Journal of Biomedical Science and Engineering
Vol. 5 No. 12A (2012) , Article ID: 26255 , 8 pages DOI:10.4236/jbise.2012.512A107
Synergy between molecular biology and imaging science toward mechanism-based biomarkers associated with prostate cancer
![]()
National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, USA
Email: setob@mail.nih.gov
Received 4 November 2012; revised 5 December 2012; accepted 15 December 2012
Keywords: Prostate Cancer; Biomarkers; Imaging; Molecular Mechanisms
ABSTRACT
Prostate cancer is a heterogeneous disease with subtypes that are characterized by different molecular profiles as a result of chromosomal rearrangements, epigenetic modifications, and activation of various signaling pathways. The subtype heterogeneity contributes to the challenges with a definitive diagnosis and biomarkers for disease progression. The current diagnostic test based on the detection of prostate specific antigen lacks sensitivity and specificity. Imaging plays an important role in characterizing biomarkers and elucidating the underlying molecular mechanisms. For example, 18F-fluoro-2-deoxy glucose is commonly used to assess cancer cell metabolism. More recently, magnetic resonance spectroscopic observations of the in vivo dynamic conversion of hyperpolarized 13Cpyruvate to lactate demonstrate that imaging enables the visualization of molecular processes. Biomarkers have also been developed that reveal aberrant cell growth and proliferation, both hallmarks of cancer. Androgen dependent and independent signaling pathways underpin prostate cancer pathogenesis as they lead to downstream effect in cell growth, proliferation, survival, and suppression of apoptosis. Molecular imaging with radiolabeled ligands and positron emission tomography/computed tomography has provided quantitative characterization of the interactions between receptors and testosterone or growth factors. These observations, along with data on genetic alterations of the receptor genes, shed light on signal transduction involved in prostate cancer. This review article highlights advances in the understanding of the molecular mechanisms of prostate cancer and the synergy of this knowledge with imaging in characterizing potential biomarkers of the disease.
1. INTRODUCTION
Understanding the molecular mechanism of disease provides an opportunity to detect disease before clinical symptoms appear. It also serves as the basis to guide drug development and treatment decisions. Thanks to decades of fundamental discoveries in recombinant DNA research in microorganisms and model animals, mechanism-based diagnosis and identification of specific genetic alterations are possible. By creating and identifying mutants, researchers can investigate the aberrant phenotypes, i.e. disease states, expressed in the mutants and the altered biological processes that contribute to the pathology. More recently, genomic, epigenetics, proteomic, and metabolomic data reveal multiple dimensions of the molecular basis of disease. Indeed discoveries of the responsible genes for diseases exploded after the sequencing of the human genome was completed [1,2]. One picture that emerged from these discoveries is the complexity of disease mechanisms, involving interactions of multiple biological processes.
This literature review examines the molecular mechanisms for prostate cancer. Much research has focused on the genetic driver for tumorigenesis and the characterization of signaling pathways and their interactions involved with prostate cancer [3,4]. A complex molecular portrait has emerged that involves multiple biological processes: transcription, signaling, inflammation, and genetic and epigenetic modifications leading to aberrant cell proliferation, differentiation, angiogenesis, and suppression of apoptosis. An approach to elucidating these biological processes is to identify the biomarkers involved. This review sheds light on the clinical applications of imaging that complement molecular profiles as biomarkers of the underlying disease mechanism for prostate cancer.
2. PROSTATE CANCER BIOMARKERS
Prostate cancer is the most common cancer and the second leading cause of cancer-related death in men, causing approximately 32,000 men to die from this cancer in 2010. The current screening method is based on prostate specific antigen (PSA). PSA is a serine protease, which is produced in an androgen-dependent mechanism at low levels in healthy prostate, but at increased levels in cancerous cells. While detection of PSA has long been used for screening of prostate cancer, the test remains controversial. Its usefulness was called into question by the recent US Preventive Services Task Force recommendation against using the PSA test to screen for prostate cancer among men regardless of age [5]. Research on alternative prostate cancer biomarkers that are mechanism-based is growing. These include molecular and metabolic biomarkers that are reflective of the aberrant metabolism and signaling pathways. Martin and her colleagues described several serum and tissue prostate cancer biomarkers that reflect cancer cell characteristics for increased transcription initiation (Src-3), evasion of cellcell adhesion (E-cadherin), activated signal transduction (interleukin-6 receptors), and stem cell differentiation (PSCA). These are up and coming potential replacements for PSA [6]. PCA3 and PSCA are described in more details to illustrate the range of potential biomarker candidates currently under development. PCA3 is a noncoding RNA that has been shown to be upregulated in more than 95% of primary prostate tumors. Because it is differentially upregulated in tumor cells among neighboring healthy cells by 66 fold, a test based on PCA3 has the potential to differentiate normal from diseased prostate tissue and thus to precisely guide biopsy procedures [7]. It is currently being developed into a urine test to identify men who pose a high risk for a positive biopsy or a follow-up re-biopsy. PSCA is an antigen found in both healthy and cancerous prostate. It is found in primary and metastatic tumors [8]. It is over-expressed in cancerous prostate and may serve as a prognosticator of cancer progression.
