Advances in Bioscience and Biotechnology
Vol.3 No.4(2012), Article ID:21870,8 pages DOI:10.4236/abb.2012.34054
Genomics of crop plant genetic resources
![]()
1Plant Genetic Resources, Scuola Superiore Sant’ Anna, Rome, Italy
2Division of Plant Sciences, University of Missouri, Columbia, USA
Email: a.dhanapal@sssup.it, dhanapala@missouri.edu
Received 15 May 2012; revised 25 June 2012; accepted 6 July 2012
Keywords: Crop plant genetic resources; genomics; next generation sequencing (NGS)
ABSTRACT
Plant genetic resources collection and utilization had made a huge impact in balancing the genetic diversity of the existing crop plant species and their application in genome based studies had also increased widely. Primarily studies were based on model species, although it now enhances the transferability of information to crops and related species. With the tremendous outbreak of new high-throughput technologies like next-generation sequencing (NGS) and reduction in their costs are bringing many more plants within the range of genome and transcriptome level analysis. The completion of reference genome sequences for many important crops and the ability to perform high-throughput resequencing are providing opportunities for improving our understanding of the crop plant genetic resources to accelerate crop improvement. The future of crop improvement will be centred on comparisons of individual crop plant genomes, and some of the best opportunities may lie in using combinations of new genetic mapping strategies and evolutionary analyses to direct and optimize the discovery and use of genetic variation. Here I review the importance of crop plant genetic resources and insights that have been emerged in recent years.
1. INTRODUCTION
Plants have always been the most important resource as food for humankind and feed for animals. The cultivation of plants species are often regarded as the starting point of modern civilization, which is some 10,000 years ago. Humans began domesticating crops as a food source in different era of civilizations. Among the available wild germplasms of plant species, the best adapted one are preferably selected for cultivation and utilization, but this also had led to a decline in the genetic diversity of the crops. Land races and traditional varieties have been replaced by less diverse modern cultivars and hybrids. In order to face the challenging demand of agricultural production with the rapid growing population and with the alarming climate change, the need for the utilization of wild relatives and underutilized crops (orphan crops) are in necessity today. Although wild ancestors have continued to persist in regions where domestication took place, there is a permanent risk of loss of the genetic variability of cultivated plants and their wild relatives in response to changing environmental conditions and cultural practices [1]. Enhancement and synergistic innovations are fundamentally important in addressing these needs for the improvement of agricultural productivity and sustainability across a broad front - food safety and security, diet and health, environmental safety and novel crops are some area of new opportunities in plant science [2]. The need to preserve and use plant genetic resources is well recognized, and the prospect of dwindling plant genetic diversity, coupled with increased demands on these resources has made them a topic of global discussion [3].
With the tremendous outbreak of new high-throughput technologies and reduction in their costs are bringing many more crops, trees and orphan plant genomes within range of analysis [4] Today entire sequence of complete crop plant genomes and transcriptomes can be well considered a newly developed genetic resource, for the information it provides on plant functions and allows us in discovering the genes from the wild species that correspond to specific phenotypes [3]. Availability of rapid techniques and the introduction of NGS and their advancement over recent years, one could foresee the very good chances of deciphering the genetic mechanism involved in novel complex traits [5] and thereby will help us dissecting the genetic variability of the selected genetic resources, their characterization and conservation, and in generating increased value for collections of crop genetics resources. Collecting rapid and accurate phenotypes in crop plants is a hindrance to integrating genomics with crop improvement, and advancement in informatics are needed to put these tools in the hands of the scientists on the ground [6].
2. PLANT GENETIC RESOURCES AND ITS IMPORTANCE
Plant genetic resources are the most important components of agrobiodiversity. They include primitive forms of cultivated crop species and land-races, modern cultivars, obsolete cultivars, breeding lines and genetic stocks and related wild species. The introgression of genes that reduced plant height and increased disease and viral resistance in wheat provided the foundation for the “Green Revolution” and demonstrated the tremendous impact that genetic resources can have on crop production [7]. Food production and security depend on the wise use and conservation of agricultural biodiversity and genetic resources [8]. Even though several thousands of plant species are globally available only few are in use (Table 1). Since importance have been given to relatively small number of crop species for global food security, it is particularly important that their genetic diversity is conserved effectively and managed wisely. So far, only a small part of the total genetic variability has been characterized and used for crop breeding purposes.
