachi and M. Ameyama, “Quinoprotein D-Glucose Dehydrogenase of the Acinetobacter calcoaceticus Respiratory Chain: MembraneBound and Soluble Forms Are Different Molecular Species,” Biochemistry, Vol. 28, No. 15, 1989, pp. 6276- 6280. doi:10.1021/bi00441a020
- V. L. Miller and J. J. Mekalanos, “A Novel Suicide Vector and Its Use in Construction of Insertion Mutations: Osmoregulation of Outer Membrane Proteins and Virulence Determinants in Vibrio Cholerae Requires toxR.,” Journal of Bacteriology, Vol. 170, 1988, p. 2575.
NOTES
*Corresponding author.

