Advances in Biological Chemistry
Vol. 2 No. 3 (2012) , Article ID: 21818 , 14 pages DOI:10.4236/abc.2012.23038
Interaction of nonsense suppressor tRNAs and codon nonsense mutations or termination codons
![]()
Department of Biotechnology, Academy of Administration State of Grain, Beijing, China
Email: luzixian@hotmail.com
Received 6 May 2012; revised 13 June 2012; accepted 25 June 2012
Keywords: Nonsense Suppression; Codon Nonsense Mutation; Suppressor tRNAs
ABSTRACT
Codon nonsense mutations include amber, ochre, or opal mutations according to termination codon consisting of three types (TAG, TAA and TGA). Codon nonsense mutations are also divided into natural and artificial mutations. We discussed the interaction of codon nonsense mutations and suppressor tRNAs in vitro and in vivo. Nonsense suppressions do not only happen in prokaryotes but also in eukaryotes. Meanwhile, the misreading of termination codon and incorporation of nonnatural amino acids into proteins are also introduced.
1. INTRODUCTION
Nonsense suppressors are alleles of tRNA genes altered in the anticodon, leading to insertion of an amino acid in response to a termination codon. Because termination codons consists of three types (TAG, TAA and TGA), nonsense mutations are divided into amber, ochre, or opal mutations (resulting in UAG, UAA or UGA). Therefore, nonsense suppressors include three kinds of amber, ochre or opal suppressor [1,2]. These suppressors have become important tools in bacterial genetics as well as in the study of recognition of tRNA by aminoacyl tRNA synthetase [3,4]. By this method, a number of genes containing amber, ochre, or opal mutations (resulting in UAG, UAA, or UGA chain-terminating codons, respectively) are expressed in mutant strains that synthesize suppressor tRNA capable of increasing an amino acid in response to the nonsense mutation [5,6]. Nonsense suppressors have been used widely in different living systems, for example, bacteria, yeast, plant, animal and human cells. This paper introduces not only the utilization of nonsense suppressors in vitro and in vivo (bacteria, yeast, zooblast, plant cell) but also discuss the relationship of nonsense suppressors and genetic diseases.
2. NONSENSE SUPPRESSORS IN VITRO ASSAY
2.1. Nonsense Suppressor Efficiency
An homologous in vitro assay was used to examine the effect of the cytoplasmically inherited genetic determinant [psi] on the efficiency of in vitro nonsense suppression for yeast nonsense suppressors. The efficiency of all yeast tRNA-mediated nonsense suppressor, consisting of ochre, amber, and opal, is much higher in cell-free lysates prepared from a sup+ [psi+] strain than in lysates prepared from an isogeneic sup+ [psi–] strain (Table 1A" target="_self">Table 1A). Lysates prepared from a [psi–] strain, into which the [psi+] determinant was reintroduced by kar1-mediated cytoduction, supply efficient suppression. Evidence is also showed that [psi–] lysates contain an inhibitor of in vitro nonsense suppression [7]. A cell-free translation system, from the yeast Saccharomyces cerevisiae (S. cerevisiae), has been utilized to develop an in vitro assay for yeast opal, ochre and amber suppressors. Amber suppression was assayed by read-through of the brome mosaic virus coat protein cistron UAG terminator, opal suppression by read-through of the rabbit β-globin UGA terminator and ochre suppression by read-through of the rabbit α-globin mRNA UAA terminator. Opal suppression was highly efficient in vitro, reaching almost 100% using a purified opal suppressor tRNA from Schizosaccharomyces pombe (Table 1B" target="_self">Table 1B). Ochre suppression was enhanced 3-fold when the globin mRNA was heat denatured prior to translation because of an increase in the synthesis of α-globin relative to β-globin [8].
The aminoacylation kinetics of 19 different variants of yeast tRNATyr with nucleotide substitutions in positions 33 - 35 were determined. Substitution of the conserved uridine-33 does not vary the rate of aminoacylation. However, substitution of the anticodon position 34 or position 35 decreases Km from 2- to 10-fold and Vmax as much as 2-fold, relying on the nucleotide inserted (Table 2). The ochre and amber suppressor tRNAsTyr both presented about a 7-fold reduction in Vmax/Km. Data from tRNATyr with different modified nucleotides at position 35 suggest
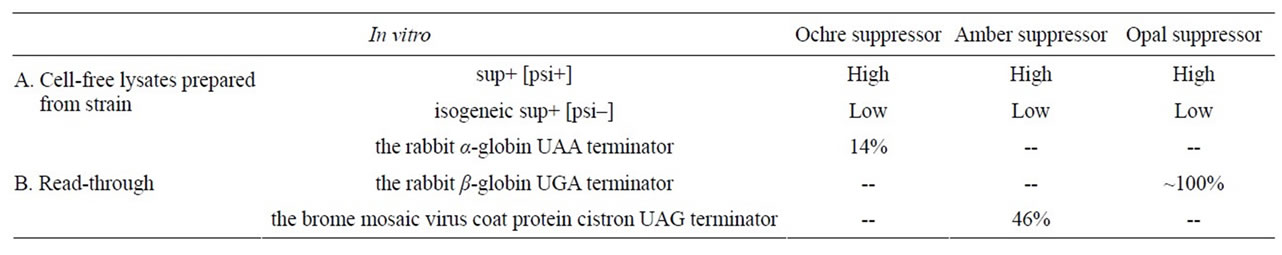
Table 1. Nonsense suppressor efficiency(I) [7,8].
that specific hydrogen bonds link between the synthetase and both the N1 and N3 hydrogens of psi-35. These data demonstrate that yeast tyrosyl-tRNA synthetase interacts with positions 34 and 35 of the anticodon of tRNATyr and opens the possibility that nonsense suppressor efficiency may be regulated by the level of aminoacylation [9].
2.2. Misreading of Codon and Incorporation of Nonnatural Amino Acids into Proteins
Effects of codon context on nonsense codon suppression may act either through release factor recognition of termination codons or aminoacyl-tRNA selection by the ribosome. The hypothesis has been researched by comparing misreading by Escherichia coli UGA suppressor tryptophan tRNA of UGU (cysteine) codons in two synthetic polymers, poly (U-G) and poly (U5, G), which differ in sequence around the UGU codons. In vitro translation of these polymers in a cell-free system from E. coli produced selection errors of 4 × 10–3 and 1.75 × 10–2 for UGU codons in poly (U-G) and poly (U5, G) respectively. This difference suggests that codon context may significantly take effect on misincorporation of amino acids into protein [10]. When RNA isolated from the Drosophila melanogaster alcohol dehydrogenase (ADH) negative mutant CyOnB was translated “in vitro” in the presence of yeast opal suppressor tRNA, a wild type size ADH protein was produced in addition to the mutant gene product. This identifies the CyOnB mutant as an opal nonsense mutant. The tryptophan codon UGG in position 234 has been altered into a UGA nonsense codon in the CyOnB mutant. Furthermore, the UAA stop codon of the wild type ADH gene is resistant to suppression by a yeast ochre suppressor tRNA [11]. To prepare a semisynthetic, nonhypermodified tRNAGly nonsense suppressor acylated with L-3-[125I]iodotyrosine, the presence of this synthetic tRNA during in vitro translation of mRNA containing a nonsense suppression site (e.g., a UAG termination codon) results in the incorporation of the nonnatural amino acid L-3-iodotyrosine into the polypeptide exclusively at the position corresponding to that site. Under optimum conditions, the unlabeled amino acid L-3-iodotyrosine was found to be incorporated with a suppression efficiency of 65%. Other nonnatural residues, including N-methylphenylalanine, D-phenylalanine, and phenyllactic acid, were
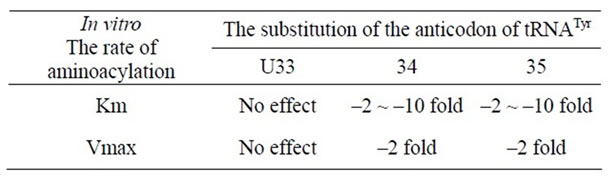
Table 2. Nonsense suppressor efficiency(II) [9].
tested in the assay under these same conditions. Suppression efficiencies for this series ranged from 0 to 72% depending on the structure of the residue incorporated [12].
