Advances in Biological Chemistry
Vol.3 No.1(2013), Article ID:28130,6 pages DOI:10.4236/abc.2013.31014
Amino terminus mutant OmpA from an isolated antibiotic resistant Escherichia coli still possess resistance to environmental stresses
![]()
School of Chemistry and Pharmaceutical Engineering, Sichuan University of Science & Engineering, Zigong, China
Email: *zhipingzhao@suse.edu.cn
Received 10 December 2012; revised 11 January 2013; accepted 30 January 2013
Keywords: Antibiotic Resistance; OmpA; Environmental Stress; E. coli
ABSTRACT
Antibiotic resistant Escherichia coli strains are becoming more common recently. OmpA is a very important antigen protein of E. coli, which consists of two separate domains, N-terminal and C-terminal domain. The N-terminal domain contains eight β- barrel regions that plays important roles in the multifaceted functions of OmpA. In the present study, we cloned a mutant OmpA gene from a multi-antibiotic resistant E. coli strain. Sequence analysis indicated that the N-terminal DNA sequence of the mutant OmpA shared 81.05% homology with the modeled OmpA from E. coli K12 and the N-terminal amino acid sequence of the mutant OmpA was 81.22% identical to that of the E. coli K12 OmpA. Moreover, several amino acids located in the β-barrel region were mutated. The mutant OmpA was expressed in BL21 suggested by SDS-PAGE. Resistance to environmental stress assay indicated that the N-terminus mutant OmpA still possessed excellent activities in pH, temperature and osmotic pressure resistance. Our present study may supply insights into better and deeper understand the relationships between OmpA N-terminal regions and its functions in environmental stress conditions and the mechanisms on antibiotic resistance of E. coli.
1. INTRODUCTION
Escherichia coli is a Gram-negative pathogenic bacterium that is more involved in hospital infection due in part to its resistance to commonly used antibiotics [1], including erythromycin, quinolones and β-lactams [2]. Mechanisms of E. coli resistance to antibiotics are very complicated and several mechanisms have been presented, including interference with cell wall synthesis, inhibition of protein synthesis, interference with nucleic acid synthesis and disruption of bacterial membrane structure [2-4]. E. coli has outer membranes that serve as a selective permeability barrier to protect cell from toxic compounds such as antibiotics [5] and β-barrel outer membrane proteins are assembled by highly conserved multi-protein complexes [6]. It has been well documented that OmpA, OmpC, OmpF are three of E. coli outer membrane proteins which play important roles in the high-permeability of antibiotics [7]. Particularly, the multifaceted protein OmpA is the most well-characterized major outer membrane protein in E. coli.
OmpA was first discovered in E. coli K12 strains and was subsequently extensively and intensively studied with a wide range of organisms, which is an abundant protein with ~105 copies per cell [8]. It has been reported that E. coli OmpA contributes greatly to the structural integrity and assembly of the outer membrane proteins [9, 10]. It functions as a receptor for colicin and a wide variety of bacteriophages, serves as an immune target and is also involved in adhesion and invasion [8,11]. OmpA is also considered to be required in bacterial conjugation and the formation of biofilm [12]. Furthermore, OmpA has been an ideal model for studies on the membrane protein structure and folding dynamics [8]. Similarly, OmpA homologs and OmpA-like proteins also have been suggested to exhibit the described roles [13-15].
The three-dimensional structure of OmpA was described by Pautsch and Arora, performed with X-ray diffraction analysis and NMR techniques, respectively [16, 17]. It can be isolated either from cytoplasmic inclusion bodies or from outer membrane and it is a 325-residue containing outer membrane protein proposed to consist of two separate domains. The N-terminal domain concluding 171 amino acid residues is thought to comprise eight transmembrane segments in the form of antiparallel β-strands, which is believed to play important roles in the assembly of OmpA [16,18]. The C-terminal domain is proposed to be comprised of 129 amino acid residues containing a high proportion of α-helices and involved to connect the outer membrane structurally to the periplasmic peptidoglycan layer. In addition, the two domains are connected by an Ala-Pro-rich hinge sequence [19]. On the other hand, OmpA is an abundant protein and the expression of OmpA is highly regulated at the transcription level and is environmentally responsive [8].
