American Journal of Computational Mathematics
Vol.05 No.02(2015), Article ID:57418,10 pages
10.4236/ajcm.2015.52017
A Survey on Semantic Similarity Measures between Concepts in Health Domain
Abdelhakeem M. B. Abdelrahman1, Ahmad Kayed2
1College of Post Graduate Studies, Sudan University of Science and Technology, Khartoum, Sudan
2CS Department, IT Faculty, Middle East University (MEU), Amman, Jordan
Email: ambashir@imamu.edu.sa, drkayed@ymail.com
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 15 April 2015; accepted 21 June 2015; published 26 June 2015
ABSTRACT
The similarity between biomedical terms/concepts is a very important task for biomedical information extraction and knowledge discovery. The measures and tests are tools used to define how to measure the goodness of ontology or its resources. The semantic similarity measuring techniques can be classified into three classes: first, measuring semantic similarity using ontology/ taxonomy; second, using training corpora and information content and third, combination between them. Some of the semantic similarity measures are based on the path length between the concept nodes as well as the depth of the LCS node in the ontology tree or hierarchy, and these measures assign high similarity when the two concepts are in the lower level of the hierarchy. However, most of the semantic similarity measures can be adopted to be used in health domain (Biomedical Domain). Many experiments have been conducted to check the applicability of these measures. In this paper, we investigate to measure semantic similarity between two concepts within single ontology or multiple ontologies in UMLS Metathesaurus (MeSH, SNOMED-CT, ICD), and compare my results to human experts score by correlation coefficient.
Keywords:
Biomedical Ontology, Semantic Similarity, Biomedical Concept, Unified Medical Language System (UMLS)

1. Introduction
Ontology is test bed of semantic web, capturing knowledge about certain area via providing relevant concept and relation between them. Quality metrics are essential to evaluate the quality. Metrics are based on structure and semantic level. At the present, the ontology evaluation is based only on structural metrics, which has not been very appropriate in providing desired results.
Semantic similarity measures are widely used in Natural Language Processing. We show how six existing domain-independent measures can be adapted to the biomedical domain. Semantic similarity techniques are becoming important components in most intelligent knowledge-based and Semantic Information Retrieval (SIR) systems [1] .
The classification of these includes semantic similarity measures for single ontology and for multiple ontologies, and the classification is based on how the semantic similarity measure is quantified [2] .
Ahmad kayed et al. [3] : proposed various methodologies to evaluate ontologies: most of them belong to one of the following categories:
o Coverage techniques between an ontology and a domain of knowledge that the ontology is created for.
o Human experts who try to assess how well the ontology meets a set of predefined criteria, standards, and requirements.
o Using the ontology in the context of an application to evaluate its effectiveness. The use of the system may detect weakness or strength points in the ontology.
o Comparing the ontology with other ontologies in the same domain.
o Studying ontology’s relationships considering some measures.
o Comparing the formal representation of the ontology with other ontologies formal representations, criterions, or measures.
Measures and tests are provided to define how we can measure the “goodness” of ontology or its resources. Many experiments have been conducted to check the applicability of these measures [3] .
General English ontology based structure similarity measures can be adopted to be used into the biomedical domain within UMLS. New approach for measuring semantic similarity between biomedical concepts using multiple ontologies is proposed by Al-Mubaid and Nguyen [2] [4] . They proposed new ontology structure based technique for measuring semantic similarity between single ontology and multiple ontologies in the biomedical domain within the frame work of Unified Medical Subject Language System (UMLS). Their proposed measure is based on three features [4] : first, cross modified path length between two concepts; second, new features of common specificity of concepts in the ontology; third, local ontology granularity of ontology cluster.
2. Health Domain Ontologies
Most of the semantic similarity work in the biomedical domain uses only ontology (e.g. MeSH, SOMED-CT) for computing the similarity between the biomedical terms [5] . However, in this work we use ICD-10 ontology as primary source to computing the similarity between concepts in biomedical domain.
New ontologies in biology and medicine continue to increase as the need for them arises. Some of the most well-studied and prominent examples are presented here.
1) MeSH: Stands for Medical Subject Headings, is the one of source vocabularies used in UMLS. Its includes about 15 high level categories, and each category is divided into sub categories and assign a letter: A for Anatomy, B for organisms, C for diseases, and so on [4] [6] .