3. NONSENSE SUPPRESSORS IN BACTERIA CELLS
3.1. In E. coli, Which Kind of Nonsense Suppressors Is Efficient?
Both the yeast tRNATyr ochre suppressor SUP6 gene and a derivative of this gene in which the 14-bp intervening sequence has been deleted, SUP6Δ, have been assayed for functional expression in E. coli. The SUP6Δ codes for a functional transfer RNA which has been presented to suppress both ochre and amber nonsense mutations. The functional expression of this mutant eukaryotic transfer RNA gene in E. coli supplies a unique opportunity for studies of expression of a gene coding for a stable RNA, in both a prokaryotic and a eukaryotic host [13]. A temperature-sensitive mutant of E. coli harbors a weak suppressing activity towards a UAG mutation, together with an inability to grow at 43˚C in rich medium. The mutation shows enhanced misreading of UAG/A, but UGA. The mutation gives an increased efficiency for some tRNA nonsense suppressors. The ochre suppressors SuB and Su5 are stimulated in their reading of both UAG/A with preference for UAG [14]. Of all the E. coli tRNA genes that can give rise to an amber or an ochre suppressor by a singlenucleotide mutation except for the tRNAGlu genes. A sitespecific mutagenesis is used to create the gene for such a tRNA in order to test the prediction that the ochre suppressor derived from tRNAGlu would act very poorly on the ribosome. The tRNAGlu-Suoc gene is cloned into a high copy number plasmid, under control of the lacUV5 promoter. The mutant tRNA suppresses both amber and ochre nonsense mutations. As predicted, it is less efficient than other suppressors expressed under similar conditions [15].
3.2. In E. coli, Synthesis of Two Selenoproteins Directed by UGA Opal Nonsense Codon and Difference of Nonsense Suppressors’ Efficiency
Sequence analysis of the genes coding for two selenoproteins, formate dehydrogenase H from E. coli and glutathione peroxidase from mammalian, demonstrated that an in-frame UGA opal nonsense codon directs the incorporation of selenocysteine. Four genes are identified whose products are necessary for selenocysteine incorporation in E. coli. One of these genes codes for a tRNA species with unique properties. It possesses an anticodon complementary to UGA and deviates in several positions from sequences, until now, considered invariant in all tRNA species. This tRNA is aminoacylated with L-serine by the seryl-tRNA ligase which also charges cognate tRNASer. Selenocysteine, therefore, is synthesized from a serine residue bound to a natural suppressor tRNA which recognizes UGA [16]. Seventeen tRNA suppressor genes are constructed from E. coli representing 13 species of tRNA. Each tRNA gene is transformed into E. coli via a plasmid vector. Some of them are not able to produce detectable levels of suppression, whereas others insert amino acids with greater than 70% efficiency. Results reported in the accompanying paper show that some of these suppressors insert the original cognate amino acid, whereas others do not (Table 3) [17]. Comparing different nonsense sites in an mRNA, not only the two bases downstream of the nonsense codon are major determinants of suppression efficiency but the nature of the contiguous upstream codon can also affect nonsense suppression, as can the third (wobble) base of the contiguous downstream codon. The non-site-specific effects of the two adjacent downstream bases may be largely at the level of the termination process, whereas more site-specific or codon-specific effects may operate primarily on the activity of the suppressor tRNA [18].
4. NONSENSE SUPPRESSORS IN YEAST CELLS
4.1. Nonsense Suppressors (sup3-e, sup3-i tRNASer and supSI) from S. pombe
The gene encoding the efficient opal suppressor sup3-e from a clone bank of EcoRI fragments from a S. pombe sup3-e strain in the hybrid yeast vector YRp17 was utilized to transform the S. cerevisiae multiple auxotroph his4-260 leu2-2 trp1-1 to prototrophy. The results demonstrate that the cloned S. pombe UGA suppressor is functional in S. cerevisiae UGA mutant strains. The inefficient suppressor sup3-i of S. pombe is an ochre suppressor. Sup3-i was derived from the efficient serine inserting UGA suppressor sup3-e. The cloning and sequencing of the sup3-i gene indicate that the suppressor is different from the parent sup3-e by a C-T substitution in the sequence coding for the middle position of the anticodon. In vitro translation assays supplemented with purified sup3-i tRNA and programmed with Xenopus globin mRNAs lead to the accumulation of a readthrough product in response to ochre termination signals, but not in response to opal termination codons. Transformation of S. cerevisiae nonsense mutant strains with plasmid DNA carrying the S. pombe sup3-i gene, led to ochre, but not amber or opal suppression in vivo. Suppression of nonsense codons in S. pombe by sup3-e tRNASer(UGA) or sup3-i tRNASer (UAA) is decreased or abolished by mutations within the suppressor locus. Twenty-five suppressor-inactive sup3-e genes and thirteen mutant sup3-i genes were isolated from S. pombe genomic clone banks by colony hybridization. Sequence analysis of these revertant alleles corroborates genetic evidence for mutational hotspots within the sup3 tRNA gene. Fifteen types of point mutations or insertions have been found and many of these replace bases are highly or completely conserved in eucaryotic tRNA genes. Transcription of the altered sup3 genes in a S. cerevisiae extract enabled the identification of mutations which affect the rate of 5’-end maturation or splicing of the tRNA precursors or both. A total of seven mutations have been found which alter transcriptional efficiencies and five of these are located outside the internal transcription control regions [19-21]. In S. cerevisiae ochre and opal, as well as amber mutations are known, whereas in the fission yeast S. pombe no amber alleles have been described. An amber allele in the trp1 locus of S. pombe has been characterized as trp1-566. Its identification as an amber allele is based on the following results: 1) the nonsense allele can be converted to an ochre allele by nitrosoguanidine mutagenesis; 2) trp1-566
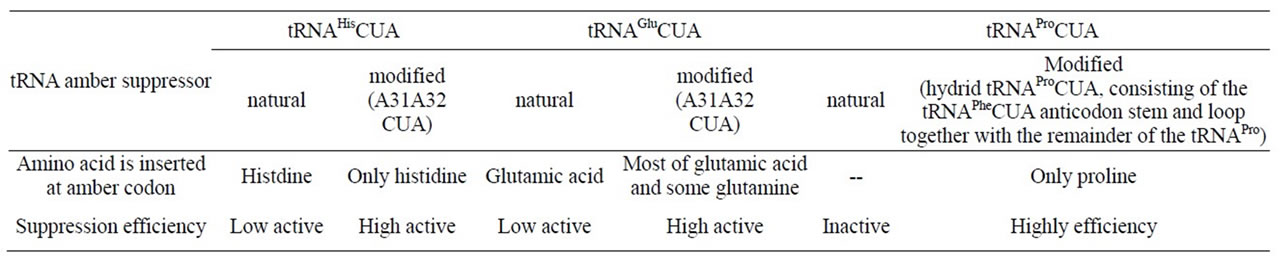
Table 3. Efficiency of modified nonsense suppressors [17].
is suppressed by a bona fide S. pombe amber suppressor tRNA, supSI. The supSI gene was obtained by primerdirected in vitro mutagenesis of a tRNASer from S. pombe. Unexpectedly, an S. cerevisiae amber suppressor tRNASer, supR21, transformed into S. pombe, failed to suppress trp1- 566. Northern analysis of S. pombe transformants reveals that this gene is not transcribed in the fission yeast [22].