3. EXPLOITATION AND MANAGEMENT OF CROP GENETIC RESOURCES
Genetic resources provide basic material for selection and improvement through breeding to ensure food security needs of world’s rapidly rising population. All aspects related to genetic resources (collection, conservation, evaluation, management and utilization) are however, needed to be done eminently. Molecular technique has proved useful in a number of ways to improve the conservation and management of PGR [9]. Molecular markers help in verification of accession identity and genetic contamination [10-12] and also been used to identify ecogeographic races within the domesticated or wild gene pools of crop species [13]. Presently, existing biotechnological approaches to overcome challenges for effective utilization and enhancement of crop genetic resources include embryo rescue and somatic hybridization [14,15]. The ability to store and exchange healthy
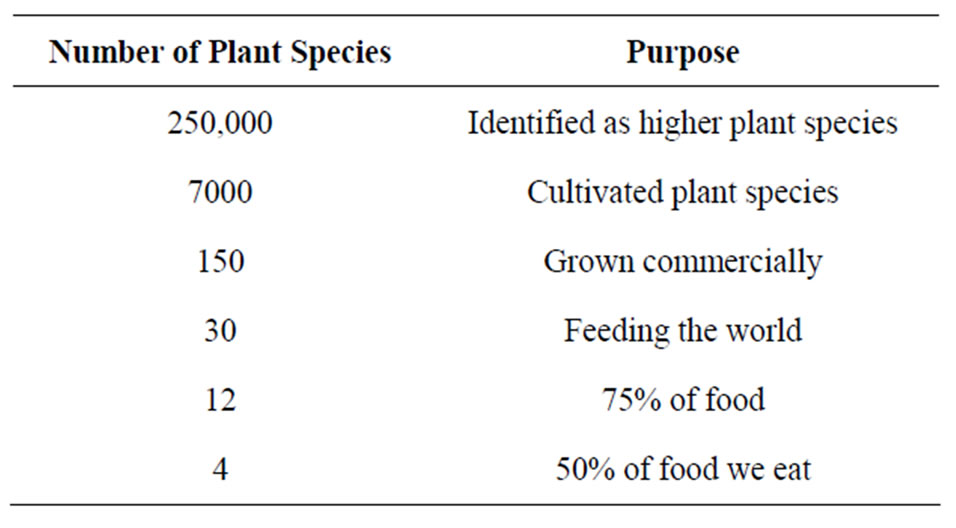
Table 1. Total number of plant species.
Germplasm is fundamental objective for effective conservation and use of crop genetic resources.
4. GENOMICS ASSISTED CROP BREEDING AND IMPROVEMENT
Genomics, the study of an organism’s entire genome is the new field of study to enhance the use of crop genetic resources. Today genomic technology has been applied in gene identification laying good foundation for functional genomics research and to aid us in understanding the gene expression and biological activity, genomics initiatives are focused on fundamental elements of plant biology with regard to growth, development, reproduction, photosynthesis and responses to environmental conditions and pathogens.
Cereal genomics potentially carries the strength to shape the future of agriculture and its sustainability [16]. The better prediction of the phenotype that a particular genotype will produce is a primary goal of genomicsbased breeding. Analysis of the crop genomes architecture and their expressed components are now possible with the development in crop genomics, and subsequently leads to an increase in our knowledge of the genes that are linked to key agronomically important complex traits particularly in major crop species. DNA-based molecular markers have played a pivotal role in detecting the genetic variation available in germplasm collections and breeding lines. Various sets of diverse molecular markers have been developed for many major crop species and are being used extensively for the development of saturated molecular, genetic and physical maps and for the identification of genes or quantitative trait loci (QTL) controlling traits of economic importance through marker assisted selection (MAS) [17,18]. Together with MAS other approaches like association mapping [19], functional genomics [20], genetical genomics [21], allele mining [17], Targeting Induced Local Lesions in Genomes (TILLING) and Eco Type TILLING (EcoTILLING) [22] have been available from past decade.
Development in cereal genomics would play key role in crop improvement in two general ways. First, a better understanding of the biological mechanisms can lead to new or improved screening methods for selecting superior genotypes more efficiently. Second, new knowledge can improve the decision-making process for more efficient breeding strategies [17]. These advances and development will provide opportunity for efficient transfer of information systems from model species and major crops to orphan crops [23]
5. COMPARATIVE GENOMICS BASED APPROACH TARGETING GENES AND TRAITS
Comparative genomics, promised to identify the functional elements in a genome based on the assumption that these elements are preferentially conserved through evolutionary time [24]. It has taken advantage of evolutionary signature that has been acted on the genome to understand the function and evolution across different crop species. With the availability of whole-genome sequences of crops and plant species, as well as other genomic resources (e.g. microarray methods, expressed sequence tag (EST) libraries, high throughput resequencing technologies), have extended the comparative method to encompass the evolution of genome structure and function [25,26].