Metabolic biomarkers are reflective of the physiologic state of cancer cells. Otto Warburg, in the 1920’s, observed that cancer cells have higher metabolic rates for glucose and faster conversion of pyruvate to lactate compared to normal cells [9,10]. Consistent with this early observation on tumor metabolism is the recent finding by Seth et al. [11] that lactate dehydrogenase A (LDH-A), an isoform of lactate dehydrogenase, which catalyzes pyruvate to lactate, is up-regulated in tumor tissues. The key to observing dynamic changes in tumor metabolism can be found through imaging. Using an imaging method called Hyperpolarized Magnetic Resonance Spectroscopy (MRS) Imaging, Chen et al. reported imaging, in vivo, of the dynamic metabolic changes of hyperpolarized 13C-pyruvate in a live transgenic adenocarcinoma mouse model for prostate cancer [12,13]. The development of such an imaging method for retaining dynamic nuclear polarization in solution allows these investigators to inject a hyperpolarized 13C compound into mice. Pyruvate, in addition to its pivotal position in glycolysis and its subsequent conversion to lactate, is a suitable metabolite because relatively high concentrations of pyruvate are not toxic to animals or humans. In order to detect the chemical shift in magnetic resonance spectroscopy (MRS) of pyruvate and lactate, the investigators needed to inject sufficiently high concentration of pyruvate in order to detect in vivo concentrations in mM range. This experimental approach was successful when the researchers overcame several major hurdles, including the intrinsically low MR signals and low (1.1%) natural abundance of 13C. Hyperpolarization creates a situation, albeit short-lived, that disturbs the equilibrium or redistributes the nuclei in a magnetic field. The result is an increase in signal in orders of magnitude. The equilibrium state of alignment of magnetic nuclei, e.g., H, C or N, with or against an applied magnetic field is proportional to the spin polarization. It is, depending upon the nucleus, typically on the order of 1 in a million. With hyperpolarization, these authors successfully increased the signals from carbons. Metabolic imaging, in vivo, of the Warburg effect is also made possible by the rapid temporal resolution, one measurement per second. Subsequently, Kurhanewicz et al. analyzed the differential glycolysis by normal cells versus prostate cancer cells in a first in-human clinical trial [14]. Using 13C-1 pyruvate, Kurhanewicz et al. detected, by MRS, increases in lactate and alanine concentrations in prostate cancer patients. The resulting higher lactate signal correlates with histological changes of early stage tumors from normal tissues and likewise from early stage tumors to late stage (Figure 1). These clinical findings are consistent with the Warburg observation that conversion of pyruvate to lactate and alanine, by lactate dehydrogenase and glutamate pyruvate aminotranferase respectively, is increased in cancer cells. Thus, it is now possible to monitor prostate cancer progression by imaging in vivo metabolic changes in patients. It is still not known, however, whether these metabolic biomarkers are relevant to the trigger for indolent prostate cancer cells to evolve into aggressive cells.
A more common approach for imaging cellular metabolism is based on the use of radiotracers and detecting them by positron emission tomography (PET)/computed tomography (CT). A frequently used radiotracer for oncology is 18F-fluoro-2-deoxy glucose (18F-FDG), an analog of glucose, which was approved for clinical indication by the Food and Drug Administration (FDA) in 2000 [15]. There are challenges to using 18F-FDG as it has a slow accumulation and clearance through the bladder. Currently, several 18F-radiotracers under studied
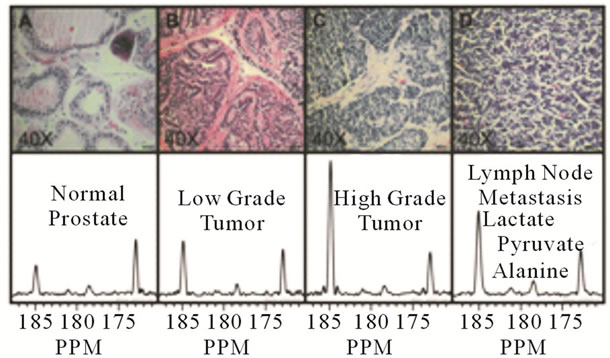
Figure 1. Representative H&E stained sections and hyperpolarized 13C spectra for one case from each of the histologically defined groups. The histology slides were processed using 5 um thick sections and photographed with 40X magnification. The hyperpolarized 13C spectra represent voxels taken from MRSI datasets and normalized to correct for differences in polarization and receiver sensitivity. The normalized spectra illustrate the strong correlation that exists between the amount of hyperpolarized 13C lactate and the progression of the disease from the normal prostates to the low grade primary tumors and the high grade primary tumors. Reprinted from Cancer Res, 2008, 68 (20), p. 8612, M.J. Albers, R. Bok, A.P. Chen, C.H. Cunningham, M.L. Zierhut, V.Y. Zhang, S.J. Kohler, J. Tropp, R.E. Hurd, Y. Yen, S.J. Nelson, D.B. Vigneron, and J. Kurhanewicz, “Hyperpolarized 13C Lactate, Pyruvate, and Alanine: Noninvasive Biomarkers for Prostate Cancer Detection and Grading,” with permission from AACR.