4.2. Nonsense Suppressors in S. cerevisiae
The S. cerevisiae leucine-inserting amber suppressor tRNA gene SUP53 (a tRNALeu3 allele) encodes a pre-tRNA which contains a 32-base intron and its mature tRNASUP53 contains a 5-methylcytosine modification of the anticodon wobble base. The SUP53 intron was mutated and the suppressor function of these mutant tRNA genes was assayed after their integration into the S. cerevisiae genome. Mutant analysis demonstrated that the totally intact precursor tRNA was necessary for efficient nonsense suppression by tRNASUP53. Less efficient suppressor activity correlated with the absence of the 5-methylcytosine modification [23]. When it is expressed in sufficient quantity, normal tRNAGln (which usually decodes the sense codon CAA) can weakly suppress the nonsense ochre codon UAA in S. cerevisiae, and this represents an example of wobble occurring at the first rather than at the third position of the codon [24]. The anticodon of the wild-type tRNA (7Ser) gene of D. melanogaster was converted and all three nonsense suppressor derivatives of the gene were constructed. These constructs were introduced into an E. coli-yeast shuttle vector (YRp7), and used to transform a S. cerevisiae strain [JG 369-3B(α)] containing an array of nonsense alleles. When assayed on appropriate omission media, the D. melanogaster suppressor genes were found to function in the yeast with strict codon specificity. Subsequent Northern hybridization analyses revealed that the D. melanogaster suppressor genes were transcribed and processed well in S. cerevisiae [25]. The anticodon of a cloned tRNATrp gene was altered by sitedirected mutagenesis from CCA to CTA with the expectation that this gene would produce tRNA molecules capable of interacting with the amber terminator codon. This form of the gene can be transcribed and spliced in vitro to produce mature tRNA with the expected base sequence. The putative suppressor gene has been transformed into several S. cerevisiae host strains by the centromere vector YCp19. Efficient suppression of amber mutations met8-1, tyr7-1, and lys2-801 occurs on the presence of the CTA form of tDNATrp. Two UAA mutants, leu2-1 and ade2-101, and the opal marker his4-260 are not suppressed. Ochre and opal suppressors from a cloned tRNATrp gene were generated by mutagenesis because of naturally occurring suppressor mutants from tRNATrp genes having never been identified in S. cerevisiae. Both forms of the gene are processed and spliced in vitro to produce tRNAs with the expected base sequences. The altered genes were subcloned into yeast vectors and transformed into yeast strains carrying a variety of amber, ochre, and opal mutations. When introduced on a CEN vector, neither the ochre suppressor forms (TRPO) or the opal suppressor (TRPOP) show suppressor activity. Deletion of the CEN region from the clones increases the copy number to 10 - 20/cell. TRPOP shows moderate suppressor activity when the gene is introduced on this vector, however, TRPO exhibits no detectable biological activity regardless of gene copy number. Northern blot analyses of the steady state levels of tRNATrp in cells containing the high copynumber clones reveal 20% - 100% increases in the abundance of tRNATrp [26,27]. Wild-type glutamine tRNAs in yeast cause low-level nonsense suppression that can be enhanced by increasing glutamine tRNA gene copy number. In order to investigate glutamine tRNA behavior further, anticodon mutations that confer nonsense suppression were identified in yeast sup70 gene, which codes for glutamine tRNA(CAG). Amber suppressor glutamine tRNA(UAG) can translate 5’-CAG-3’ glutamine codons with low efficiency in the presence of an A/C mismatch at the first position of the codon, suggesting that reading may occur at a low level by a two-out-of-three reading mechanism. In addition, when glutamine tRNA(CAA) is over-expressed in vivo, it translates 5’-CAG-3’ codons using a mechanism that resembles prokaryotic-like U/G wobble, which normally does not occur in yeast. The yeast glutamine tRNA suppressors could potentially be exploited to express ciliated protozoan genes that normally contain internal 5’-UAG-3’ and 5’-UAA-3’ codons [28]. In yeast S. cerevisiae nonsense-mutants containing premature stop-codon in mRNA of the essential SUP45 gene were obtained. These strains are viable in the absence of mutant suppressor tRNA, therefore it is supposed that there are alternative mechanisms providing nonsense-suppression and mutants viability. Analysis of transformants obtained by transformation of strain bearing nonsense-mutant allele of SUP45 gene with multicopy yeast genomic library revealed three genes encoding wild type tRNATyr and four genes encoding wild type tRNAGln that improve nonsense-mutants viability. Moreover, overexpression of these genes leads to the increase in the amount of fulllength eRF1 protein in cell and compensates nonsensemutants sensitivity to high temperature [29].
5. NONSENSE SUPPRESSORS IN ANIMAL CELLS
5.1. Nonsense Suppressors in D. melanogaster
The suppressor tRNA genes created by sd-type tRNATyr gene were subcloned into a pUChsneo transformation vector providing heat-shock controlled neomycin resistance. The seven xanthine dehydrogenase and cross-reacting material negative D. melanogaster rosy stocks were germline transformed with amber and ochre tDNATyr, and the G1 generation was screened for Geneticin resistance. Surviving rosy516 flies transformed with the amber suppressor showed an eye colour intermediate between the original ry516 stock and the wild-type, suggesting that ry516 is an amber nonsense mutant. The analysis of the ry516 gene sequence revealed a C-to-T transition in a CAG glutamine codon at nucleotide 1522 of the wildtype rosy gene [30]. Ochre, opal and amber nonsense mutations were placed at two leucine codons (Leu205 and Leu309) in Drosophila’s major rhodopsin gene (ninaE) by site-directed mutagenesis, and then the corresponding mutants was created by P element-mediated transformation of a ninaE deficiency strain. In the absence of a genetic suppressor, flies harboring any of the nonsense mutations at the 309 site, but not the 205 site, show increased rhodopsin activity. The presence of leucine-inserting tRNA nonsense suppressors DtLa Su+ and DtLb Su+ in the mutant strains produced a small increase (less than 0.04%) in functional rhodopsin. The opal (UGA) suppressor derived from the DtLa tRNA gene is more efficient than the amber (UAG) or opal suppressor derived from the DtLb gene, and both DtLa and DtLb derived suppressors are more efficient at site 205 than 309 [31].
5.2. Nonsense Suppressors and Xenopus laevis
Amber, ochre, and opal nonsense suppressor tRNAs isolated from yeast were introduced into Xenopus laevis oocytes together with purified mRNAs (globin mRNA from rabbit, tobacco mosaic virus-RNA). Yeast opal suppressor tRNA is able to read the UGA stop codon of the rabbit β-globin mRNA, thus producing a readthrough protein. A large readthrough product is also obtained upon coinjection of yeast amber or ochre suppressor tRNA with TMV-RNA. The Xenopus laevis oocyte provides a simple, sensitive, and well buffered in vivo screening system for all three types of eukaryotic nonsense suppressor tRNAs. After nonsense mutants in the herpes simplex virus thymidine kinase (HSV tk) gene, the E. coli xanthine-guanine phosphoribosyl transferase (Eco-gpt) gene and the aminoglycoside 3’ phosphotransferase gene of the Tn5 transposon (NPT-II) were isolated and characterized, each gene was engineered with the appropriate control signals to allow expression in mammalian cells respectively. Xenopus laevis amber suppressor genes, prepared by site-specific mutagenesis of a normal X. laevis tRNA gene, were microinjected into the above cell lines and selected for the expression of one or more of the amber mutant gene products. The resulting cell lines, containing functional amber suppressor genes, are stable and exhibit normal growth rates [32,33]. The anticodon of a Xenopus laevis tyrosine tRNA gene is altered by site-specific mutagenesis for recognizing ochre codons. This tRNA gene is expressed when amplified in monkey cells as part of a SV40 recombinant and efficiently suppresses termination at both the ochre codon separating the adenovirus 2 hexon genes from a 23-kd downstream gene and the ochre codon at the end of the NS1 gene of influenza virus A/Tex/1/68 [34]. The conditional lethal amber nonsense mutants of vesicular stomatitis virus (VSV) were isolated from a chemically mutagenized stock of wild-type virus by their ability to grow on genetically engineered cells which express a Xenopus laevis amber suppressor tyrosine tRNA gene (su+ cells) but not on the non-suppressor parental cells (su– cells) [35].
5.3. Nonsense Suppressors in Mammalian Cells
A nonsense mutation (UAG) in the thymidine kinase gene of herpes simplex virus type 1 can be suppressed in vivo to produce active thymidine kinase by prior infection with a defective simian virus 40 stock which acts as a vector to introduce a functional suppressor tRNA gene into mammalian cells in culture. The suppression is specific for UAG, but not UGA or missense, mutants and restores thymidine kinase activity to 20% to 40% of the wild-type level. Through genetic engineering techniques, it is now possible to introduce suppressor (Su+) tRNA molecules into mammalian cells. Suppressor activity in L cells continuously expressing Su+ (tRNAtyr) was 3.5 percent and that in CV-1 cells infected with an SV40– Su+ (tRNAtyr) recombinant was 22.5 percent [36,37]. Serine codon 27 of the E. coli chloramphenicol acetyltransferase (cat) gene has been converted to three nonsense codons by oligonucleotide-directed site-specific mutagenesis. The mutant cat genes, under transcriptional control of the Rous sarcoma virus long terminal repeat, were then introduced into mammalian cells by DNA transfection along with UAG, UAA, and UGA suppressor tRNA genes derived from a human serine tRNA. Assay for CAT enzymatic activity in extracts from such cells allowed to detect and quantitate nonsense suppression in monkey CV-1 cells and mouse NIH3T3 cells. Using such an assay, an opal suppressor tRNA gene is functional in mammalian cells as the first direct evidence. The pattern of suppression of the three cat nonsense mutations in bacteria suggests that the serine at position 27 of CAT can be replaced by a wide variety of amino acids without loss of enzymatic activity. A human amber suppressor tRNASer gene linked to the SV40 origin of replication and a second DNA carrying a temperature-sensitive SV40 large T antigen gene were cotransfected into monkey cells. Cell lines having stably integrated the DNAs were isolated. Shifting the cells from the nonpermissive temperature to a lower permissive temperature caused the amplification of the suppressor tRNA gene, which resulted in suppression efficiencies at amber codons of 50% - 70%, as measured by suppression of an amber codon in the E. coli cat gene [38,39].