Rapid progress in crop genomics is making possible to undertake detailed structural and functional comparisons of genes involved in various biological processes among important crops and other plant species. This flow of research was promising and would provide important information for future work on the evolution of higher crop plants. Comparisons at varying levels of evolutionary divergence are likely to reveal functional regions characteristic of different plant groups, Moreover intraspecific genomic approaches have been shown to be useful in predicting functional sequence motifs [27]. Since significant genomic colinearity has been reported in cereal species [28] the comparative genomics approach using bioinformatics tools might therefore provide an opportunity for efficient transfer of information from model species and major crops to minor and orphan crops, and particularly crops such as pearl millet, small millets and tef that are often considered orphan crops for poor genomic research shelter, and these crops are regionally or locally important for nutrition and income, particularly in developing countries [23,27,29].
6. TRANSLATIONAL GENOMICS FROM MODEL SPECIES TO CROPS
Plant translational genomics, a challenge faced by the plant genomics research to develop applications in crop plants which imply the translation of gene functions from a model to a crop species. Candidate gene approach (CGA) renown as a tool for translational genomics has been considered for successful application in crops with determined factors such as the type of crop, the complexity of the trait and the type of genes involved. The CGA is based on the assumption that genes with a proven or predicted function in a “model” species (functional candidate genes) or genes that are co-localized with a traitlocus (positional candidate genes) could control a similar function or trait in an arbitrary crop of interest (target crop) [30]. Studying the sequence variation among alleles (paralogs and orthologs) of candidate genes may provide conserved sequence motifs or conserved SNPs associated with a trait [31]. The extrapolations of gene function from a model crop to a more distant species have been well discussed [32,33].
7. NUTRIGENOMICS
In recent years crop genomics based research has been providing the means to uncover the genetic basis of crop characteristics with significance to human life and health. Crop-plant genome research integrated with human genome analysis, nutritional science and medicine constitutes a novel discipline of research in support of human welfare [34,35]. This existing new multidisciplinary approach is called nutrigenomics. This approach focuses on the highly complex interplay between human genetic predisposition and nutrition, in regard to both, food nutritional quality and disease prevention [36]. Strong