Most of the outer membrane proteins are specifically porins allowing them to serve as channels into the cell, that are essential to bacterial survival since through which the ions and molecules to be imported to and exported from the cell as well as preventing entry of toxic compounds [20]. It has been reported that E. coli OmpA forms ion channels with similar size compared to that of formed by OmpF and OmpC, but the penetration rates of small solutes through the OmpA channel are dramatically slower than that through the OmpF channel [21]. In addition, the N-terminal domain of OmpA is closely related with the formation of ion channel [22]. Since OmpA is an abundant protein, it has been developed as an important target in defense against bacterial pathogens [14,23]. Increasing literatures regarding to antibiotic resistant E. coli have been reported [7]. However, the mechanisms involved are still poorly understood. Moreover, it is currently unknown whether mutations within the OmpA β-barrel regions will make strong effects on OmpA functions in environmental stress resistance. In the present study, we cloned a mutant OmpA from an isolated antibiotic resistant E. coli strain where several mutant amino acids occurred in the β-barrel domain. Experimental observations suggested that the mutant OmpA still possessed the resistance to environmental stress. Our present study may supply insights into better and deeper understand the relationships between OmpA N-terminal regions and its functions in environmental stress conditions.
2. MATERIALS AND METHODS
2.1. Bacterial Strains Growth Conditions
The isolated multi-antibiotic resistant E. coli strain from diarrhoeic rabbit intestine was grown at 37˚C in LB media containing 50 μg/ml ampicilline, 30 μg/ml streptomycin. E. coli strains were grown at 37˚C in LB media supplement with 50 μg/ml ampicilline if necessary.
2.2. Isolation of the Multi-Antibiotic Resistant E. coli Strain
Dead rabbits caused by E. coli which was identified by pathology, were collected and consequently anatomized in aseptic conditions. The stained bacteria were harvested and incubated at 37˚C on LB agar medium plate after dilution. Single colony was incubated in EMB agar medium at 37˚C. Suspected colonies that possessed E. coli morphological properties were finally obtained after microscopical tests. Antibiotic resistance of the isolated organisms was determined by the disc diffusion method. Gentamicin, streptomycin, kanamycin and ampicilline were employed in the antibiotic resistance examination.
2.3. Clone of the Mutant OmpA and Construction of the Expression Vector
Genomic DNA was purified from 1 ml of an overnight grown cell culture of the isolated multi-antibiotic resistant E. coli strain using the Bacterial DNA Extraction Kit (Omega, USA) according to the manufacture’s instructions. Mutant OmpA forward primer sequences (GAATTCATGAAAAAGACAGCTATCGCG) and reverse primer sequences (AAGCTTAGCCTGCGGCTGAGTTACAAC) were designed according to the reported OmpA sequence (Accession No: 7437746). Hind III and EcoR I restriction site were added to the 5’-terminus of the forward and reverse primer, respectively. PCR was carried out using Pfu DNA polymerase under the following conditions: 30 seconds at 98˚C, 30 seconds at 55˚C, 2 minutes at 72˚C, repeated for 29 times. The amplified mutant OmpA product was purified with Omega DNA Purification Kit and digested with Hind III and EcoR I and subsequently ligated into pET32a cut with the same restriction enzymes, resulting in the mutant OmpA expression vector pET32a-mOmpA. The expression vector was then sequenced by the Shenzhen Huada Genomics Institute (China) and analyzed with DNAMAN software. After sequence, the expression vector was consequently transformed into E. coli BL21 (DE3) strain.