are applicable to different types of cancer including prostate cancer: fluoride, 3-Deoxy-3-[18F] fluorothymidine (18F-FLT) [16] and [18F]fluoromethyl-[1,2-2 H4]- choline [17].
Similar to 18F-FDG, these two radiotracers are intermediates in metabolic or biosynthetic pathways: deoxyglucose in glycolysis, fluorothymidine in DNA synthesis, and fluoromethylcholine in phospholipid biosynthesis. Because these are radiolabeled analogs of natural metabolites, they are transported into cells, but not metabolized. They exist as stable intermediates in the pathways, enabling their imaging in tumor metabolism. As metabolism increases in cancer cells generally, quantitative imaging of these radiotracers provides distinguishing features of cancer from normal cells. For example, increased accumulation of 18F-FDG can be imaged and quantified in cancer cells. This technique is useful for staging cancer, detecting bone metastases, and monitoring treatment response. Not all metastasis, however, can be detected by 18F-FDG uptake, e.g, lymph nodes metastasis in breast cancer.
There are several near-infrared (NIR) devices that can be used to monitor changes of the hemoglobin oxygen saturation in prostate lesions. In a canine model, Jiang et al. used an integrated NIR and ultrasound for transrectal imaging to detect hemoglobin oxygen saturation measurements and blood flow [18]. This combined imaging technique enabled monitoring of intra-lesional oxygenation dynamics and the hemoglobin content of prostate lesions. The advantage of such techniques is that they do not require contrast agents and are potentially much cheaper.
In 1971, Folkman made a seminal observation that tumor growth requires angiogenesis [19]. Angiogenic processes involve signaling pathways and the interactions of the endothelial cells with the extracellular matrix (ECM). Angiogenic endothelial cells, when stimulated by vascular endothelial cell growth factors (VEGF) [20- 22] or other growth factors, show increased expression of a number of proteins, including transcription factors, which in turn upregulate the expression of integrins. The result of upregulation is increased cell proliferation and invasion of tissues, both hallmarks of tumor growth.
Among the receptors involved in angiogenesis, integrins have been extensively studied [23]. Integrins are heterdimers of various combinations of the 18 different α and 8 different β subunits. They play an important role in endothelial cell attachment to the ECM. The observation that both ECM components and integrins are upregulated for angiogenesis has led to research and exploration for cancer drug therapies that target the integrins. The hypothesis is that integrin inhibitors should impede angiogenesis, resulting in reduced blood supply to the tumors and thus reduce tumor growth. Signaling by integrins has been implicated in the regulation of protease expression [24]. Protease plays an important role in the invasion of the matrix surrounding prostate cancer cells and it is particularly relevant to bone metastasis.
Phosphocholine plays an important role in transmembrane signaling in tumor cell growth [25,26]. Choline is transported into the cell and subsequently phosphorylated by choline kinase. Choline kinase is over-expressed, resulting in increased enzymatic activities in cancer cells. Leyton et al. [17] developed 18F-fluoromethyl choline as a more stable analog of 11C-choline to measure the elevated phosphocholine biosynthesis found in prostate cancer cells. The longer half-life of 18 fluorine, 109.8 min, as compared to 11 carbon, 20.4 min, is a clear advantage for PET imaging, especially in the absence of an available cyclotron. PET images of 18F-fluoromethyl choline should shed light on the role of phospholipids in tumor cell growth.