To assess the role that extragenic flanking sequences may have on the expression of mammalian tRNA genes in vivo, deletion/substitutions ending in the 5’-flanking sequence or 3’-flanking sequence of a cloned human serine amber suppressor tRNA gene were constructed. The phenotypic expression of these mutant genes was examined by transfection in mammalian cells. In in vivo competition studies, the 5’-flanking region up to nucleotide position -18 deletion clone was less able to compete out the activity of a second suppressor tRNA gene than was the wild-type corresponding gene, suggesting that the upstream region plays a role in the formation of active transcription complexes in vivo. Therefore, the human serine tRNA gene contains an upstream regulatory region that plays a positive role in modulating expression of this gene in vivo [40]. The E. coli lac operator/repressor system has been exploited as a means to regulate the expression of a mammalian tRNA gene in vivo and in vitro. An oligonucleotide containing a lac operator (lac O) site was cloned immediately upstream of a human serine amber suppressor (Su+) tRNA gene. Insertion of a single lac repressor binding site at position –1 or –32 relative to the coding region had no effect on the amount of functional tRNA made in vivo, as measured by suppression of a nonsense mutation in the E. coli cat gene following cotransfection of mammalian cells. Inclusion of a plasmid expressing the lac repressor in the transfections resulted in 75% to 98% inhibition of suppression activity of lac operator-linked tRNA genes but had no effect on expression of the wild-type gene. Inhibition could be quantitatively relieved with the allosteric inducer isopropylthio-beta-D-galactoside (IPTG). These results demonstrate that the bacterial lac operator/repressor system can be used to reversibly control the expression of mammalian genes that are transcribed by RNA polymerase III [41]. As an approach to inducible suppression of nonsense mutations in mammalian and in higher eukaryotic cells, the expression of an E. coli glutamineinserting amber suppressor tRNA gene has been analyzed in COS-1 and CV-1 monkey kidney cells. The tRNA gene used has the suppressor tRNA coding sequence flanked by sequences derived from a human initiator methionine tRNA gene and has two changes in the coding sequence. The transcript of this tRNA gene is processed to yield the mature tRNA in COS-1 and CV-1 cells. The tRNA is not aminoacylated in COS-1 cells by any of the endogenous aminoacyl-tRNA synthetases and is therefore not functional as a suppressor. Concomitant expression of the E. coli glutaminyl-tRNA synthetase gene results in aminoacylation of the suppressor tRNA and its functioning as a suppressor. These results open up the possibility of attempts at regulated suppression of nonsense codons in mammalian cells by regulating expression of the E. coli glutaminyl-tRNA synthetase gene in an inducible, celltype specific, or developmentally regulated manner [42].
6. NONSENSE SUPPRESSORS IN PLANT CELLS
6.1. Nonsense Suppressors in Model Lower Plants
Photosynthesis-deficient mutants of the green alga Chlamydomonas reinhardtii come from nonsense mutations within the chloroplast rbcL gene, which encodes the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (EC 4.1.1.39). Photosynthesis-competent revertants of these nonsense mutants are stably heteroplasmic, giving rise to both mutant and revertant cells during cell divisions under nonselective growth conditions. Therefore, the stable heteroplasmicity might arise from a balanced polymorphism of suppressor and wild-type alleles of a tRNA gene maintained within the polyploid chloroplast genome. Under nonselective conditions, the suppressor mutation was lost from cells that also lost the revertant phenotype. Stable heteroplasmicity can arise as a balanced polymorphism of organellar alleles. An additional tRNA suppressor may be identified due to their heteroplasmic nature within polyploid genomes [43]. Numerous nuclear gene products are required for the correct expression of organellar genes. One such gene in the green alga C. reinhardtii is MCD1, whose product is required for stability of the chloroplast-encoded petD mRNA. In mcd1 mutants, which are non-photosynthetic, petD mRNA is degraded by a 5’-3’ exonuclease activity, resulting in a failure to synthesize its product, subunit IV of the cytochrome b 6/f complex. The sequence of the wild-type MCD1 gene encodes a large and novel putative protein. Analysis of three mutant alleles showed that two harbored large deletions, but that one allele, mcd1-2, had a single base change resulting in a nonsense codon near the N-terminus. This same mutant allele can be suppressed by a second-site mutation in the nuclear MCD2 gene, whereas mcd2-1 cannot suppress the deletion in mcd1-1. The mutation of the cloning of mcd2-1 lies in a tRNASer (CGA), which has been modified to translate the nonsense codon in mcd1-2. How may the existence of a large tRNASer gene family permit this suppression without pleiotropic consequences [44,45]?
6.2. Nonsense Suppressors in Model Higher Plants
An Arabidopsis thaliana L. DNA containing the tRNATrp (UGG) gene was isolated and altered to encode the amber suppressor tRNATrp (UAG) or the ochre suppressor tRNATrp (UAA). These DNAs were electroporated into carrot protoplasts and tRNA expression was demonstrated by the translational suppression of amber and ochre nonsense mutations in the cat reporter gene. DNAs encoding tRNATrp (UAG) and tRNATrp (UAA) nonsense suppressor tRNAs caused suppression of their cognate nonsense codons in CAT mRNAs, with the tRNATrp (UAG) gene exhibiting the greater suppression under optimal conditions for expression of CAT. The development of these translational suppressors which function in plant cells facilitates the study of plant tRNA gene expression and will make possible the manipulation of plant protein structure and function. Chloroplast and cytoplasmic tRNATrp CmCA have been identified as the first natural UGA suppressor tRNAs in plants. The interaction of these tRNAs with UGA involves a Cm: A mismatch at the first anticodon position. Here, tRNATrp CmCA is incapable of misreading UAA and UAG codons in vitro, implying that unconventional base pairs are not tolerated in the middle anticodon position. A pronounced influence on suppression by tRNATrp CmCA was seen only if 2 or 3 nt at the 3’-side of the UGA codon had been simultaneously replaced. As a consequence of the flexible codon context accepted by tRNATrp CmCA, this tRNA is able to misread the UGA in a number of plant and animal viral RNAs that use translational read-through for expression of some of their genes [46,47]. A nuclear tRNALys gene from Arabidopsis thaliana was cloned and mutated so as to express tRNAs with altered anticodons which bind to a UAG nonsense (amber) codon and to the Arg (AGG), Asn (AAC,AAT), Gln (CAG) or Glu (GAG) codons. Concomitantly, a codon in the firefly luciferase gene for a functionally important Lys was altered to an amber codon, or to Arg, Asn, Gln, Glu, Thr and Trp codons, so as to construct reporter genes reliant upon incorporation of Lys. The altered tRNALys and luciferase genes were introduced into Nicotiana benthamiana protoplasts and expression of the mutated tRNAs was verified by translational suppression of the mutant firefly luciferase genes. Expression of the amber suppressor tRNALys (CUA) from nonreplicative vectors promoted 10% - 40% suppression of the luciferase nonsense reporters while expression of the amber and missense tRNALys suppressor genes from a geminivirus vector capable of replication promoted 30% - 80% suppression of the luciferase nonsense reporter and up to 10% suppression of the luciferase missense reporters with Arg, Asn, Gln and Glu codons [48]. The transient expression of three novel plant amber suppressors derived from a cloned Nicotiana tRNASer (CGA), an Arabidopsis intron-containing tRNATyr (GTA) and an Arabidopsis intron-containing tRNAMet (CAT) gene, respectively, was studied in a homologous plant system by transforming each of them to Arabidopsis hypocotyl explants. This versatile system allows the detection of β-glucuronidase (GUS) activity by histochemical and enzymatic analyses. The activity of the suppressors was demonstrated by the ability to suppress a premature amber codon in a modified GUS gene. Co-transformation of Arabidopsis hypocotyls with the amber suppressor tRNASer gene and the GUS reporter gene resulted in ~10% of the GUS activity found in the same tissue transformed solely with the functional control GUS gene. Amber suppressor tRNAs derived from intron-containing tRNATyr or tRNAMet genes were functional in vivo only after some additional gene manipulations. The improved amber suppressors tRNATyr and tRNAMet were subsequently employed for studying various aspects of the plant-specific mechanism of pre-tRNA splicing as well as for demonstrating the influence of intron-dependent base modifications on suppressor activity [49]. Oligonucleotide-directed mutagenesis was used to generate amber, ochre and opal suppressors from cloned Arabidopsis and Nicotiana tRNATyr genes. The nonsense suppressor tRNATyr genes were efficiently transcribed in HeLa and yeast nuclear extracts, however, intron excision from all mutant pre-tRNAsTyr was severely impaired in the homologous wheat germ extract as well as in the yeast in vitro splicing system. The change of one nucleotide in the anticodon of suppressor pre-tRNAs leads to a distortion of the potential intron-anticodon interaction. In order to demonstrate that this caused the reduced splicing efficiency, a point mutation was created in the intron of Arabidopsis tRNATyr which affected the interaction with the wild-type anticodon. As expected, the resulting pre-tRNA was also inefficiently spliced. Another mutation in the intron, which restored the basepairing between the amber anticodon and the intron of pre-tRNATyr, resulted in an excellent substrate for wheat germ splicing endonuclease. This type of amber suppressor tRNATyr gene which yields high levels of mature tRNATyr should be useful for studying suppression in higher plants [50].