2.4. Expression of the Mutant OmpA
10-ml culture of LB media supplement with 50 μg/ml ampicilline were incubated with a colony of BL21/ pET32a-mOmpA and grown overnight at 37˚C with vigorous shaking. 1 ml of the overnight grown cell cultures were used to inoculate a 100-ml culture of LB media containing 50 μg/ml ampicilline. The culture were incubated for about 2 h when the OD600 reached 0.6 - 0.8, at which point the culture were induced by the addition of IPTG to a final concentration of 1 mM. 1 ml of sample was collected at 1, 2, 3, 4, and 5 h post-induction under the same growing conditions. The induced samples were collected by centrifugation at 7000 g for 1 min at 4˚C. The samples were boiled for 5 min and subsequently analyzed by SDS-PAGE with 12% gel.
2.5. Mutant OmpA Resistance to Environmental Stress
After correct expression, mutant OmpA functions in environmental stress were consequently identified based on previous literatures with slight modification [24-26]. For all the experimental analysis, the expressed cell cultures were adjusted OD600 to 0.7 and subsequently diluted 105-fold with LB media. For acid resistance assay, 100 μl of the suspensions were plated on LB agar media (pH 5.0) and incubated at 37˚C overnight. For alkali resistance assay, 100 μl of the suspensions were plated on LB agar media (pH 8.5) and incubated at 37˚C overnight. For osmotic pressure resistance assay, 100 μl of the suspendsions were plated on LB agar media with 500 mM NaCl and without NaCl, and incubated at 37˚C overnight. For high temperature resistance assay, the diluted suspendsions were incubated at 42˚C for 3 h and then 100 μl of the suspensions were plated on LB agar media and incubated at 37˚C overnight. For low temperature resistance assay, the diluted suspensions were incubated at −80˚C for 3 h and unfreeze, repeat for several times and then 100 μl of the suspensions were plated on LB agar media and incubated at 37˚C overnight. Control samples were treated with the same methods and grown under the same conditions.
3. RESULTS
3.1. Clone of the Mutant OmpA Gene
Mutant OmpA gene was amplified from the genomic DNA isolated from the multi-antibiotic resistant E. coli strain and subsequently sequenced. The DNA sequence analysis with DNAMAN software indicated that the N-terminus of the mutant OmpA shared 81.05% homology with the OmpA from E. coli K12. Large portion of the mutant OmpA sequence was mutated. Furthermore, the full-length of the N-terminal of the mutant OmpA was slight longer than that of the E. coli K12 OmpA, as revealed in Figure 1.
Similarly, the amino acid sequence analysis by DNAMAN software suggested that the N-terminus of the mutant OmpA was 81.22% identical to that of the E. coli K12 OmpA. The N-terminus of the mutant OmpA possessed 181 amino acid residues with 10 amino acids longer than that of E. coli K12 OmpA. A high number of substitutions were observed within the variable domains of the mutant OmpA regions. Moreover, mutations were apparently observed in periplasmic loop1 (L1) to L4. Particularly, five additional amino acid residues were located in L1 and L3 of mutant OmpA compared to that of E. coli K12 OmpA, respectively. Three mutations were found in the third periplasmic turns (T3). It has to be figured out that mutations were also found in the β-barrel regions of the N-terminal of OmpA. Thr9, Ala11, Val49, Tyr72, Thr106, Asn114 and Tyr129 were substituted by Ala9, Gly11, Leu49, Phe72, Ser106, Glu114 and Trp129. Two mutations were observed in β1 and β6, respectively. Furthermore, one mutation was observed in β3, β4 and β5, respectively. Protein structure predication analysis through Swiss-Model suggested that the loops orientation of mOmpA N-termunus was dramatically changed compared with OmpA from E. coli K12 (data not shown).