Mena et al. [27] reported the differential uptake of 11C-acetate, as imaged by PET/CT, by prostate cancer, benign prostate hyperplasia, and normal prostate tissues in 39 men in comparison with multiparametric MRI and histology. They observed higher 11C-acetate uptake in tumor foci than in normal prostate tissues. However, there was no observed difference between tumors and benign prostate hyperplasia.
Prostate-specific membrane antigen (PSMA) is a transmembrane glycoprotein of 100 kDa molecular weight and exhibits enzymatic activity, N-acetyl α-linked acidic dipeptidase [28]. It is over-expressed on prostate cancer cell membrane and in the neovasculature of prostate tumors, but not in the vasculature of normal tissues [29, 30]. However, its role in prostate cancer biology is not known. It should be noted that PSMA is also expressed in other tissues such as the kidneys, the proximal small intestine, and the salivary glands. Because of the possibility that the over-expression may be a biomarker for prostate cancer disease, PSMA has been a therapeutic target. A number of monoclonal antibodies have been developed. Radiolabeled monoclonal antibodies, 111Incapromabpendetide [31,32] was approved by the FDA for Phase 1 trial in patients with Gleason scores greater than 6 and rising PSA after prostatectomy. Single-photon emission computed tomography (SPECT) and SPECT/ CT imaging were used but the inadequate sensitivity presented challenges for broad adoption. More recent developments of alternative monoclonal antibodies against the exposed, extracellular domain of PSMA have yielded possible candidates for radioimmunotherapy [33,34]. This strategy holds promise, especially when the radiolabeled antibodies are coupled to cytotoxic drugs in targeted drug delivery.
Recognizing the clinical value in extracting quantitative data from images, the National Institute of Biomedical Imaging and Bioengineering commissioned the Radiological Society of North America to form partnership with industry and academic institutions to achieve cross-industry consensus on and adoption of quantitative imaging biomarkers. Achieving reliable extraction of quantitative results from imaging scans requires both scientific understanding of the sources of variance and multidisciplinary consensus around how best to minimize and compensate for variance. The goal is to achieve reliable imaging quantification that will lead to validation and qualification of imaging biomarkers for use in clinical trials, drug development, and clinical decisionmaking. These activities are ongoing.
3. MOLECULAR PROFILES OF PROSTATE CANCER
Genomic profiles of prostate cancer patients provide molecular portraits of the various subtypes of disease. Patient subtypes can be characterized by genetic alterations: deletions, chromosomal rearrangements, and single nucleotide polymorphism (SNP). Recent findings from several research groups [35,36] shed light on a subtype of prostate cancer that involves chromosomal fusion of TMPRSS2 with the v-ets erythroblastosis virus E26 oncogene homolog (ERG). TMPRSS2 is an androgen-regulated gene that codes for serine 2 protease. As a result of the fusion, androgen receptor (AR) driven expression of the oncogene is over-expressed in malignant prostate epithelial cells. These researchers have focused on fine mapping of these fused genes with the aim of understanding the predictive value of such fusion in response to antiandrogen treatment.
Using a herringbone design, Stott et al. [37] developed a microfluidic chip to isolate and enumerate circulating tumor cells (CTCs) by optical imaging of fluorescence labels in whole blood from prostate cancer patients. These cells are fluorescence labeled for PSA, DAPI for nuclei, and Ki-67 for cell proliferation. The authors deployed an isolation strategy of antibodies and cell surface proteins binding. Antibodies against a number of protein biomarkers that are over-expressed in cancer cells were used. Initial studies were based on prostate circulating tumor cells adhered to antibodies against epithelial cell adhesion molecules. The TEMPRSS2/ERG fusion was identified in the CTCs of some prostate cancer patients, following RNA isolation and RT-PCR amplification. RT-PCR amplification was performed using primers flanking exon 1 of TMPRSS2 and exon 6 of ERG to detect the TMPRSS2:ERG fusion transcripts. Using gel electrophoresis of patient samples and a positive control, the VCaP cell line, which is known to harbor the translocation, these authors identified the characteristic T1:E4 breakpoint. Liu et al. [38] provided evidence that multiple genomic alterations, including recurrent genomic rearrangements, deletions and translocations, contributed to various TMPRSS:ERG fusion transcripts. The authors used GeneChip 500K SNP array to precisely map the start and end of each deletion and located them to specific introns of these two genes. The authors further identified consensus sequences in these fusion transcripts that were homologous to the human Alu-Sq and Alu-Sp families. The significance of the consensus sequence in prostate cancer is unknown, but may shed light on tumorigenesis.