priorities are now to focus on the genes that determine characteristics supporting the production of crops in an environmentally, friendly and sustainable manner. Millions of poor children in the world particularly in semiarid tropics of Asia and Africa suffer from vitamin A deficiency, which causes blindness, and reduces the bioavailability of other important dietary micro-nutrients, including iron that were important for human health. This serious public health problem is addressed by genetically increasing the levels of pro-vitamin A (primarily green and yellow vegetables) in dietary staples like rice, maize, sorghum, and pearl millet. Besides the genes that control the accumulation of bulk nutrients, efforts are taken to uncover the genes that determine the content of valuable compounds such as potential pharmaceuticals, health promoting pro-biotics, flavor and fragrance compounds, protectants, biocides, fine chemicals, etc. The rapid development and innovations in crop plant genomics are expected to provide newer knowledge in these areas and will open up ways for targeted crop improvement, both through the direct use of natural genetic diversity and via genetic engineering [36].
8. CROP PLANT WHOLE GENOME SEQUENCING—PRESENT STATUS
Revolution in DNA sequencing technology has brought down the cost of DNA sequencing and made the sequencing of an increased number of genomes both feasible and cost effective [37]. The first plant genome Arabidopsis was completely sequenced in December 2000, and it was the third complete genome of a higher eukaryote and further studies were carried in recent years on Arabidposis thaliana and Arabidopsis lyrata [38,39].
Subsequently after Arabidopsis, several other cropplants have been sequenced [38-64] (Table 2). Genomes of other crop plant species like Tomato (Solanum lycopersicum), monkey flower (Mimulus guttatus), rose gum tree (Eucalyptus grandis) Cassava (Manihot esculenta), Peaches (Prunus persica), Columbine (Aquilegia
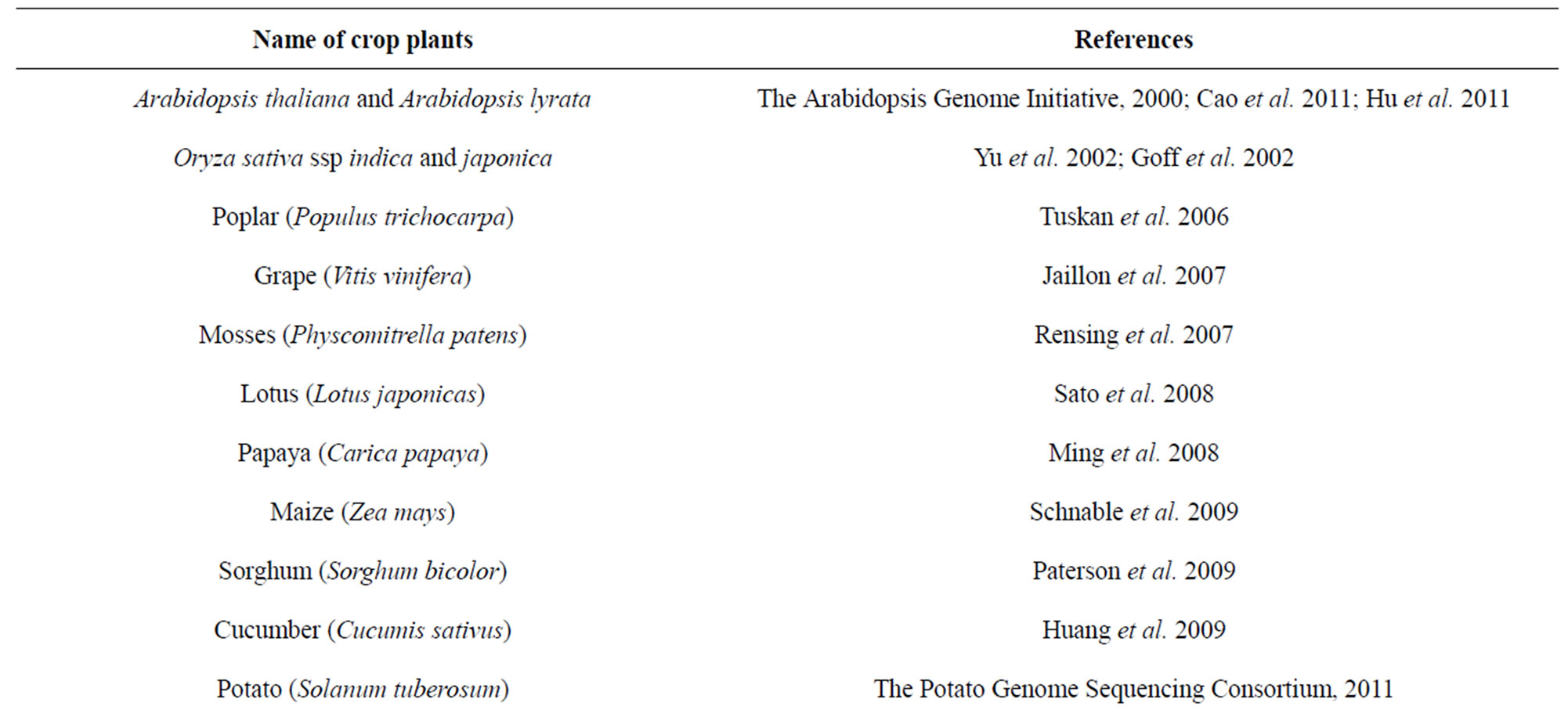
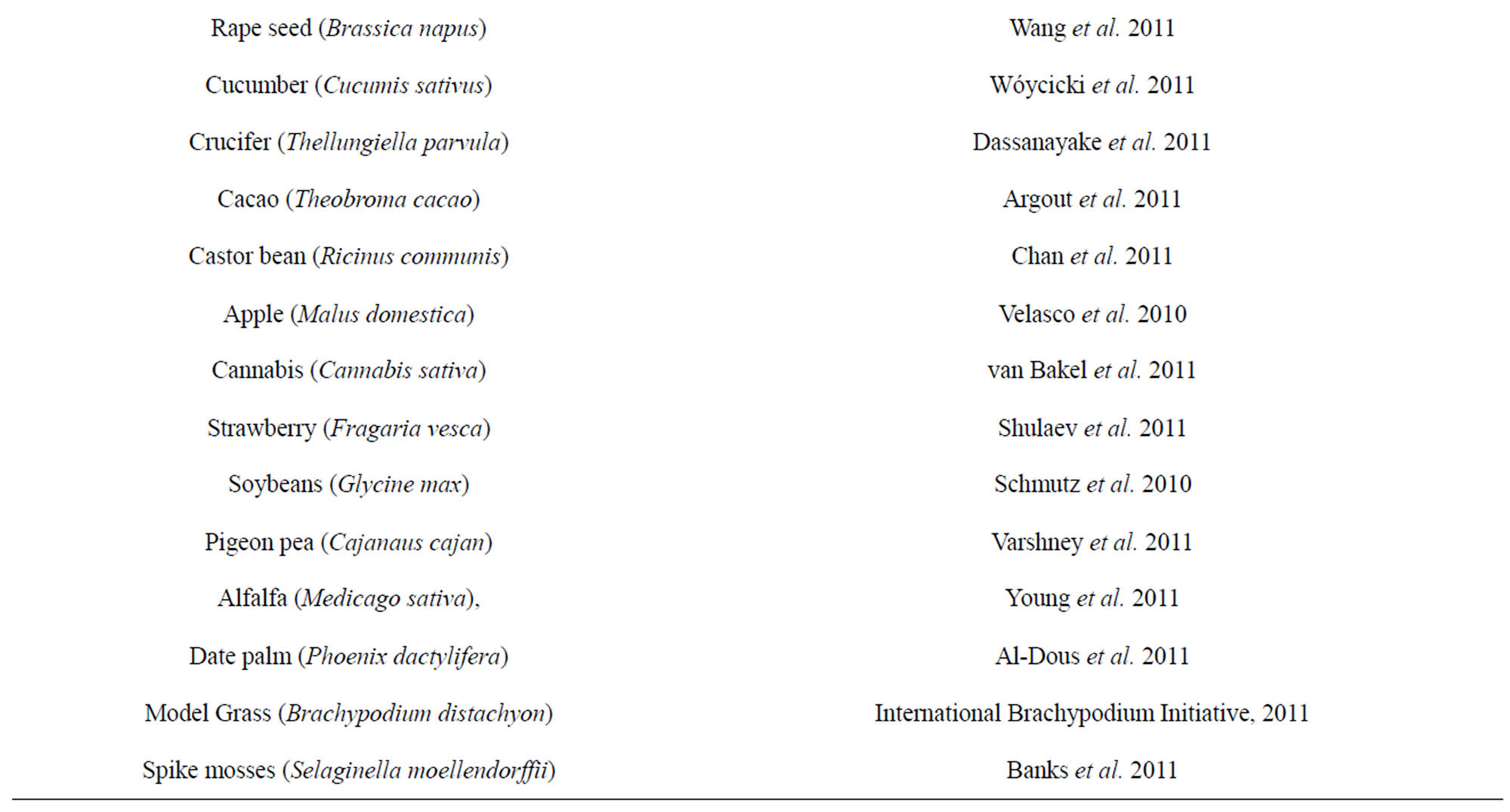
Table 2. Genome sequencing of crop plants.
sp.) and Foxtail Millet (Setaria italica) are still not yet completely published. These genomes reveals numerous species-specific details, including genome size, gene number, patterns of sequence duplication, a catalog of transposable elements, and syntenic relationships. To understand the complex instructions contained in all these raw sequence information of the plant genome, largescale functional genomics projects are required. Progress towards a complete understanding of gene regulatory networks shared among many crop plants is important for improving cultivated species and for understanding plant evolution.
9. SEQUENCING TECHNOLOGIES