3.2. Expression of the Mutant OmpA
After correct construction of the expression vector pET32a-mOmpA, it was transformed into the BL21 strain where to be expressed. SDS-PAGE implied that the target mutant OmpA was expressed since the protein band was observed, as revealed in Figure 2. On the other hand, the control sample containing pET32a plasmid only did not produce the suitable size protein after the induction with IPTG, indicating that the mutant OmpA protein was expressed as expectation, which was also further determined by its resistance to environmental stress as described in the next part. In addition, to determine the induction expression time of the mutant OmpA, samples induced for 1, 2, 3, 4 and 5 h was examined. Clearly, 3 h was the optimal induction time to be used to induce the expression of mutant OmpA as the protein

Figure 1. Alignment of the N-terminal of the amplified mutant OmpA from isolated multiantibiotic resistant E. coli with that of the OmpA from E. coli K12 with the identity of ~81%. Total seven amino acids that located in β1, β3, β4, β5 and β6 region of the OmpA β-barrel were mutated. Over twenty amino acids were substituted. “*” symbol indicated the mutations within the β-barrel. “•” symbol implied the mutations within the T3 region and “^” symbol suggested the mutations in variable regions of OmpA.
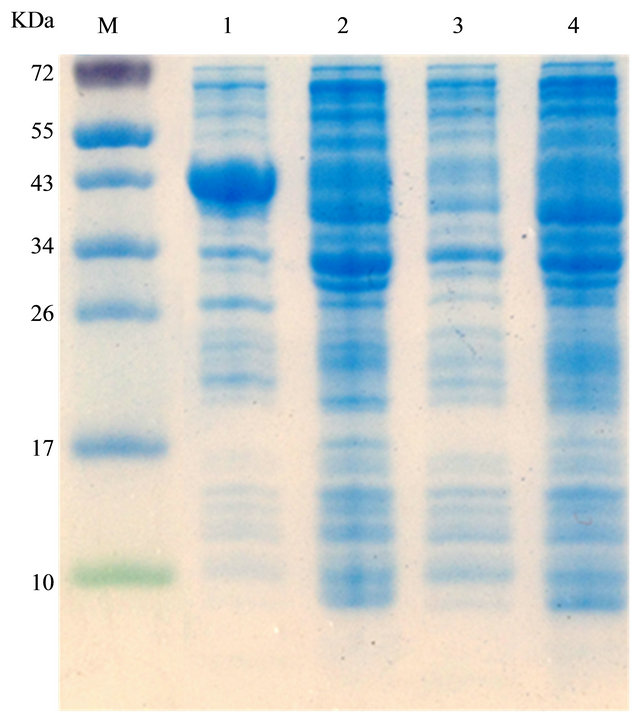
Figure 2. Expression of the mutant OmpA. 1 and 2 represented the expression of pET32a-mOmpA with and without the addition of IPTG, respectively. 3 and 4 revealed the expression of pET32a with and without the addition of IPTG, respectively.
band induced for 3 h was as bright as the samples that induced for 4 h and 5 h, as shown in Figure 3. Expression of the mutant OmpA implied that the plenty mutations in N-terminal may not make effects on the assembly of the OmpA, although the variable mutations may affect the protein structure suggested through protein structure predication by Swiss-Model (data not shown).