To further correlate the clinical significance of the TMPRSS:ERG fusion, Stott et al. identified these transcripts in 9 of 20 cases, or 45%, which is consistent with the prevalence rate of the fusion product as reported in the literature [37]. They confirmed the prevalence rates by both RT-PCR and fluorescence in situ hybridization (FISH) on matched archival primary tumor specimens, and found a similar prevalence of the TMPRSS2: ERG fusion product by both assays. Danila et al. [39] reported finding fusion transcripts in 15 out of 41 (37%) prostate cancer patients who had a median baseline CTC count of 17 (interquartile range: 7 - 103 cells/7.5 ml). Presence of these fusion transcripts, however, does not correlate with PSA. It is not known whether fusion is a predictor for clinical outcomes. In contrast, CTC counts seem to be a better predictor for survival.
4. SIGNALING PATHWAYS
Androgen binding to its receptor triggers downstream signaling that activates the transcription genes involved in cell proliferation, differentiation, and survival [40]. The mechanism for transcription initiation involves multiple factors. Upon hormone binding, AR is released from heat-shock protein and moves to the nucleus where it binds to androgen-response elements in the chromatin (Figure 2). Once inside the nucleus, the machinery for transcription is poised to be turned on along with histone acetyl transferase, steroid receptor coactivator (SRC), chromatin remodeling complexes and RNA polymerase II forming the transcription initiation complex [41]. The rationale for androgen ablation therapy is to lower the androgen level so as to prevent upregulation of AR signaling. Resistance to androgen ablation may be due to aberrant downstream signaling. It would be advantageous to be able to detect androgen receptor and its interaction with testosterone by in vivo imaging techniques. Such technique should shed light on the signaling pathways which in turn should facilitate identification of patients who are on the path toward castrate resistant prostate cancer. Ulmert et al. [42] have developed a novel radiotracer, 89Zr-5A10 that targets free PSA and it is accumulated in bone lesions in an androgen-receptor dependent manner. Their preliminary results are encouraging for quantitative imaging biomarker for bone metastasis.
The TMPRSS2:ERG fusion gene occurs in approximately 40% of prostate cancer [43,44]. Over-expression of the ERG oncogene activates a number of signaling

Figure 2. Androgen-dependent signaling pathway.
pathways including WNT signaling which induces epithelial to mesenchymal transition, loss of cell adhesion and gaining migratory ability [45,46]. ERG interacts with phosphatase tensin-(PTEN) induced phospha-tidylinositol 3-kinase (PI3K)/AKT activation, resulting in prostate cancer cell invasion. These processes underpin prostate cancer cell invasion and dissemination leading to metastases at a distant site. ERG over-expression also modulates AR signaling and initiates epigenetic silencing resulting in cellular dedifferentiation [47]. Epigenetic processes have implications for prostatic stem or progenitor cells that play a role in tumorigenesis.
A number of other signaling pathways interact with the AR signaling. For example, the epidermal growth factor receptor (EGFR) is expressed in 40% to 80% of malignant prostate cancer cells [41]. EGFR is a family of four structurally related receptor tyrosine kinases: HER1, HER2, HER3, and HER4. HER1 and HER2 are overexpressed on many solid tumor cells [48-50]. Receptor tyrosine kinases, of which there are 58 distinct proteins, are all transmembrane with a general structure consisting of an extracellular ligand-binding domain and an intracellular kinase domain [51,52]. HER receptor tyrosine kinases are expressed in prostate cancer cells. The intracellular kinase phosphorylates tyrosine amino acid with the transfer of phosphate from ATP. Phosphorylation, a reversible reaction, is a key control mechanism for a number of cellular processes including cell proliferation, signaling, and cell survival. Biological processes can be switched on or off, serving as pivotal regulatory controls. For example, excessive signaling is associated with the development of a wide variety of solid tumors, and it may be a critical factor in the malignancy of these tumors
5. CONCLUSION
Advances in our understanding of the molecular mechanisms of prostate cancer have been striking. These have been made through the synergy of imaging and the molecular approaches to characterize the different prostate cancer subtypes. This report described examples of qualitative and quantitative imaging as biomarkers for prostate cancer detection and disease mechanism. Understanding of the molecular mechanisms has led to mechanistically driven therapeutics development. Components of the signaling pathways are promising drug targets. However, challenges remain due to the intricate regulatory network of signaling and the cross-talks among pathways, including androgen dependent and independent signaling. They are regulated by multiple control points, modulators, and on/off switches. Biomarkers that reflect the activation/deactivation of the signaling pathways will be useful tools to elucidate prostate disease mechanism and enable targeted therapeutics development. Prostate cancer is a complex and heterogeneous disease and advances will need to build on the convergence of imaging and molecular biology.