7. IMPACT FACTORS OF NONSENSE SUPPRESSOR
7.1. Impact of rRNA, mRNA and the First 3’ Base Codon Context
The in vivo expression of mutations constructed within helix 34 of 16S rRNA has revealed two novel results: Firstly, some of the rRNA mutations affected suppression at UAA and UAG nonsense codons. Secondly, both an increase and a decrease in the efficiency of the suppressor tRNA were induced by the mutations. The data are interpreted as there being competition between the two release factors (RF-1 and RF-2) for an overlapping domain and that helix 34 influences this interaction [51]. The abundance of the mRNA for human triosephosphate isomerase (TPI) is decreased to 20% - 30% of normal by frameshift and nonsense mutations that prematurely terminate translation within the first three-quarters of the reading frame. The decrease has been shown to be attributable to a reduced level of TPI mRNA that copurifies with nuclei. 1) A suppressor tRNA, which acts in trans to suppress an amber nonsense codon within TPI mRNA; and 2) a hairpin structure in the 5’ untranslated region of TPI mRNA, which acts exclusively in cis to inhibit initiation of TPI mRNA translation, were found, individually, and to a greater extent, together, to abrogate the decrease in mRNA. These results show that tRNA and ribosomes coordinately mediate the effect of a nonsense codon on the level of newly synthesized TPI mRNA. The premature termination of TPI mRNA translation in the cytoplasm can reduce the level of TPI mRNA that fractionates with nuclei [52]. The effect of 3’ context on the efficiency of nonsense suppressor tRNAs in E. coli and human cells has been compared. Plasmids containing amber (UAG) termination codons were constructed in the vector pRSV β gal by oligonucleotide insertion at an N-terminal location in a lacZ fusion. A family of identical vectors was prepared with either A, C, G or U as the first 3’ base following the stop codon. Nonsense suppression was monitored using enzyme assays for β-galactosidase. In E. coli the efficiency of a plasmid-borne bacterial tRNAtrp (UAG) suppressor varied A > G > C = U. When the same lacZ reporter vectors were cotransfected with a human tRNAser (UAG) suppressor plasmid into human cells, context effects of a different nature were detected. Double reciprocal analysis of dose-response experiments were used to show that the efficiency of suppression varied C > G > U = A. Therefore, the interaction between mammalian tRNAs or release factors and their target codons may have different characteristics from those in bacteria [53].
7.2. Impact of 5’ Flanking Region of Nonsense Suppressor tRNA and Release Factors
The consequences of altering the 5’ flanking region of a human amber suppressor tRNAser gene on phenotypic expression in vivo and transcription in vitro was examined by constructing a series of upstream deletion and substitution mutants. Mutant genes in which the 18 nucleotides 5’ proximal to the coding region were deleted and replaced with heterologous sequences were 2 to 5 fold more active in vivo in comparison to the wild type gene. In certain cases, introduction of an oligonucleotide encompassing this 18 nucleotide element upstream of more active tRNA genes reduced both the levels of suppression and template activity. These results indicate that the immediate 5’ contiguous sequence of this tRNA gene negatively modulates expression both in vivo and in vitro [54]. Eukaryotic release factor 1 (eRF1) catalyzes the release of the polypeptide chain without any stop codon specificity. The GTP-binding protein eRF3 confers GTP dependence to the termination process and stimulates eRF1 activity. In a cat assay, the competition between the suppressor tRNA and the human release factors has been measured when a stop codon was present in the ribosomal A site. Whatever the stop codon (UAA, UAG, or UGA) present in the cat open reading frame, the overexpression of human eRF1 alone markedly decreased translational readthrough by suppressor tRNA. Thus, like the procaryotic release factors RF1 and RF2 in E. coli, eRF1 seems to have an intrinsic antisuppressor activity in human cells. Levels of antisuppression of overexpression of both eRF3 and eRF1 were almost the same as those of overexpression of eRF1 alone, suggesting that eRF1-eRF3 complex-mediated termination may be controlled by the expression level of eRF1. Surprisingly, when overexpressed alone, eRF3 had an inhibitory effect on cat gene expression. The results of cat mRNA stability studies suggest that eRF3 inhibits gene expression at the transcriptional level. This indicates that in vivo, eRF3 may perform other functions, including the stimulation of eRF1 activity. Using a sensitive and versatile readthrough assay in conjunction with RNA interference technology, the effects of depleting eukaryotic releases factors 1 and 3 (eRF1 and eRF3) were assessed on the termination reaction in human cell lines. Consistent with the established role of eRF1 in triggering peptidyl-tRNA hydrolysis, depletion of eRF1 enhances readthrough at all three stop codons in 293 cells and HeLa cells. Depletion of eRF3 has little or no effect on readthrough in 293 cells but does increase readthrough at all three stop codons in HeLa cells. These results support a direct role for eRF3 in translation termination in higher eukaryotes and also highlight the potential for differences in the abundance or activity of termination factors to modulate the balance of termination to readthrough reactions in a cell-type-specific manner [55,56]. The site-specific incorporation of non-natural amino acids into proteins by nonsense suppression has been widely used to investigate protein structure and function. The intracellular concentration of the eukaryotic release factor 1 (eRF1) is a critical parameter influencing the efficiency of amino acid incorporation by nonsense suppression. RNA interference could be used to lower eRF1 gene expression significantly. It is a five times higher level of amino acid incorporation that was achieved as compared with non-treated control cells, as demonstrated by enhanced green fluorescent protein (EGFP) fluorescence recovery after importing a mutated reporter mRNA together with an artificial amber suppressor tRNA [57].
7.3. Impact of the GAL7 Signal and N2,N2-Dimethylguanosine-26
A 26-nt sequence from the 3’ UTR of the yeast GAL7 mRNA directs accurate and efficient cleavage and polyadenylation to form the 3’ end of the GAL7 mRNA in vivo and in vitro. Whether can this polyadenylation signal function within the context of a tRNA? Insertion of the GAL7 signal into the intron of the dominant SUP4 nonsense suppressor allowed to judge the effect of the insert on SUP4 function by observation of nonsense suppression efficiency in vivo. The GAL7 signal impairs the function of SUP4 in an orientation-dependent manner in vivo, consistent with its ability to specify cleavage and polyadenylation in this context in vitro. Mutation of a UA repeat within the GAL7 signal restores SUP4 function partially, consistent with the role of this repeat as an efficiency element in polyadenylation. Mutations that impair the mRNA 3’ end-processing factors Rna14p and Rna15p restore suppressor function partially. Molecular biological analysis show that the GAL7 signal directs polyadenylation within the body of pre-SUP4 and within the terminator, suggesting that polyadenylation inhibits 5’ and 3’ end processing, as well as removal of the pre-tRNA intron. These results indicate that the GAL7 polyadenylation signal is capable of targeting a pre-tRNA to the mRNA processing pathway [58]. In the majority of eukaryotic tRNAs, the guanosine at position 26 is modified by a dimethyl group, but so far a function of this modification has not been detected. The S. pombe gene (trm1) has been isolated to encoding the tRNA N2,N2-dimethylguanosine-26 methyltransferase. Strains having the gene deleted completely lack N2,N2-dimethylguanosine. In strains carrying the weak ochre tRNA suppressor sup3-i, deletion of trm1 abolishes suppression indicating that the trm1 deletion acts as an antisuppressor mutation. The result suggests that in vivo N2,N2-dimet-hylguanosine- 26 increases the capacity of the sup3-i serine tRNA to translate the UAA (ochre) codon [59].