2) SNOMED-CT: Stand for Systemized Nomenclature of Medicine Clinical Term, was included in UMLS in May 2004 [4] [7] . is a comprehensive clinical Ontology maintained by the International Health Terminology Standards Development Organization (IHTSDO) [8] .
3) FMA: Stand for Foundational Model of Anatomy: One of the most coherently structured ontologies in biomedicine is the Foundational Model of Anatomy, domain ontology of the classes and relationships that pertain to the structural organization of the human body [7] .
4) GALEN: Stand for Generalized Architecture for Languages, Encyclopedias and Nomenclatures (GALEN), is Common Reference Model. The goal of the GALEN project is to provide re-usable terminology resources for clinical systems [7] [9] .
5) Medical Entities Dictionary: The Medical Entities Dictionary (MED) is a concept-oriented terminology developed and used in Columbia University and the New York Presbyterian Hospital (NYPH) [10] . It currently contains approximately 97,000 concepts organized into a semantic network of frame-based term descriptions, encompassing those terms used in laboratory, pharmacy, radiology, and billing systems. It includes knowledge about synonyms, taxonomic and other types of relations, and mappings to other terminologies.
6) National Cancer Institute Thesaurus. The NCI Thesaurus is a description logic-based terminology that is a component of the US National Cancer Institute (NCI) Bioinformatics ca CORE distribution [9] .
7) Unified Medical Language System: Many studies evaluating the usefulness of the UMLS as a terminology and knowledge resource for tasks ranging from terminology translation to domain ontology construction have been published in recent years [9] .
8) International Classification of Diseases (ICD): The newest edition (ICD-10) is divided into 21 chapters: (Infections, Neoplasm, Blood Diseases, Endocrine Diseases, etc.), and denote about 14,000 classes of diseases and related problems. The first character of the ICD code is a letter, and each letter is associated with a particular chapter, except for the letter D, which is used in both Chapter II, Neoplasm, and Chapter III, Diseases of the blood and blood-forming organs and certain disorders involving the immune mechanism, and the letter H, which is used in both Chapter VII, Diseases of the eye and adnexa and Chapter VIII, Diseases of the ear and mastoid process. Four chapters (Chapters I, II, XIX and XX) use more than one letter in the first position of their codes. Each chapter contains sufficient three-character categories to cover its content; not all available codes are used, allowing space for future revision and expansion. Chapters I-XVII relate to diseases and other morbid conditions, and Chapter XIX to injuries, poisoning and certain other consequences of external causes. The remaining chapters complete the range of subject matter nowadays included in diagnostic data. Chapter XVIII covers Symptoms, signs and abnormal clinical and laboratory findings, not elsewhere classified. Chapter XX, External causes of morbidity and mortality, was traditionally used to classify causes of injury and poisoning, but, since the Ninth Revision, has also provided for any recorded external cause of diseases and other morbid conditions. Finally, Chapter XXI, Factors influencing health status and contact with health services, is intended for the classification of data explaining the reason for contact with health-care services of a person not currently sick, or the circumstances in which the patient is receiving care at that particular time or otherwise having some bearing on that person’s care. The chapters are subdivided into homogeneous “blocks” of three-character categories. most of the three character categories are subdivided by means of a fourth, numeric character after a decimal point, allowing up to 10 subcategories [9] [11] .
3. Semantic Similarity Measures Challenges in the Health Domain
Most of the semantic similarity work in the biomedical domain uses only ontology (e.g. MeSH, SOMED-CT) for computing the similarity between the biomedical terms.
Most of existing semantic similarity measures that used ontology structure as the primary source can’t measure the similarity between terms/concepts using single ontology or multiple ontologies in the biomedical domain within frame work Unified Medical Language System (UMLS).
Some of the semantic similarity measures have been adopted to biomedical field by incorporating domain information extracted from clinical data or medical ontologies.
4. Semantic Similarity Measures Classification
Figure 1 and Figure 2 [12] : illustrate the semantic similarity classification for single ontology and cross ontologies. To find Semantic similarity between two terms in ontology, by find shortest path length between them in the ontology (shortest path length) giving the length are is-a/part of. A number of approaches have been developed using ontology as primary information sources, and mostly applied in the general English domain using for example WordNet. However, most of the semantic similarity techniques such as general English ontology based structure similarity measures can be adopted to be used into the biomedical domain within UMLS framework.
5. Semantic Similarity Measures for Single Ontology
In this paper, we focus only on these semantic similarity measures that used ontology as primary information source.