6. ACKNOWLEDGEMENTS
I am indebted to Ms. Christine Rogers for her help with preparing this manuscript. I would also like to thank the following individuals for their review: Drs. Richard Conroy, Alan Mclaughlin, Anthony Sastre, Richard Leapman, Christina Liu, and Guoying Liu.
REFERENCES
- Imanishi, T., Itoh, T., Suzuki, Y., O’Donovan, C., Fukuchi, S., Koyanagi, K.O., et al. (2004) Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biology, 6, e162. doi:10.1371/journal.pbio.0020162
- Collins, F.S., Green, E.D., Guttmacher, A.E. and Guyer, M.S. (2003) A vision for the future of genomics research. Nature, 422, 835-847. doi:10.1038/nature01626
- Mimeault, M. and Batra, S.K. (2011) Frequent gene products and molecular pathways altered in prostate cancerand metastasis-initiating cells and their progenies and novel promising multitargeted therapies. Molecular Medicine, 17, 949-964.
- Altieri, D.C., Languino, L.R., Lian, J.B., Stein, J.L., Leav, I., van Wijnen, A.J., et al. (2009) Prostate cancer regulatory networks. Journal of Cellular Biochemistry, 107, 845-852. doi:10.1002/jcb.22162
- US Preventive Services Task Force (2012) Screening for prostate cancer. http://www.uspreventiveservicetaskforce.org/prostatecancerscreening/prostatefinalrs.htm
- Martin, S.K., Vaughan, T.B., Atkinson, T., Zhu, H. and Kyprianou, N. (2012) Emerging biomarkers of prostate cancer (Review). Oncology Reports, 409-417. doi:10.3892/or.2012.1832
- Hessels, D., van Gils, M.P., van Hooij, O., Jannink, S.A., Witjes, J.A., Verhaegh, G.W., et al. (2010) Predictive value of PCA3 in urinary sediments in determining clinico-pathological characteristics of prostate cancer. Prostate, 70, 10-16. doi:10.1002/pros.21032
- Gu, Z., Thomas, G., Yamashiro, J., Shintaku, I.P., Dorey, F., Raitano, A., et al. (2000) Prostate stem cell antigen (PSCA) expression increases with high gleason score, advanced stage and bone metastasis in prostate cancer. Oncogene, 19, 1288-1296. doi:10.1038/sj.onc.1203426
- Warburg, O., Wind, F. and Negelein, E. (1927) Themetabolism of tumors in the body. Journal of General Physiology, 6, 519-530. doi:10.1085/jgp.8.6.519
- Warburg, O. (1956) On the origin of cancer cells. Science, 123, 309-314. doi:10.1126/science.123.3191.309
- Seth, P., Grant, A., Tang, J., Vinogradov, E., Wang, X., Lenkinski, R., et al. (2011) On-target inhibition of tumor fermentative glycolysis as visualized by hyperpolarized pyruvate. Neoplasia, 13, 60-71.
- Chen, A.P., Albers, M.J., Cunningham, C.H., Kohler, S.J., Yen, Y.F., Hurd, R.E., et al. (2007) Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T-initial experience. Magnetic Resonance in Medicine, 58, 1099-1106. doi:10.1002/mrm.21256
- Chen, A.P., Kurhanewicz, J., Bok, R., Xu, D., Joun, D., Zhang, V., et al. (2008) Feasibility of using hyperpolarized [1-13C]lactate as a substrate for in vivo metabolic 13C MRSI studies. Magnetic Resonance Imaging, 26, 721-726. doi:10.1016/j.mri.2008.01.002
- Kurhanewicz, J., Vigneron, D.B., Brindle, K., Chekmenev, E.Y., Comment, A., Cunningham, C.H., et al. (2011) Analysis of cancer metabolism by imaging hyperpolarized nuclei: Prospects for translation to clinical research. Neoplasia, 13, 81-97.
- Vallabhajosula, S. (2007) 18F-labeled positron emission tomographic radiopharmaceuticals in oncology: An overview of radiochemistry and mechanisms of tumor localization. Seminars in Nuclear Medicine, 37, 400-419. doi:10.1053/j.semnuclmed.2007.08.004
- Turcotte, E., Wiens, L.W., Grierson, J.R., Peterson, L.M., Wener, M.H. and Vesselle, H. (2007) Toxicology evaluation of radiotracer doses of 3'-deoxy-3'-[18F]fluorothymidine (18F-FLT) for human PET imaging: Laboratory analysis of serial blood samples and comparison to previously investigated therapeutic FLT doses. BMC Nuclear Medicine, 7, 3. doi:10.1186/1471-2385-7-3
- Leyton, J., Smith, G., Zhao, Y., Perumal, M., Nguyen, Q.D., Robins, E., et al. (2009) [18F]fluoromethyl-[1,2- 2H4]-choline: A novel radiotracer for imaging choline metabolism in tumors by positron emission tomography. Cancer Research, 69, 7721-7728. doi:10.1158/0008-5472.CAN-09-1419
- Jiang, Z., Piao, D., Bartels, K.E., Holyoak, G.R., Ritchey, J.W., Ownby, C.L., et al. (2011) Transrectal ultrasoundintegrated spectral optical tomography of hypoxic progression of a regressing tumor in a canine prostate. Technology in Cancer Research and Treatment, 10, 519-531.