7.4. The Aminoacylation Specifity of tRNAs and Their Nonsense Suppressor
The exquisite specificity of aminoacylation of tRNA is critically dependent on the correct recognition of tRNA by aminoacyl-tRNA synthetase in protein biosynthesis. The relative concentrations of a tRNA and its cognate aminoacyl-tRNA synthetase are normally well balanced and crucial for maintenance of accurate aminoacylation. When E. coli tRNAGln synthetase is overproduced in vivo, it incorrectly acylates the supF amber suppressor tRNATyr with Gln. This effect is abolished when the intracellar concentration of the cognate tRNAGln2 is also elevate [60]. The aminoacylation specificity (“acceptor identity”) of tRNAs has previously been associated with the position of particular nucleotides, as opposed to distinctive elements of three-dimensional structure. The contribution of a G.U wobble pair in the accepter helix of tRNAAla to acceptor identity was examined with synthetic amber suppressor tRNAs in E. coli. The acceptor identity was not affected by replacing the G.U wobble pair in tRNAAla with a G.A, C.A, or U.U wobble pair. A tRNAAla acceptor identity was conferred on tRNALys when the same site in the acceptor helix was replaced with any of several wobble pairs. When the G.U wobble pair was translocated to another site in acceptor helix of tRNAAla, a substantial acceptor identity was retained [61]. Using synthetic oligonucletides, a collection of E. coli amber suppressor tRNA genes were constructed. In order to determine their specificities, these tRNAs were each used to suppress an amber (UAG) nonsense mutation in the E. coli dihydrofolate reductase gene fol. The mutant proteins were subjected to N-terminal sequence analysis to determine which amino acid had been inserted by the suppressor tRNAs at the position of the amber codon. The suppressors can be classified into three groups on the basis of the protein sequence information (Table 4) [62].
An opal suppressor system has been constructed in E. coli to complement an existing amber suppressor system to study the structural basis of tRNA acceptor identity, particularly the role of middle anticodon nucleotide at position 35. Opal suppressor tRNAs of two tRNAArg isoacceptor sequences each gave arginine in the suppressed protein, while the corresponding amber suppressors with U35 in their CUA anticodons each gave arginine plus a second amino acid in the suppressed protein. Since C35 but not U35 is present in the anticodon of wild-type tRNAArg molecules, while the first anticodon position contains either C34 or U34, these results establish that C35

Table 4. Different amber suppressors insert various kinds of amino acid [62].
contributes to tRNAArg acceptor identity. Initial characterizations of opal suppressor tRNAArg mutants by suppression efficiency measurements suggest that the fourth nucleotide from the 3’ end of tRNAArg (A73 or G73 in different isoacceptors) also contributes to tRNAArg acceptor identity. Wild-type and mutant versions of opal and amber tRNALys suppressors were examined, revealing that U35 and A73 are important determinants of tRNALys acceptor identity [63]. An aminoacylated amber suppressor tRNA (supF) derived from the E. coli tyrosine tRNA can be imported into COS1 cells and acts as a suppressor of amber codons, whereas the same suppressor tRNA imported without prior aminoacylation does not, suggesting that the supF tRNA is not a substrate for any mammalian aminoacyl-tRNA synthetase. These results open the possibility of using the supF tRNA aminoacylated with an amino acid analogue as a general approach for the site-specific insertion of amino acid analogues into proteins in mammalian cells [64].
8. NONSENSE SUPPRESSORS AND GENETIC DISEASES
The characterization of a molecular lesion of β thalassemia in Sardinia was found in 1981. β thalassemia in this area is predominantly the β O type with low levels of β-globin mRNA. Double-stranded complementary DNA prepared from reticulocyte mRNA from a Sardinian patient was cloned in a bacterial plasmid and a β-globin complementary DNA containing clone was isolated and sequenced. At the position corresponding to amino acid number 39, a single nucleotide mutation converted a glutamine codon (CAG) to an amber termination codon (UAG). An amber nonsense mutation at amino acid 17 as a cause of Chinese β O thalassemia was ever report. Thus, β O thalassemia in Sardinia represents the second example of a nonsense mutation, and it was predicted that other β O thalassemias with mutations at various points along the β-globin chain would be found to form a discrete subgroup of β O thalassemia. These experiments further illustrate the heterogeneity of lesions that lead to defective globin chain synthesis in β thalassemia. A human tRNALys gene was converted to an amber suppressor by site-specific mutagenesis of the anticodon. The mutated tRNALys gene directed synthesis of a tRNA that suppressed the UAG amber nonsense mutation in β O thalassemia mRNA. Such genes may be used to detect other nonsense mutations in mammalian cells and may provide an approach to gene therapy for β O thalassaemia due to nonsense mutations [65,66]. Nonsense suppressor tRNAs have been suggested as potential agents for human somatic gene therapy. 179 events of mutations have been surveyed to nonsense codons which cause human germline or somatic disease. The analysis revealed a ratio of approximately 1:2:3 for mutation to UAA, UAG and UGA respectively. This pattern is similar, but not identical, to that of naturally occurring stop codons. The 3’ contexts of new mutations to stop were also analysed. Once again, the pattern was similar to the contexts surrounding natural termination signals. These results imply there will be little difference in the sensitivity of nonsense mutations and natural stop codons to suppression by nonsense suppressor tRNAs. Analysis of the codons altered by nonsense mutations suggests that efforts to design human UAG suppressor tRNAs charged with Trp, Gln, and Glu; UAA suppressors charged with Gln and Glu, and UGA suppressors which insert Arg, would be an essential step in the development of suppressor tRNAs as agents of human somatic gene therapy [67]. Ablation of specific cells can be achieved through the transcriptionally regulated expression of the diphtheria toxin Achain (DT-A) gene in both cell culture and transgenic mice. Such targeted toxin gene expression provides a novel approach to cancer and acquired immunodeficiency syndrome (AIDS) therapy. The effects of mutating selected serine codons to amber (TAG) nonsense codons in the DT-A coding sequence have been investigated. In transient transfection of HeLa cells, DT-A activity was markedly reduced by the introduction of a single amber codon and was virtually eliminated by two amber mutations. Cotransfection of a serine inserting suppressor tRNA expression plasmid substantially restored DT-A expression from both single and double amber mutants. Expression of the same suppressor tRNA also suppressed a previously described amber mutation at the tyrosine codon 28 in DT-A. Thus, nonsense suppression can be used to control the expression of DT-A in mammalian cells, potentially allowing binary control over the targeting of tissues for selective ablation [68]. Nonsense mutations in the dystrophin gene are the cause of Duchenne muscular dystrophy (DMD) in 10% - 15% of patients. In such an event, one approach to gene therapy for DMD is the use of suppressor tRNAs to overcome the premature termination of translation of the mutant mRNA. It was found that the number of cells producing β-galactosidase depends inversely on the dose of the suptRNA gene. A single in vivo injection of the construct providing for expression of the suptRNAochre gene into mdx mouse muscle resulted in the production of dystrophin in 2.5% of fibers. This suggests that suppressor tRNAs are applicable in gene therapy for hereditary diseases caused by nonsense mutations. The mdx mouse is an animal model for human DMD. The lack of dystrophin in mdx mice is caused by an ochre mutation in exon 23 of the dystrophin gene. This study tested the feasibility of inhibiting translational termination as an approach for genetic therapy for diseases caused by nonsense mutations. The first target was a CAT reporter gene bearing an ochre mutation at the 5’ end (CATochre). The second target was the dystrophin gene in mdx mice. The readthrough efficiencies were about 20% in COS cells and 5.5% in rat hearts. At four weeks after a direct injection of plasmid DNA encoding the tRNA suppressor into mdx mice, dystrophin positive fibers were detected by sarcolemmal immunostaining. This is the first convincing data that a tRNA suppressor gene might be a useful in vivo treatment for the genetic disorders caused by nonsense mutations [69,70]. Multicopy tRNA expression plasmids were directly injected into skeletal muscle and into the hearts of transgenic mice carrying a reporter gene with an ochre mutation. A strategy for modulation of suppressor efficiency, applicable to diverse systems and based on tandem multimerization of the tRNA gene, is developed. The product of suppression (chloramphenicol acetyltransferase) accumulates linearly with increases in suppressor tRNA concentration to the point where the ochre-suppressing tRNA(Ser) is in fourto five-fold excess over the endogenous tRNA(Ser). The subsequent suppressor activity plateau seems to be attributable to accumulation of unmodified tRNAs. These results define many salient variables for suppression in vivo, for example, for tRNA suppression employed as gene therapy for nonsense defects [71].
9. SUMMARY AND FUTURE PRESPECTIVES
Positions 34 and 35 of the anticodon of tRNA are very important for its aminoacylation. Some nonnatural amino acids can be aminoacylated by tRNA nonsense suppressor into proteins. A functional transfer RNA suppressor from an eukaryotic yeast can be aminoacylated and inserted an amino acid at a termination codon in prokaryotic host. The efficiency of nonsense codon suppression may be affected by a specific base sequence around nonsense codons and the context of a nonsense suppressor tRNA. No matter which is an eukaryotic host, for example: fruit fly, Xenopus laevis or mammalian cell, tRNA nonsense suppressors are universal. Not only in model lower plants but also higher plants can suppression of nonsense codons happen in their cells? Future work will undoubtedly focus on which kind of living being nonsense suppressors still exist besides known prokaryotic and eukaryotic host. Especially, the application of nonsense suppressors on medicine will attract attention.