3.3. Mutant OmpA Resistance to Environmental Stress
After correct expression of the mutant OmpA, activities in environmental stress resistance were subsequently assayed, as shown in Figure 4. The number of the BL21/ pET32a-mOmpA clones that grown in acid, alkali, high and low temperature, low and high osmotic pressure environmental stress conditions was 36, 49, 73, 50, 60 and 38, respectively. However, the number of the BL21/ pET32a clones was 23, 25, 20, 22, 16 and 13, which was significantly less than that of the BL21/pET32a-mOmpA at the same conditions. Moreover, the environmental stress made different effects on the growth of the clone implied by the different number of the clone. The acid and high osmotic pressure stress environment may put the strongest effects on the growth of the clone since the number of the clones grown in the condition were least compared to other environmental stress. However, all the experimental observations suggested that N-terminus
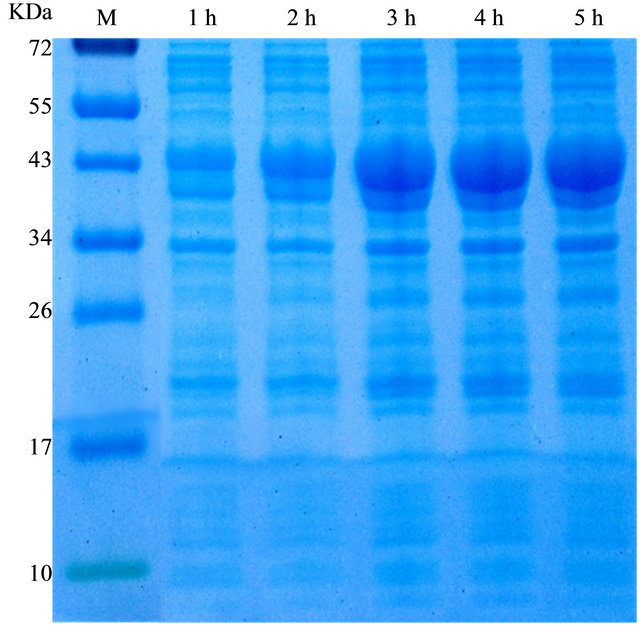
Figure 3. Determination of the optimal induction time for the expression of the mutant OmpA. Induction for 3 h after the addition of IPTG made the highest expression levels.
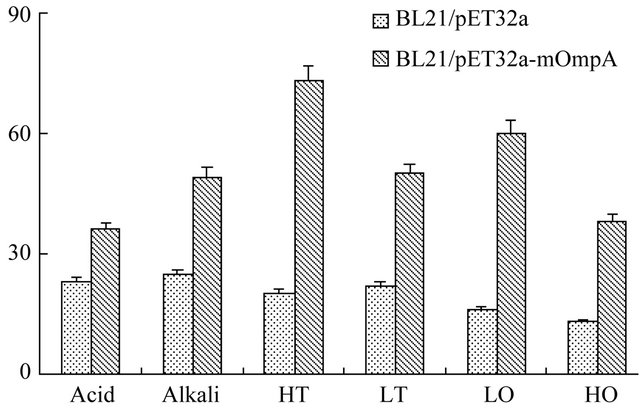
Figure 4. Mutant OmpA activities in environmental stresses resistance. The number of the BL21/pET32a-mOmpA clones was much more than that of the BL21/pET32a at the same conditions. HT, LT, LO and HO represented high temperature, low temperature, low osmotic pressure and high osmotic pressure, respectively.
mutant OmpA still possessed the resistance to environmental stress.
4. DISCUSSION
In the present study, we cloned a N-terminus mutant OmpA gene from an isolated multi-antibiotic resistant E. coli strain. Sequence analysis revealed that a high number of mutations observed within the variable regions of the mutant OmpA. Activity assay in the environmental stress resistance suggested that the mutant OmpA still exhibited excellent activities in pH, temperature and osmotic pressure resistance. The present study may supply insights into better and deeper understand the relationships between OmpA N-terminal regions and its functions in environmental stress conditions and the mechanisms on the antibiotic resistance of E. coli.
It has been well documented that E. coli outer membrane protein OmpA is considered as the excellent model for studies on the membrane protein structure [8]. On the other hand, the N-terminal domain including eight β- barrel regions plays important roles in the assembly of OmpA [16,18]. However, a high number of acid amino residues mutations were observed in the OmpA we cloned from isolated multi-antibiotic resistant E. coli in the present study, as shown in Figure 1. Experimental observations demonstrated that the mutant OmpA still possessed expected activities in environmental stress resistance, as revealed in Figure 4. On the other hand, OmpA is highly conserved outer membrane protein [27]. It will be concluded that the mutated sites were not necessary for the functions of OmpA in environmental stress conditions or other domains besides the β-barrel regions also plays important roles in the functions of OmpA and it is suggested the seven amino acid residues, Thr9, Ala11, Val49, Tyr72, Thr106, Asn114 and Tyr129 should not play very important roles in the assembly and functions of OmpA in environmental stress conditions. Similarly, the three mutated amino acid residues within the T3 region, Ile131, Pro133 and Glu134 were proposed not to affect the refold of OmpA. However, Swiss-Model protein structure predication indicated that the orientation of Loops changed significantly due to the plenty of mutations in the N-terminal of the mutant OmpA. Moreover, the function domain may not be changed significantly since the mutant OmpA still possessed the expected activities in environmental stress growth conditions.