- Folkman, J. (1995) Angiogenesis in cancer, vascular, rheumatoid and other disease. Nature Medicine, 1, 27-31. doi:10.1038/nm0195-27
- Risau, W. (1997) Mechanisms of angiogenesis. Nature, 386, 671-674.
- Costa, L.J. and Drabkin, H.A. (2007) Renal cell carcinoma: New developments in molecular biology and potential for targeted therapies. Oncologist, 12, 1404-1415. doi:10.1634/theoncologist.12-12-1404
- Samlowski, W.E., Wong, B. and Vogelzang, N.J. (2008) Management of renal cancer in the tyrosine kinase inhibitor era: A view from 3 years on. BJU International, 2, 162-165. doi:10.1111/j.1464-410X.2008.07670.x
- Lim, E.H., Danthi, N., Bednarski, M. and Li, K.C. (2005) A review: Integrin alphavbeta3-targeted molecular imaging and therapy in angiogenesis. Nanomedicine, 1, 110-114. doi:10.1016/j.nano.2005.03.008
- Munshi, H.G. and Stack, M.S. (2006) Reciprocal interactions between adhesion receptor signaling and MMP regulation. Cancer and Metastasis Reviews, 25, 45-56. doi:10.1007/s10555-006-7888-7
- Zeisel, S.H. (1993) Choline phospholipids: Signal transduction and carcinogenesis. FASEB Journal, 7, 551-557.
- Zeisel, S.H. (1995) Nutrients, signal transduction and carcinogenesis. Advances in Experimental Medicine and Biology, 369, 175-183. doi:10.1007/978-1-4615-1957-7_16
- Mena, E., Turkbey, B., Mani, H., Adler, S., Valera, V.A., Bernardo, M., et al. (2012) 11C-Acetate PET/CT in localized prostate cancer: A study with MRI and histopathologic correlation. Journal of Nuclear Medicine, 53, 538-545. doi:10.2967/jnumed.111.096032
- Leung, K. (2004) Quenched indocyanine green-anti-prostate-specific membrane antigen antibody J591. Molecular Imaging and Contrast Agent Database (MICAD), Bethesda.
- Nanus, D.M., Milowsky, M.I., Kostakoglu, L., Smith-Jones, P.M., Vallabahajosula, S., Goldsmith, S.J., et al. (2003) Clinical use of monoclonal antibody HuJ591 therapy: Targeting prostate specific membrane antigen. Journal of Urology, 170, S84-S88.
- Nargund, V., Al Hashmi, D., Kumar, P., Gordon, S., Otitie, U., Ellison, D., et al. (2005) Imaging with radiolabelled monoclonal antibody (MUJ591) to prostate-specific membrane antigen in staging of clinically localized prostatic carcinoma: Comparison with clinical, surgical and histological staging. BJU International, 95, 1232-1236. doi:10.1111/j.1464-410X.2005.05511.x
- Kahn, D., Williams, R.D., Haseman, M.K., Reed, N.L., Miller, S.J. and Gerstbrein, J. (1998) Radioimmunoscintigraphy with In-111-labeled capromabpendetide predicts prostate cancer response to salvage radiotherapy after failed radical prostatectomy. Journal of Clinical Oncology, 20, 284-289.
- Kahn, D., Williams, R.D., Manyak, M.J., Haseman, M.K., Seldin, D.W., Libertino, J.A., et al. (1998) 111Indiumcapromab pendetide in the evaluation of patients with residual or recurrent prostate cancer after radical prostatectomy. Journal of Urology, 16, 2041-2046.
- Holmes, E.H. (2001) PSMA specific antibodies and their diagnostic and therapeutic use. Expert Opinion on Investigational Drugs, 10, 511-519. doi:10.1517/13543784.10.3.511
- Wolf, P., Freudenberg, N., Buhler, P., Alt, K., SchultzeSeemann, W., Wetterauer, U., et al. (2010) Three conformational antibodies specific for different PSMA epitopes are promising diagnostic and therapeutic tools for prostate cancer. Prostate, 70, 562-569.