Sanger dideoxy sequencing [65] and its modifications dominated the DNA sequencing field for nearly 30 years and in the past 10 years the length of Sanger sequence reads has increased from 450 bases to more than 1 kb [37]
Currently, with the rapid demand, novel innovations at the technological perspectives are being made and the important innovation was the introduction of Next-Generation Sequencing (NGS).
The term NGS is used to collectively describe technologies other than Sanger sequencing that have the potential to sequence the human genome in coming years for US $1000 [66] and such technologies are either already commercially available [67]. Present manufacturers of sequencing machine are constantly increasing sequence output in terms of number of reads (bp-base pair), increasing read length, as well as working to improve read quality [4,68,69].
10. DATA STORAGE AND MANAGEMENT
A greater challenge for sequence storage and management is solved by redundant data being generated by the new sequencing technologies. Several tools offer various levels of sophistication and simplicity for accessing the reference genome sequences available, genome viewers had been developed to allow users for visualization and assessment of data through common browsers such as EnsEMBL [70-72], Gbrowse and the University of California, Santa Cruz genome browser [73]. The International Nucleotide Sequence Databases consisting of GenBank [74], the DNA Databank of Japan (DDBJ) [75], and European Molecular Biological Laboratory (EMBL) [76] has been the principle repositories for DNA sequence data. The collection of large volumes of structured phenotypic data and its integration with the abundant genome data will add dimensions and challenges for the storage, management, and visualization of this information. Generation of new online servers like cloud computing and Galaxy has allowed researchers to perform comparative biology at an unprecedented level, providing insights into the foundations of life and the evolutionary processes that shape biological processes [77-79].
11. CONCLUSIONS
Crop evolution under domestication has led increased productivity of crop species, but at the same time has narrowed their genetic basis. Fortunately, wild relatives of crop plants exhibit vast genetic diversity for adaptation to stressful environments such as frost, drought and high salt and metal. Genomics is a brand new science that promises to revolutionize genetics, plant breeding and biotechnology using molecular characterization, transcript profiling and cloning of whole genomes to understand the structure, function and evolution of genes as well as to answer fundamental biological questions. Combining comprehensive sequence information with knowledge of the morphological and physiological diversity of crop plant and well-understood phylogeny promises to answer many questions about crop genome evolution and function.