OmpA is one of the most important outer membrane proteins responds to stress factors and plays important roles in the survival of E. coli [9]. In the present study, the importance of mutant OmpA to the survival of E. coli grown in the environmental stress was demonstrated by its critical contribution to the resistance to pH, temperature and osmotic pressure, as shown in Figure 4. On the other hand, the numbers of BL21/pET32a and BL21/ pET32a-mOmpA strains grown in the media were different under the same environmental stress growth conditions suggesting that environmental stress growth conditions affected the survival of the E. coli strains differently. Currently, there is no literature to demonstrate the relationships between the mutation of outer membrane protein and E. coli evolution. The mutation in OmpA and other outer membrane protein may increase the complexity of E. coli antibiotic resistance, which thus also increase the difficulties in the defense of the E. coli pathogen through genetic engineering vaccine. Our present study may supply insights into better understand the functions of OmpA in environmental stress conditions and much more need to be done to further illustrate the structures of OmpA and its functions and mechanisms on E. coli antibiotic resistance.
5. ACKNOWLEDGEMENTS
This work was supported by the Scientific Research Foundation of the Education Department of Sichuan Province, China (No. 11ZB102) and the Talent Project of Sichuan University of Science & Engineering (No. 2011RC12).
REFERENCES
- Nicholson, T.F., Watts, K.M. and Hunstad, D.A. (2009) OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infection and Immunity, 12, 5245-5251. doi:10.1128/IAI.00670-09
- Martinez, J.L. and Baquero, F. (2002) Interactions among strategies associated with bacterial infection: Pathogenicity, epidemicity, and antibiotic resistance. Clinical Microbiology Reviews, 4, 647-679. doi:10.1128/CMR.15.4.647-679.2002
- Gohre, V., Jones, A.M., Sklenar, J., et al. (2012) Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Molecular Plant-Microbe Interactions, 8, 1083-1092. doi:10.1094/MPMI-11-11-0301
- Saenz, Y., Brinas, L., Dominguez, E., et al. (2004) Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrobial Agents and Chemotherapy, 10, 3996- 4001. doi:10.1128/AAC.48.10.3996-4001.2004
- Desclos-Theveniau, M., Arnaud, D., Huang, T.Y., et al. (2012) The Arabidopsis lectin receptor kinase LecRKV.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLOS Pathogens, 2, e1002513. doi:10.1371/journal.ppat.1002513
- Hagan, C.L., Kim, S. and Kahne, D. (2010) Reconstitution of outer membrane protein assembly from purified components. Science, 5980, 890-892. doi:10.1126/science.1188919
- Sugawara, E. and Nikaido, H. (2012) OmpA is the principal nonspecific slow porin of Acinetobacter baumannii. Journal of Bacteriology, 15, 4089-4096. doi:10.1128/JB.00435-12
- Smith, S.G., Mahon, V., Lambert, M.A., et al. (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiology Letters, 1, 1-11. doi:10.1111/j.1574-6968.2007.00778.x
- Yagi, K. (2007) Applications of whole-cell bacterial sensors in biotechnology and environmental science. Applied Microbiology and Biotechnology, 6, 1251-1258. doi:10.1007/s00253-006-0718-6
- Kleinschmidt, J.H. (2003) Membrane protein folding on the example of outer membrane protein A of Escherichia coli. Cellular and Molecular Life Sciences, 8, 1547-1558. doi:10.1007/s00018-003-3170-0