- Tomlins, S.A., Aubin, S.M., Siddiqui, J., Lonigro, R.J., Sefton-Miller, L., Miick, S., et al. (2011) Urine TMPRSS2: ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Science Translational Medicine, 3, 72.
- Tomlins, S.A., Bjartell, A., Chinnaiyan, A.M., Jenster, G., Nam, R.K., Rubin, M.A., et al. (2009) ETS gene fusions in prostate cancer: From discovery to daily clinical practice. European Urology, 56, 275-286. doi:10.1016/j.eururo.2009.04.036
- Stott, S.L., Lee, R.J., Nagrath, S., Yu, M., Miyamoto, D.T., Ulkus, L., et al. (2010) Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Science Translational Medicine, 2, 25-23.
- Liu, W., Ewing, C.M., Chang, B.L., Li, T., Sun, J., Turner, A.R., et al. (2007) Multiple genomic alterations on 21q22 predict various TMPRSS2/ERG fusion transcripts in human prostate cancers. Genes Chromosomes Cancer, 46, 972-980. doi:10.1002/gcc.20482
- Danila, D.C., Anand, A., Sung, C.C., Heller, G., Leversha, M.A., Cao, L., et al. (2011) TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. European Urology, 60, 897-904. doi:10.1016/j.eururo.2011.07.011
- Dahlman, K.B., Parker, J.S., Shamu, T., Hieronymus, H., Chapinski, C., Carver, B., et al. (2012) Modulators of prostate cancer cell proliferation and viability identified by short-hairpin RNA library screening. PLoS One, 7, e34414. doi:10.1371/journal.pone.0034414
- Kaarbo, M., Klokk, T.I. and Saatcioglu, F. (2007) Androgen signaling and its interactions with other signaling pathways in prostate cancer. BioEssays, 29, 1227-1238. doi:10.1002/bies.20676
- Ulmert, D., Evans, M.J., Holland, J.P., Rice, S.L., Wongvipat, J., Pettersson, K., et al. (2012) Imaging androgen receptor signaling with a radiotracer targeting free prostate-specific antigen. Cancer Discovery, 2, 320-327. doi:10.1158/2159-8290.CD-11-0316
- Tomlins, S.A., Mehra, R., Rhodes, D.R., Smith, L.R., Roulston, D., Helgesson, B.E., et al. (2006) TMPRSS2: ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Research, 66, 3396-3400. doi:10.1158/0008-5472.CAN-06-0168
- Tomlins, S.A., Rhodes, D.R., Perner, S., Dhanasekaran, S.M., Mehra, R., Sun, X.W., et al. (2006) Recurrent fusion of TMPRSS2 and ETS transcription factors in prostate cancer. FASEB Journal, 20, A1327-A1327.
- Gupta, S., Iljin, K., Sara, H., Mpindi, J.P., Mirtti, T., Vainio, P., et al. (2010) FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymaltransition in human prostate Cancer Cells. Cancer Research, 70, 6735-6745. doi:10.1158/0008-5472.CAN-10-0244
- Iljin, K., Wolf, M., Edgren, H., Gupta, S., Kilpinen, S., Skotheim, R.I., et al. (2006) TMPRSS2 fusions with oncogenic ETS factors in prostate cancer involve unbalanced genomic rearrangements and are associated with HDAC1 and epigenetic reprogramming. Cancer Research, 66, 10242-10246. doi:10.1158/0008-5472.CAN-06-1986
- Yu, J.D., Yu, J.J., Mani, R.S., Cao, Q., Brenner, C.J., Cao, X.H., et al. (2010) Anintegrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell, 17, 443-454. doi:10.1016/j.ccr.2010.03.018
- Zwick, E., Bange, J. and Ullrich, A. (2001) Receptor tyrosine kinase signalling as a target for cancer intervenetion strategies. Endocrine-Related Cancer, 8, 161-173. doi:10.1677/erc.0.0080161
- Pawson, T. (1995) Protein modules and signalling networks. Nature, 373, 573-580. doi:10.1038/373573a0
- Bublil, E.M. and Yarden, Y. (2007) The EGF receptor family: Spearheading a merger of signaling and therapeutics. Current Opinion in Cell Biology, 19, 124-134.
- Johnson, L.N. (2009) Protein kinase inhibitors: Contributions from structure to clinical compounds. Quarterly Reviews of Biophysics, 42, 1-40. doi:10.1017/S0033583508004745
- Robinson, D.R., Wu, Y.M. and Lin, S.F. (2000) The protein tyrosine kinase family of the human genome. Oncogene, 19, 5548-5557.