![]()
![]()
REFERENCES
- Kohli, J., Kwong, T., Altruda, F., et al. (1979) Characterization of a UGA-suppressing serine tRNA from Schizosaccharomyces pombe with the help of a new in vitro assay system for eukaryotic suppressor tRNAs. Journal of Biological Chemistry, 254, 1546-1551.
- Raftery, L.A., Egan, J.B., Cline, S.W., et al. (1984) Defined set of cloned termination suppressors: In vivo activity of isogenetic UAG, UAA, and UGA suppressor tRNAs. Journal of Bacteriology, 158, 849-859.
- Grundy, F.J. and Henkin, T.M. (1993) tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell, 74, 475-482. doi:10.1016/0092-8674(93)80049-K
- Ryu, Y. and Schultz, P.G. (2006) Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nature Methods, 3, 263-265. doi:10.1038/nmeth864
- Cremer, K.J., Bodemer, M., Summers, W.P., et al. (1979) In vitro suppression of UAG and UGA mutants in the thymidine kinase gene of herpes simplex virus. Proceedings of the National Academy of Sciences USA, 76, 430-434. doi:10.1073/pnas.76.1.430
- Köhrer, C., Yoo, J.H., Bennett, M., et al. (2003) A possible approach to site-specific insertion of two different unnatural amino acids into proteins in mammalian cells via nonsense suppression. Chemistry & Biology, 10, 1095-1102. doi:10.1016/j.chembiol.2003.10.013
- Tuite, M.F., Cox, B.S. and McLaughlin, C.S. (1983) In vitro nonsense suppression in [psi+] and [psi-] cell-free lysates of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences USA, 80, 2824-2828. doi:10.1073/pnas.80.10.2824
- Tuite, M.F., Cox, B.S. and McLaughlin C.S. (1981) An homologous in vitro assay for yeast nonsense suppressors. Journal of Biological Chemistry, 256, 7298-304.
- Bare, L.A. and Uhlenbeck, O.C. (1986) Specific substitution into the anticodon loop of yeast tyrosine transfer RNA. Biochemistry, 25, 5825-5830. doi:10.1021/bi00367a072
- Carrier, M.J. and Buckingham, R.H.(1984)An effect of codon context on the mistranslation of UGU codons in vitro. Journal of Molecular Biology, 175, 29-38. doi:10.1016/0022-2836(84)90443-1
- Kubli, E., Schmidt, T., Martin, P.F., et al. (1982) In vitro suppression of a nonsense mutant of Drosophila melanogaster. Nucleic Acids Res, 10, 7145-7152. doi:10.1093/nar/10.22.7145
- Bain, J.D., Diala, E.S., Glabe, C.G., et al. (1991) Chamberlin AR. Site-specific incorporation of nonnatural residues during in vitro protein biosynthesis with semisynthetic aminoacyl-tRNAs. Biochemistry, 30, 5411-5421. doi:10.1021/bi00236a013
- Rossi, J.J., Schold, M., Larson, G.P., et al. (1982) Functional expression of a yeast ochre suppressor tRNA gene in Escherichia coli. Gene, 20, 423-432. doi:10.1016/0378-1119(82)90211-6
- Rydén, S.M. and Isaksson, L.A. (1984) A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Molecular and General Genetics, 193, 38-45. doi:10.1007/BF00327411
- Raftery, L.A., and Yarus, M. (1985) Site-specific mutagenesis of Escherichia coli gltT yields a weak, glutamic acid-inserting ochre suppressor. Journal of Molecular Biology, 184, 343-345. doi:10.1016/0022-2836(85)90385-7
- Leinfelder, W., Zehelein, E., Mandrand-Berthelot, M.A., et al. (1988)Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature, 331, 723-725. doi:10.1038/331723a0
- Kleina, L.G., Masson, J.M., Normanly, J., et al. (1990) Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. Journal of Molecular Biology, 213, 705-717. doi:10.1016/S0022-2836(05)80257-8
- Buckingham, R.H., Sörensen, P., Pagel, F.T., et al. (1990) Third position base changes in codons 5’ and 3’ adjacent UGA codons affect UGA suppression in vivo. Biochimica et Biophysica Acta, 1050, 259-262. doi:10.1016/0167-4781(90)90177-4
- Hottinger, H., Pearson, D., Yamao, F., et al. (1982) Nonsense suppression in Schizosaccharomyces pombe: The S. pombe Sup3-e tRNASerUGA gene is active in S. cerevisiae. Molecular and General Genetics, 188, 219-224. doi:10.1007/BF00332678
- Hottinger, H., Stadelmann, B., Pearson,D.et al. (1984) The Schizosaccharomyces pombe sup3-i suppressor recognizes ochre, but not amber codons in vitro and in vivo. EMBO Journal, 3, 423-428.
- Pearson, D., Willis, I., Hottinger, H., et al. (1985) Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Molecular and Cellular Biology, 5, 808-815.
- Krupp, G., Thurianx, P., Willis, I., et al. (1985) First identification of an amber nonsense mutation in Schizosaccharomyces pombe: Major differences in the efficiency of homologous versus heterologous yeast suppressor tRNA genes. Molecular and General Genetics, 201, 82-87. doi:10.1007/BF00397990
- Strobel, M.C. and Abelson, J. (1986) Effect of intron mutations on processing and function of Saccharomyces cerevisiae SUP53 tRNA in vitro and in vivo. Molecular and Cellular Biology, 6, 2663-2673.
- Pure, G.A., Robinson, G.W., Naumovski, L., et al. (1985) Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. Journal of Molecular Biology, 183, 31-42. doi:10.1016/0022-2836(85)90278-5
- Pappu, S.S., Roy, K.L. and Bell J.B. (1990) Drosophila melanogaster tRNA(Ser) suppressor genes function with strict codon specificity when introduced into Saccharomyces cerevisiae. Gene, 91, 255-259. doi:10.1016/0378-1119(90)90096-A
- Kim, D. and Johnson, J. (1988) Construction, expression, and function of a new yeast amber suppressor, tRNATrpA. Journal of Biological Chemistry, 263, 7316-7321.
- Kim, D., Raymond, G.J., Clark, S.D., et al. (1990) Yeast tRNATrp genes with anticodons corresponding to UAA and UGA nonsense codons. Nucleic Acids Research, 18, 4215-4221. doi:10.1093/nar/18.14.4215
- Edelman, I. and Culbertson, M.R. (1991) Exceptional codon recognition by the glutamine tRNAs in Saccharomyces cerevisiae. EMBO Journal, 10, 1481-1491.
- Murina, O.A., Moskalenko, S.E. and Zhuravleva, G.A. (2010) Overexpression of genes encoding tRNA(Tyr) AND tRNA(Gln) improves viability of nonsense mutants in SUP45 gene in yeast Saccharomyces cerevisiae. Molecular Biology, 44, 301-310.
- Doerig, R.E., Suter, B., Gray, M., et al. (1988) Identification of an amber nonsense mutation in the rosy516 gene by germline transformation of an amber suppressor tRNA gene. EMBO Journal, 7, 2579-2584.
- Washburn, T. and O’Tousa, J.E. (1992) Nonsense suppression of the major rhodopsin gene of Drosophila. Genetics, 130, 585-595.
- Bienz, M., Kubli, E., Kohli, J., et al. (1980) Nonsense suppression in eukaryotes: The use of the Xenopus oocyte as an in vivo assay system. Nucleic Acids Research, 8, 5169-5178. doi:10.1093/nar/8.22.5169
- Hudziak, R.M., Laski, F.A., RajBhandary, U.L., et al. (1982) Establishment of mammalian cell lines containing multiple nonsense mutations and functional suppressor tRNA genes. Cell, 31, 137-146. doi:10.1016/0092-8674(82)90413-5
- Laski, F.A., Belagaje, R., Hudziak, R.M.,et al. (1984) Synthesis of an ochre suppressor tRNA gene and expression in mammalian cells. EMBO Journal, 3, 2445-2452.
- White, B.T. and McGeoch, D.J. (1987) Isolation and characterization of conditional lethal amber nonsense mutants of vesicular stomatitis virus. Journal of General Virology, 68, 3033-3044. doi:10.1099/0022-1317-68-12-3033
- Summers, W.P., Summers, W.C, Laski, F.A., et al. (1983) Functional suppression in mammalian cells of nonsense mutations in the herpes simplex virus thymidine kinase gene by suppressor tRNA genes. Journal of Virology, 47, 376-379.