5.1. Ontology Structure-Based Similarity Measures
Most of the measures that are based on the structure of the ontology are actually based on: path length/distance (shortest path length) between the two concept nodes, and depth of concept nodes in the ontology/is-a hierarchy tree, e.g. some of the measures are based on WordNet include: path length, Wu & palmer, leacock & chodorow, and Li et al. [4] [12] .
Figure 1. Classification of semantic similarity measures for single ontology.
Figure 2. Classification of semantic similarity measures for cross ontology.
5.1.1 Path Length Based Measures
The similarity measurement among concepts is based on the path distance separating the concepts. These measures compute similarity in terms of the shortest path between the target synsets (group of synonyms) in the taxonomy.
Rada et al. [12] in this measure the semantic distance is computed by counting the number of edges between concepts in the taxonomy. The experiments were conducted using MeSH (Medical Subject Headings-Biomedi- cal ontology) ontology. They assume two concept c1, c2 as shortest path linking them (sp (c1, c2)) as estimate distance.
 (1)
(1)
Figure 3 [2] [4] [13] : show the shortest path between two concepts a5 and b1 is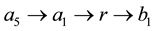 .
.
Also simple edge-counting measure proposed by Rada [14] :
 (2)
(2)
where N1 and N2 are the minimum number of taxonomical links from c1 to c2 to their LCS, respectively.
5.1.2. Depth Relative Measures
Depth relative measures are basically the shortest path approaches, but they consider the depth of the edges connecting the two concepts in the overall structure of the ontology to quantify similarity. It calculates the depth
Figure 3. Hierarchy tree of concepts.
from the root of the taxonomy to the target concept.
1) Wu and Palmer Similarity Measure [12] proposed a new method which define the semantic similarity measure between concepts C1 and C2 as
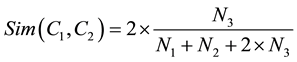 (3)
(3)
where N1 is the length given as number of nodes in the path from C1 to C3 which is the least common super concept of C1 and C2, and N2 is the length given in number of nodes on a path from C2 to C3. N3 represents the global depth of the hierarchy and it serves as the scaling factor. For example: (LCS (G00.1, G00.3) = G00 and LCS (G00, G01) = G) of two concept nodes and N1, N2 are the path lengths from each concept node to LCS, respectively.
2) Leacok and Chodorow [12] are proposed non linear adaptation of Rada’s distance:
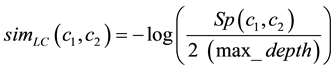 (4)
(4)
max_depth is longest of the shortest path linking concept to concept, which subsumed all others. The Least Common Ancestor (LCA) of concept a5 and b1 is r in Figure 3.
5.2. Information Content-Based Similarity Measure
These measures use Information Content (IC) of concept nodes drive from ontology hierarchy structure and corpus statistics. Some of Information Content-based similarity measures in WordNet include: [2] [4] .
5.2.1. Resnik Similarity Measure
Resnik [7] the similarity between a pair of concepts (c1 and c2) is estimated as the amount of taxonomical information they share. In a taxonomy, this information is represented by the least common subsumer of both terms (LCS (c1, c2)), which is the most specific taxonomical ancestor common to c1 and c2 in a given ontology. Formally:
 (5)
(5)
5.2.2. Lin Similarity Measure
This measure depends on the relation between information content (IC) of the LCS of two concepts and the sum of the information content of the individual concepts [7] [12] [15] .
Formally:
 (6)
(6)
5.2.3. Jiang and Conrath Similarity Measure
Jiang and Conrath [12] [16] define the measure as subtract the information content of the LCS from the sum of the information content of the individual concepts, formally:
 (7)
(7)
5.3. Hybrid-Based Similarity Measures
Combine the above ideas [8] : term similarity is computed by matching synonyms, term neighborhoods and term features.
5.4. Feature-Based Similarity Measure
Measure similarity between two terms as a function of their properties (e.g., their description or “glosses” in WordNet or “scope notes” in MeSH) or based on their relationships to other similar terms in the taxonomy [8] . Analyzing the amount of common and non-common knowledge features to estimate similarity between two concepts [17] .
6. Semantic Similarity Measures for Cross Ontology
In this case, the concepts for which similarity is to be assessed belong to two different ontologies. The secondary ontology is connected to the primary ontology through the common nodes. Two nodes in two ontologies are equivalent if they refer to the same concept.