Applications of genome resequencing, high-density genetic markers and a new generation of experimental designs that more readily relate mutational diversity to agronomic phenotypes, comparative genomics will become increasingly relevant to crop improvement. A combination of Genome Wide Association Studies (GWASs) and next-generation-mapping populations will improve our ability to connect phenotypes and genotypes, and genomic selection can take advantage of this data for rapid selection and breeding. The combination of these approaches with the promise of improved genomic technologies provides an opportunity for comparative genomics to apply our understanding of the past to the future of crop improvement.
12. ACKNOWLEDGEMENTS
I thank Shalini Narasimhan for helping me in editing and finalizing this manuscript.
REFERENCES
- Borner, A. (2006) Preservation of plant genetic resources in the biotechnology era. Biotechnology Journal, 1, 1393- 1404. doi:10.1002/biot.200600131
- Porceddu, E. (1999) Agricultural production and natural resources. In: Scarascia Mugnozza, G.T., et al., Eds., Genetics and Breeding for Quality and Resistance, Kluwer Academic Publishers, Amsterdam, 377-396.
- Monti, L. and Carputo, D. (2006) Genetic resources and genomics: Two sides of the same coin. Proceedings of the 50th Italian Society of Agricultural Genetics Annual Congress, Ischia, 10-14 September 2006.
- Metzker, M. (2010) Sequencing technologies—The next generation. Nature Review Genetics, 11, 31-46. doi:10.1038/nrg2626
- Edwards, D. and Batley, J. (2010) Plant genome sequencing: applications for crop improvement. Plant Biotechnology Journal, 8, 2-9. doi:10.1111/j.1467-7652.2009.00459.x
- Jackson, S., Iwata, A., Lee, S., Schmutz, J. and Shoemaker, R. (2011) Sequencing crop genomes: Approaches and applications. New Phytologist, 191, 915-925. doi:10.1111/j.1469-8137.2011.03804.x
- Hoisington, D., Khairallah, M., Reeves, T., Ribaut, J.M., Skovmand, B., Taba, S. and Warburtan, M. (1999) Plant genetic resources: What can they contribute toward increased crop productivity? Proceedings of the National Academy of Sciences of the USA, 96, 5937-5943. doi:10.1073/pnas.96.11.5937
- Esquinas-Alcázar, J. (2005) Science and society: Protecting crop genetic diversity for food security: Political, ethical and technical challenges. Nature Review Genetics, 6, 946-953. doi:10.1038/nrg1729
- Hodgkin, T., Roviglioni, R., De Vicente, M.C., Dudnik, N. (2001) Molecular methods in the conservation and use of plant genetic resources. Acta Horticultura, 546, 107- 118.
- Collard, B.C.Y., Jahufer, M.Z.Z., Brouwer, J.B. and Pang, E.C.K. (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica, 142, 169-196. doi:10.1007/s10681-005-1681-5
- Spooner, D., van Treuren, R. and de Vicente, M.C. (2005) Molecular markers for genebank management. IPGRI Technical Bulletin No. 10, International Plant Genetic Resources Institute, Rome.
- Weising, K., Nybom, H., Wolff, K. and Kahl, G. (2005) DNA fingerprinting in plants: Principles, methods, and applications. CRC Press, Boca Raton. doi:10.1201/9781420040043
- Yu, S.B., Xu, W.J., Vijayakumar, C.H.M., Ali, J., Fu, B.Y., Xu, J.L., Jiang Y.Z., Marghirang, R., Domingo, R., Aquino, C., Virmani, S.S. and Li, Z.K. (2003) Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theoretical and Applied Genetics, 108, 131-140. doi:10.1007/s00122-003-1400-3
- Rao, N.K., Reddy, L.J., Bramel, P. (2003) Potential of wild species for genetic enhancement of some semi-arid food crops. Genetic Resources and Crop Evolution, 50, 707-721. doi:10.1023/A:1025055018954
- Zimnoch-Guzowska, E., Lebecka, R., Kryszczuk, A., Maciejewska, U., Szczerbakowa, A. and Wielgat, B. (2003) Resistance to Phytophthora infestens in somatic hybrids of Solanum nigrum L. and diploid potato. Theoretical and Applied Genetics, 107, 43-48.
- Tuberosa, R., Gill, B.S. and Quarrie, S.A. (2002) Cereal genomics: Ushering in a brave new world. Plant Molecular Biology, 48, 445-449. doi:10.1023/A:1014818417927
- Varshney, R.K., Graner, A. and Sorrells, M.E. (2005) Genomics-assisted breeding for crop improvement. Trends in Plant Science, 10, 621-630. doi:10.1016/j.tplants.2005.10.004
- Varshney, R.K., Hoisington, D.A. and Tyagi, A.K. (2006) Advances in cereal genomics and applications in crop breeding. Trends in Biotechnology, 24, 490-499.