- Young, J.F., Capecchi, M., Laski, F.A., et al. (1983) Measurement of suppressor transfer RNA activity. Science, 221, 873-875. doi:10.1126/science.6308765
- Capone, J.P., Sedivy,. J.M., Sharp, P.A., et al. (1986) Introduction of UAG, UAA, and UGA nonsense mutations at a specific site in the Escherichia coli chloramphenicol acetyltransferase gene: Use in measurement of amber, ochre, and opal suppression in mammalian cells. Molecular and Cellular Biology, 6, 3059-3067.
- Sedivy, J.M., Capone, J.P., RajBhandary, U.L., et al. (1987) An inducible mammalian amber suppressor: Propagation of a poliovirus mutant. Cell, 50, 379-389. doi:10.1016/0092-8674(87)90492-2
- Capone, J.P. (1988) Modulation of the phenotypic expression of a human serine tRNA gene by 5’-flanking sequences. DNA, 7, 459-468. doi:10.1089/dna.1.1988.7.459
- Syroid, D. E., Tapping, R.I., and Capone, J.P. (1992) Regulated expression of a mammalian nonsense suppressor tRNA gene in vivo and in vitro using the lac operator/repressor system. Molecular and Cellular Biology, 12, 4271-4278.
- Drabkin, H.J., Park, H.J. and RajBhandary, U.L. (1996) Amber suppression in mammalian cells dependent upon expression of an Escherichia coli aminoacyl-tRNA synthetase gene. Molecular and Cellular Biology, 16, 907- 913.
- Yu, W. and Spreitzer, R.J. (1992) Chloroplast heteroplasmicity is stabilized by an amber-suppressor tryptophan tRNA(CUA). Proceedings of the National Academy of Sciences USA, 89, 3904-3907. doi:10.1073/pnas.89.9.3904
- Esposito, D., Higgs, D.C., Drager, R.G., et al. (2001) A nucleus-encoded suppressor defines a new factor which can promote petD mRNA stability in the chloroplast of Chlamydomonas reinhardtii. Current Genetics, 39, 40-48. doi:10.1007/s002940000179
- Murakami, S., Kuehnle, K. and Stern, D.B. (2005) A spontaneous tRNA suppressor of a mutation in the Chlamydomonas reinhardtii nuclear MCD1 gene required for stability of the chloroplast petD mRNA. Nucleic Acids Research, 33, 3372-3380. doi:10.1093/nar/gki651
- Franklin. S., Lin, T.Y. and Folk, W.R. (1992) Construction and expression of nonsense suppressor tRNAs which function in plant cells. The Plant Journal, 2, 583-588.
- Urban, C., Zerfass, K., Fingerhut, C., et al. (1996) UGA suppression by tRNACmCATrp occurs in diverse virus RNAs due to a limited influence of the codon context. Nucleic Acids Research, 24, 3424-3430. doi:10.1093/nar/24.17.3424
- Chen, Z., Ulmasov, B. and Folk, W.R. (1998) Nonsense and missense translational suppression in plant cells mediated by tRNA(Lys). Plant Molecular Biology, 36, 163- 170. doi:10.1023/A:1005996125011
- Akama, K. and Beier, H. (2003) Translational nonsense codon suppression as indicator for functional pre-tRNA splicing in transformed Arabidopsis hypocotyl-derived calli. Nucleic Acids Research, 31, 1197-207. doi:10.1093/nar/gkg220
- Prescott, C.D. and Kornau, H.C. (1992) Mutations in E. coli 16s rRNA that enhance and decrease the activity of a suppressor tRNA. Nucleic Acids Research, 20, 1567- 1571. doi:10.1093/nar/20.7.1567
- Szweykowska-Kulinska, Z. and Beier, H. (1991) Plant nonsense suppressor tRNA(Tyr) genes are expressed at very low levels in vitro due to inefficient splicing of the intron-containing pre-tRNAs. Nucleic Acids Research, 19, 707-712. doi:10.1093/nar/19.4.707
- Belgrader, P., Cheng, J. and Maquat, L.E. (1993) Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proceedings of the National Academy of Sciences USA, 90, 482-486. doi:10.1073/pnas.90.2.482
- Phillips-Jones, M.K., Watson, F.J. and Martin, R. (1993) The 3’ codon context effect on UAG suppressor tRNA is different in Escherichia coli and human cells. Journal of Molecular Biology, 233, 1-6. doi:10.1006/jmbi.1993.1479
- Tapping, R.I., Syroid, D.E., Bilan, P.T., et al. (1993) The 5’ flanking sequence negatively modulates the in vivo expression and in vitro transcription of a human tRNA gene. Nucleic Acids Research, 21, 4476-4482. doi:10.1093/nar/21.19.4476
- Le Goff, X., Philippe, M. and Jean-Jean, O. (1997) Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Molecular and Cellular Biology, 17, 3164-3172.
- Janzen, D.M. and Geballe, A.P. (2004) The effect of eukaryotic release factor depletion on translation termination in human cell lines. Nucleic Acids Research, 32, 4491-502. doi:10.1093/nar/gkh791
- Ilegems, E., Pick, H.M. and Vogel, H.(2004) Downregulation of eRF1 by RNA interference increases misacylated tRNA suppression efficiency in human cells. Protein Engineering, Design and Selection, 17, 821-827. doi:10.1093/protein/gzh096
- Liang, S., Briggs, M.W. and Butler, J.S. (1997) Regulation of tRNA suppressor activity by an intron-encoded polyadenylation signal. RNA, 3, 648-659.
- Niederberger, C., Gräub, R., Costa, A., et al. (1999) The tRNA N2,N2-dimethylguanosine-26 methyltransferase encoded by gene trm1 increases efficiency of suppression of an ochre codon in Schizosaccharomyces pombe. FEBS Letters, 464, 67-70. doi:10.1016/S0014-5793(99)01679-8
- Swanson, R., Hoben, P., Sumner-Smith, M., et al. (1988) Accuray of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science, 242, 1548-1551. doi:10.1126/science.3144042
- McClain, W.H., Chen, Y.M., Foss, K., et al. (1988) Association of transfer RNA acceptor identity with a helical irregularity. Science, 243, 1256. doi:10.1126/science.243.4896.1256-b
- Normanly, J., Kleina, L.G., Masson, J.M., et al. (1990) Construction of E. coli amber suppressor tRNA genes. III. Determination of tRNA specifity. Journal of Molecular Biology, 213, 719-726. doi:10.1016/S0022-2836(05)80258-X
- McClain, W.H., Foss, K., Jenkins, R.A., et al. (1990) Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proceedings of the National Academy of Sciences USA, 87, 9260-9264. doi:10.1073/pnas.87.23.9260
- Köhrer, C., Xie, L., Kellerer, S., et al. (2001) Import of amber and ochre suppressor tRNAs into mammalian cells: A general approach to site-specific insertion of amino acid analogues into proteins. Proceedings of the National Academy of Sciences USA, 98, 14310-14315. doi:10.1073/pnas.251438898
- Trecartin, R.F., Liebhaber, S.A., Chang, J.C., et al. (1981) beta zero thalassemia in Sardinia is caused by a nonsense mutation. J Clin Invest, 68, 1012-1017. doi:10.1172/JCI110323
- Temple, G.F., Dozy, A.M., Roy, K.L., et al. (1982) Construction of a functional human suppressor tRNA gene: An approach to gene therapy for beta-thalassaemia. Nature, 296, 537-540. doi:10.1038/296537a0
- Atkinson, J. and Martin, R. (1994) Mutations to nonsense codons in human genetic disease: Implications for gene therapy by nonsense suppressor tRNAs. Nucleic Acids Research, 22, 1327-1334. doi:10.1093/nar/22.8.1327
- Robinson, D.F. and Maxwell, I.H. (1995) Suppression of single and double nonsense mutations introduced into the diphtheria toxin A-chain gene: A potential binary system for toxin gene therapy. Human Gene Therapy, 6, 137-143. doi:10.1089/hum.1995.6.2-137
- Kiselev, A.V., Ostapenko, O.V., Rogozhkina, E.V., et al. (2002) Suppression of nonsense mutations in the Dystrophin gene by a suppressor tRNA gene. Molecular Biology, 36, 43-47.
- Li, K., Zhang, J., Buvoli, M., et al. (1997) Ochre suppressor transfer RNA restored dystrophin expression in mdx mice. Life Sciences, 61, 205-209. doi:10.1016/S0024-3205(97)00714-5
- Buvoli, M., Buvoli, A. and Leinwand, L.A. (2000) Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Molecular and Cellular Biology, 20, 3116-3124. doi:10.1128/MCB.20.9.3116-3124.2000