6.1. Al-Mubaid and Nguyen Similarity Measure
They are combined both features, in one measure, i.e. they will take the specificity of concept node into account by utilizing the depth features of concepts. The LCS of two nodes determine the common specificity of two concept according to ontology structure only. Further more local densities such as link strength/weight also affect the similarity. One way for measuring local density is using IC of concept based on corpus statistics, and since there is no standard corpus in biomedical domain. They used only ontology based features as properties of semantic similarity. Their proposed method don’t required ontology essentially with formal semantic relation between terms, it can be applied onto any terminology structure, structure vocabulary or in general Direct Acyclic Graph (DAG) with length between the nodes.
In this measure they are put rules and assumptions which satisfied their proposed measure. They wont to combine all semantic features in one measure in an effective and logical way.
Rule 1: The semantic similarity scale system reflects the degree of similarity of pairs of concepts comparably in single ontology or in cross-ontology. This rule ensures that the mapping of one ontology (called secondary ontology) to another ontology (called primary ontology) does not deteriorate the similarity scale of the primary ontology [2] [4] .
Rule 2: The semantic similarity must obey local ontology’s similarity rule as follow:
Rule 2.1: The shorter the distance between two concept nodes in the ontology, the more they are similar.
Rule 2.2: Lower level pairs of concept nodes are more similar than higher level pairs.
Rule 2.3: The maximum similarity is arises when the two concept nodes are the same node in the ontology.
Assumptions
1) They used logarithms (inverse of exponential for semantic distance). In rule 2.3, the semantic similarity reached higher similarity when the two concept nodes are in the same node regardless of any other features, hence, should used non linear approach to combine the features.
2) Non linear function is universal combination low of semantic similarity features.
6.2. New Common Specificity Features
Proposed by [2] [4] , they used path length and depth of concept nodes to improved performance. The least common subsumer (LCS) of two concepts node in the ontology is lowest node that connect pairs of concepts. It used to determine common specificity of two concept nodes in the cluster. So finding the depth of their LCS node and then scaling this depth by depth D of the cluster as follow:
 (8)
(8)
where D is depth of the cluster. The smaller common specificity of two concept nodes means that they are more similar and share more information.
Single cluster similarity: Their proposed measure for single cluster is:
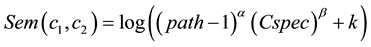 (9)
(9)
where α > 0 and β > 0, k constant and must be (k ≥ 1), and Cspec calculate in Equation (8).
Sem = 0 when depth = 1 regardless of (Cspec).
6.3. Cross-Cluster Semantic Similarity
The cluster has largest depth is main cluster (primary cluster) and all remaining cluster is secondary.
Case 1 similarity within primary cluster:
When two concept nodes in the primary cluster, used Equation (9) to measure similarity.
Case 2 cross cluster similarity:
The LCS of two concept node is global root node, which belong to the two clusters, and one of the two cluster belong to the primary cluster while another belong to secondary cluster. Then the common specificity is given as follow:
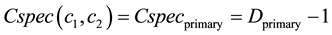 (10)
(10)
where Dprimary is the depth of the primary cluster.
Case 3: Similarity within a single secondary ontology: when two concept nodes are in single secondary cluster.
 (11)
(11)
 (12)
(12)

where Path(C1, C2) secondary and CSpec(C1, C2) secondary are the Path and CSpec between C1 & C2 in the secondary cluster.
Case 4 Similarity within multiple secondary ontology:
One of the secondary clusters acts temporarily as the primary cluster to calculate Cspec and path using cross- cluster approach as in case 2 above. Then semantic distance is computed using case 3.
Hisham Al-Mubaid & Nguyen [13] [18] proposed measure take the depth of their least common subsumer (LCS) and the distance of the shortest path between them. The higher similarity arises when the two concept are in the lower level of the hierarchy. Their similarity measure is:

where
L(c1, c2) is shortest distance between c1 and c2.
Depth L(c1, c2) is depth of L(c1, c2) using node counting.
L(c1, c2) lowest common subsumer of c1 and c2.
D is maximum depth of the taxonomy.
The similarity equal 1, where two concept nodes are in the same cluster/ontology. The maximum value of this measure occurs when one of the concept is the left most leaf node, and the other concept is right leaf node in the tree.
Path distance between two concepts, when two pairs of two concepts have the same path distance, they have the same value of semantic similarity. In Figure 3, similarity (n1, n5) = similarity (n2, n4) but (n2, n4) share more information and attributes, so they are more similar than (n1, n5). In this measure the high numeric similarity result between (c1, c2) means the lower semantic similarity between two concept.