- Ersoz, E.S. (2007) Applications of linkage disequilibrium and association mapping in crop plants. In: Varshney, R.K. and Tuberosa, R.T., Eds., Genomics Assisted Crop Improvement: Genomics Approaches and Platforms. Springer, Berlin, 97-120.
- Schena, M. (1998) Microarrays: Biotechnology’s discovery platform for functional genomics. Trends in Biotechnology, 16, 301-306. doi:10.1016/S0167-7799(98)01219-0
- Jansen, R.C. and Nap, J.P. (2001) Genetical genomics: The added value from segregation. Trends in Genetics, 17, 388-391. doi:10.1016/S0168-9525(01)02310-1
- Till, B.J. (2007) TILLING and EcoTILLING for crop improvement. In: Varshney, R.K. and Tuberosa, R.T., Eds., Genomics Assisted Crop Improvement: Genomics Approaches and Platforms. Springer, Berlin, 333-349.
- Naylor, R.L., Falcon, W.P., Goodman, R.M., Jahn, M.M., Sengooba, T., Tefera, H. and Nelson, R.J. (2004) Biotechnology in the developing world: A case for increased investments in orphan crops. Food Policy, 29, 15-44. doi:10.1016/j.foodpol.2004.01.002
- Morrell, P., Buckler, E. and Ross-Ibarra, J. (2011) Crop genomics: Advances and applications. Nature Review Genetics, 13, 85-96.
- Fredslund, J., Madsen, L.H., Hougaard, B.K., Nielsen, A.M., Bertioli, D., Sandal, N., Stougaard, J. and Schauser, L. (2006) A general pipeline for the development of anchor markers for comparative genomics in plants. BMC Genomics, 14, 207. doi:10.1186/1471-2164-7-207
- Paterson, A.H. (2006) Leafing through the genomes of our major crop plants: Strategies for capturing unique information. Nature Review Genetics, 7, 174-184. doi:10.1038/nrg1806
- Boffelli, D., Weer, C.V., Weng, L., Lewis, K.D., Shoukry, M.I., Pachter, L., Keys D.N. and Rubin E.M. (2004) Intraspecies sequence comparisons for annotating genomes. Genome Research, 14, 2406-2411. doi:10.1101/gr.3199704
- Devos, K.M. (2005) Updating the “crop circle”. Current Opinion in Plant Biology, 8, 155-162. doi:10.1016/j.pbi.2005.01.005
- Nelson, R.J., Naylor, R.L. and Jahn, M.M. (2004) The role of genomics research in improvement of “orphan” crops. Crop Science, 44, 1901-1904. doi:10.2135/cropsci2004.1901
- Salentijn, E.M.J., Pereira, A., Angenent, G.C., van der Linden, C.G., Krens, F., Smulders, M.J.M. and Vosman, B. (2007) Plant translational genomics: From model species to crops. Molecular Breeding, 20, 1-13. doi:10.1007/s11032-006-9069-3
- Caicedo, A.L. and Purugganan, M.D. (2005) Comparative plant genomics. Frontiers and prospects. Plant Physiology, 2, 545-547. doi:10.1104/pp.104.900148
- Gutterson, N. and Zhang, J.Z. (2004) Genomics applications to biotech traits: A revolution in progress? Current Opinion in Plant Biology, 7, 226-230. doi:10.1016/j.pbi.2003.12.002
- Laurie, D.A., Griffiths, S., Dunford, R.P., Christodoulou, V., Taylor, S.A., Cockram, J., Beales, J. and Turner, A. (2004) Comparative genetic approaches to the identification of flowering time genes in temperate cereals. Field Crops Research, 90, 87-99. doi:10.1016/j.fcr.2004.07.007
- Bouchard, C. and Ordovas, J.M. (2012) Fundamentals of nutrigenetics and nutrigenomics. Progress in Molecular Biology and Translational Science, 108, 1-15.
- Parnell, L.D. (2012) Advances in technologies and study design. Progress in Molecular Biology and Translational Science, 108, 17-50.
- Atanassov, A., Batchvarova, R. and Djilianov, D. (2007) Strategic vision for plant biotechnology and genomics development. Biotechnology and Biotechnological Equipment, 21, 1-7.