- Mohan Nair, M.K. and Venkitanarayanan, K. (2007) Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatric Research, 6, 664-669. doi:10.1203/PDR.0b013e3181587864
- Barrios, A.F., Zuo, R., Ren, D., et al. (2006) Hha, YbaJ, and OmpA regulate Escherichia coli K12 biofilm formation and conjugation plasmids abolish motility. Biotechnology and Bioengineering, 1, 188-200. doi:10.1002/bit.20681
- Bartra, S.S., Gong, X., Lorica, C.D., et al. (2012) The outer membrane protein A (OmpA) of Yersinia pestis promotes intracellular survival and virulence in mice. Microbial Pathogenesis, 1, 41-46. doi:10.1016/j.micpath.2011.09.009
- Pore, D., Mahata, N. and Chakrabarti, M.K. (2012) Outer membrane protein A (OmpA) of Shigella flexneri 2a links innate and adaptive immunity in a TLR2-dependent manner and involvement of IL-12 and nitric oxide. Journal of Biological Chemistry, 15, 12589-12601. doi:10.1074/jbc.M111.335554
- Maruvada, R. and Kim, K.S. (2012) IbeA and OmpA of Escherichia coli K1 exploit Rac1 activation for invasion of human brain microvascular endothelial cells. Infection and Immunity, 6, 2035-2041. doi:10.1128/IAI.06320-11
- Pautsch, A. and Schulz, G.E. (1998) Structure of the outer membrane protein A transmembrane domain. Nature Structural & Molecular Biology, 11, 1013-1017. doi:10.1038/2983
- Arora, A., Abildgaard, F., Bushweller, J.H., et al. (2001) Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nature Structural & Molecular Biology, 4, 334-338. doi:10.1038/86214
- Ried, G., Koebnik, R., Hindennach, I., et al. (1994) Membrane topology and assembly of the outer membrane protein OmpA of Escherichia coli K12. Molecular Genetics and Genomics, 2, 127-135.
- Sugawara, E., Steiert, M., Rouhani, S., et al. (1996) Secondary structure of the outer membrane proteins OmpA of Escherichia coli and OprF of Pseudomonas aeruginosa. Journal of Bacteriology, 20, 6067-6069.
- Tamm, L.K., Arora, A. and Kleinschmidt, J.H. (2001) Structure and assembly of beta-barrel membrane proteins. Journal of Biological Chemistry, 35, 32399-32402. doi:10.1074/jbc.R100021200
- Sugawara, E. and Nikaido, H. (1994) OmpA protein of Escherichia coli outer membrane occurs in open and closed channel forms. Journal of Biological Chemistry, 27, 17981-17987.
- Saint, N., El Hamel, C., De, E., et al. (2000) Ion channel formation by N-terminal domain: A common feature of OprFs of Pseudomonas and OmpA of Escherichia coli. FEMS Microbiology Letters, 2, 261-265. doi:10.1111/j.1574-6968.2000.tb09296.x
- Soulas, C., Baussant, T., Aubry, J.P., et al. (2000) Outerman macrophages. Journal of Immunology, 5, 2335-2340.
- Sato, M., Machida, K., Arikado, E., et al. (2000) Expression of outer membrane proteins in Escherichia coli growing at acid pH. Applied and Environmental Microbiology, 3, 943-947. doi:10.1128/AEM.66.3.943-947.2000
- Begic, S. and Worobec, E.A. (2006) Regulation of Serratia marcescens ompF and ompC porin genes in response to osmotic stress, salicylate, temperature and pH. Microbiology, Pt 2, 485-491. doi:10.1099/mic.0.28428-0
- Hutsul, J.A. and Worobec, E. (1997) Molecular characterization of the Serratia marcescens OmpF porin, and analysis of S. marcescens OmpF and OmpC osmoregulation. Microbiology, 143, 2797-2806. doi:10.1099/00221287-143-8-2797
- Beher, M.G., Schnaitman, C.A. and Pugsley, A.P. (1980) Major heat-modifiable outer membrane protein in gramnegative bacteria: Comparison with the ompA protein of Escherichia coli. Journal of Bacteriology, 2, 906-913.
NOTES
*Corresponding author.