In the ICD tree, let us consider an example in ICD10 terminology. The category tree is “Inflammatory diseases of the central nervous system” and is assigned letter G in ICD10 terminology version 2015 [19] at the link (http://www.apps.who.int/classifications/icd10/browse/2015/en). This tree looks as follows:
Inflammatory diseases of the central nervous system [G]
The similarity between “bacterial meningitis, not elsewhere classified [G00]” and “meningitis in bacterial diseases classified [G01]” is less similarity than the similarity between “Pneumococcal meningitis [G00.1]” and “Staphylococcal meningitis [G00.3]”. However, in this measure they take into account the depth of the LCS of two concepts, in path length and leacock & chodorwo produce semantic similarity for two pairs [(G00, G01) and (G00.1, G00.3)] in sim (c1, c2) measure Equation (14) give high similarity in lower level in the ontology hierarchy ([G00.1, G00.3]).
Choi & Kim (C.K.) [2] [4] [13] semantic similarity measure was developed to measure concept similarity in the Yahoo category tree. However, it doesn’t use LCS in the measure so like the case of the example above, it gives the same similarity value for the two pair (a1, b1) and (a2, a5), see Table 1. With this analysis, we see that Al-Mubaid & Nyguan Measure can surpass the other measures.
The higher numeric similarity result between (G00, G01) means the lower semantic similarity between (G00, G01).
7. Semantic Similarity Measures in Health Domain
7.1. Rada et al.
Proposed semantic distance as a potential measure for semantic similarity between two concepts in MeSH, and
Table 1. Measure comparison.
implemented the shortest path length measure, called CDist, based on the shortest distance between two concept nodes in the ontology. They evaluated CDist on UMLS Metathesaurus (MeSH, SNOMED, ICD9), and then compared the CDist similarity scores to human expert scores by correlation coefficients.
7.2. Caviedes and Cimino
[7] implemented shortest path based measure, called CDist, based on the shortest distance between two concepts nodes in the ontology. They evaluated CDist on UMLS Metathesaurus (MeSH, SNOMED, ICD9), and then compared the CDist similarity scores to human expert scores by correlation coefficient.
7.3. Pedersen et al.
[1] proposed semantic similarity and relatedness in the biomedicine domain, by applied a corpus-based context vector approach to measure similarity between concepts in SNOMED-CT. Their context vector approach is ontology-free but requires training text, for which, they used text data from Mayo Clinic corpus of medical notes.
8. Evaluation
8.1. Datasets
There are no standard human rating sets for semantic similarity in biomedical domain. Thus, Al-Mubaid and Nguyen [2] [13] used dataset from Pedersen et al. [1] in Table 2, which was annotated by 3 physician and 9 medical index experts to evaluate their proposed measure in biomedical domain.
Table 2. Dataset.
8.2. Experiments and Results
Table 2. Test set of 30 medical term pairs sorted in the order of the averaged physicians’ scores (taken from Pedersen et al. 2005 [1] ). Al-Mubaid and Nguyen [13] [18] find only 25 out of the 30 concept pairs in MeSH using MeSH browser version 2006.
Pedersen et al. [1] tested 29 out of the 30 concept pairs as one pair was not found in SNOMED-CT). The concept pairs in bold, in Table 2, are the ones that contains a term that was not found in MeSH and we did not include in their experiments. Some terms have more than one position in MeSH tree, for example, the term “Acne” has three different positions: C17.800.030.150, C17.800.271.125.200, and C17.800.794.111. They use in this case the minimum semantic similarity distance between the two concepts.
9. Conclusions & Future Work
In this work, we discuss the basics of semantic similarity measures, the classification of single ontology similarity measures and cross ontologies similarity measures. We prepare a brief introduction of the various semantic similarity measures in health domain. However, from all the above, we can used SemDist as semantic similarity measures in the health domain.
In future work, we intend to explore the semantic similarity measures in health domain (ICD, MeSH, and SNOMED-CT) within UMLS frame work. We also prepare to implement a web-based user interface for all these semantic similarity measures and to make it available freely to researchers over the Internet. That will be much helpful for interested researchers in the field of bioinformatics text mining.
References
- Pedersen T., Pakhomov, S.V.S., Patwardhan, S. and Chute, C.G. (2007) Measures of Semantic Similarity and Relatedness in the Biomedical Domain. Journal of Biomedical Informatics, 40, 288-299. http://dx.doi.org/10.1016/j.jbi.2006.06.004
- Al-Mubaid, H. and Nguyen, H.A. (2009) Measuring Semantic Similarity between Biomedical concepts within Multiple Ontologies. IEEE Transactions on Systems, Man, and Cybernetics―Part C: Applications and Reviews, 39, 389-397.
- Kayed, A., et al. (2013) Ontology Evaluation: Which Test to Use. 2013 5th International Conference on Computer Science and Information Technology (CSIT), Amman, 27-28 March 2013, 45-48. http://dx.doi.org/10.1109/csit.2013.6588756
- Al-Mubaid, H. and Nguyen, H.A. (2006) A Cluster-Based Approach for Semantic Similarity in the Biomedical Domain. Proceedings of the 28th IEEE, EMBS Annual International Conference, New York, 30 Augugst-3 September 2006, 2713-2717. http://dx.doi.org/10.1109/iembs.2006.259235
- Al-Mubaid, H. and Nguyen, H.A. (2006) Using MEDLINE as Standard Corpus for Measuring Semantic Similarity in the Biomedical Domain. 6th IEEE Symposium on BionInformatics and BioEngineering (BIBE’06), Arlington, 16-18 October 2006, 315-318. http://dx.doi.org/10.1109/bibe.2006.253295
- Tartir, S. (2009) Ontology-Driven Question Answering and Ontology Quality Evaluation. PhD Thesis, University of Georgia, Athens.
- Batet, M., Sánchez, D. and Valls, A. (2011) An Ontology-Based Measure to Compute Semantic Similarity in Biomedicine. Journal of Biomedical Informatics, 44, 118-125. http://dx.doi.org/10.1016/j.jbi.2010.09.002
- Garla1, V.N. and Brandt, C. (2012) Semantic Similarity in the Biomedical Domain: An Evaluation across Knowledge Sources. Garla and Brandt BMC Bioinformatics, 13, 261. http://dx.doi.org/10.1186/1471-2105-13-261
- Ivanovic, M. and Budimac, Z. (2014) An Overview of Ontologies and Data Resources in Medical Domains. Expert Systems with Applications, 41, 5158-5166. http://dx.doi.org/10.1016/j.eswa.2014.02.045
- Nguyen, H. and Al-Mubaid, H. (2006) New Semantic Similarity Techniques of Concepts Applied in the Biomedical Domain and WordNet. MS Thesis, University of Houston Clear Lake, Houston.
- World Health Organization (2010) International Statistical Classification of Diseases and Related Health Problems. 10th Revision Edition, WHO, Geneva.
- Elavarasi, S.A., Akilandeswari, J. and Menaga, K. (2014) A Survey on Semantic Similarity Measure. International Journal of Research in Advent Technology, 2, 389-398.
- Batet, M. (2010) Ontology-Based Semantic Clustering. Ph.D. Thesis, University of Rovira I Vergili.
- Sanchez, D., Solé-Ribalta, A., Batet, M. and Serratosa, F. (2012) Enabling Semantic Similarity Estimation across Multiple Ontologies: An Evaluation in the Biomedical Domain. Journal of Biomedical Informatics, 45, 141-155. http://dx.doi.org/10.1016/j.jbi.2011.10.005
- Hliaoutakis, A., Varelas, G., Voutsakis, E., Petrakis, E.G.M. and Milios, E. (2006) Information Retrieval by Semantic Similarity. International Journal on Semantic Web and Information Systems, 3, 55-73.
- Sathya, D. and Uthayan, K.R. (2011) Proposal for Semantic Metric to Assess the Quality of Ontologies. Proceedings of the 2011 International Conference on Signal Processing, Communication, Computing and Networking Technologies, Thuckafay, 21-22 July 2011, 754-756. http://dx.doi.org/10.1109/ICSCCN.2011.6024651
- Lin, D.K. (1998) An Information-Theoretic Definition of Similarity. Proceedings of the 15th International Conference on Machine Learning, Madison, 24-27 July 1998, 296-304.
- Al-Mubaid, H. and Nguyen, H.A. (2006) New Ontology-Based Semantic Similarity Measure for the Biomedical Domain. Proceedings of the 2006 IEEE International Conference on Granular Computing, Atlanta, 10-12 May 2006, 623-628.
- http://apps.who.int/classifications/icd10/browse/2015/en#/VI