- Varshney, R.K., Nayak, S.N., May, G.D. and Jackson, S.A. (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends in Biotechnology, 9, 522-530. doi:10.1016/j.tibtech.2009.05.006
- Cao, J., Schneeberger, K., Ossowski, S., Günther, T., Bender, S., Fitz, J., Koenig, D., Lanz, C., Stegle, O., Lippert, C., Wang, X., Ott, F., Müller, J., Alonso-Blanco, C., Borgwardt, K., Schmid, K.J. and Weigel, D. (2011) Wholegenome sequencing of multiple Arabidopsis thaliana populations. Nature Genetics, 43, 956-965. doi:10.1038/ng.911
- Hu, T.T., Pattyn, P., Bakker, E.G., Cao, J., Cheng, J.F., Clark, R.M. (2011) Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nature Genetics, 43, 476-481. doi:10.1038/ng.807
- Al-Dous, E.K., George, B., Al-Mahmoud, M.E., Al-Jaber, M.Y., Wang, H., Salameh, Y.M., Al-Azwani, E.K., Chaluvadi, S., Pontaroli, A.C., Debarry, J., Arondel, V., Ohlrogge, J., Saie, I.J., Suliman-Elmeer, K.M., Bennetzen, J.L., Kruegger, R.R. and Malek, J.A. (2011) De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nature Biotechnology, 29, 521-527. doi:10.1038/nbt.1860
- Argout, X., et al. (2011) The genome of Theobroma cacao. Nature Genetics, 43, 101-108. doi:10.1038/ng.736
- Banks, J.A., et al. (2011) The selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science, 332, 960-963. doi:10.1126/science.1203810
- Chan, A.P., et al. (2010) Draft genome sequence of the oil-seed species Ricinus communis. Nature Biotechnology, 28, 951-956. doi:10.1038/nbt.1674
- Dassanayake, M., et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nature Genetics, 43, 913-918. doi:10.1038/ng.889
- Goff, S.A., et al. (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science, 296, 92- 100. doi:10.1126/science.1068275
- Huang, S. (2009) The genome of the cucumber, Cucumis sativus L. Nature Genetics, 12, 1275-1283. doi:10.1038/ng.475
- Initiative, I.B., et al. (2010) Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature Biotechnology, 463, 763-768.
- Arabidopsis thaliana Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature Biotechnology, 408, 796-815.
- Jaillon, O., et al. (2007) Characterization French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature Biotechnology, 449, 463-467.
- Ming, R., et al. (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature Letters, 24, 991-997. doi:10.1038/nature06856
- Rensing, S.A., et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science, 319, 64-69. doi:10.1126/science.1150646
- Sato, S., et al. (2008) Genome structure of the legume, Lotus japonicus. Genome structure of the legume, Lotus japonicus. DNA Research, 15, 227-239. doi:10.1093/dnares/dsn008
- Schmutz, J., et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature, 463, 178-183. doi:10.1038/nature08670
- Schnable, P.S., et al. (2009) The B73 maize genome: Complexity, diversity, and dynamics. Science, 326, 1112. doi:10.1126/science.1178534
- Shulaev, V., et al. (2011) The genome of woodland strawberry (Fragaria vesca). Nature Genetics, 43, 109-116. doi:10.1038/ng.740
- Tuskan, G.A., et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313, 1596-1604. doi:10.1126/science.1128691
- van Bakel, H., Stout, J.M., Cote, A.G., Tallon, C.M., Sharpe, A.G., Hughes, T.R. and Page, J.E. (2011) The draft genome and transcriptome of Cannabis sativa. Genome Biology, 12, 102. doi:10.1186/gb-2011-12-10-r102
- Varshney, R.K., et al. (2011) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotechnology, 30, 83-89. doi:10.1038/nbt.2022
- Velasco, R., et al. (2010) The genome of the domesticcated apple (Malus × domestica Borkh.). Nature Genetics, 42, 833-839. doi:10.1038/ng.654
- Wang, X., et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics, 43, 1035- 1039. doi:10.1038/ng.919
- Wóycicki, R., et al. (2011) The genome sequence of the North-European cucumber (Cucumis sativus L.) unravels evolutionary adaptation mechanisms in plants. PLoS ONE, 6, 7.
- Young, N.D., et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature, 480, 520-524.
- Yu, J., (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science, 296, 79-92. doi:10.1126/science.1068037
- The Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature, 475, 189-195. doi:10.1038/nature10158
- Sanger, F., Nicklen, S. and Coulson, A.R. (1997) DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the USA, 74, 5463-5467. doi:10.1073/pnas.74.12.5463
- Service, R.F. (2006) Gene sequencing: The race for the $1000 genome. Science, 311, 1544-1546. doi:10.1126/science.311.5767.1544
- Kling, J. (2005) The search for sequencing thoroughbred. Nature Biotechnology, 23, 1333-1335. doi:10.1038/nbt1105-1333
- Mardis, E. (2008) The impact of next-generation sequenceing technology on genetics. Trends in Genetics, 24, 133- 141. doi:10.1016/j.tig.2007.12.007
- Mardis, E.R. (2010) The $1000 genome, the $100,000 analysis? Genome Medicine, 2, 84. doi:10.1186/gm205
- Birney, E., et al. (2004) An overview of Ensembl. Genome Research, 14, 925-958. doi:10.1101/gr.1860604
- Chen, Y., et al. (2010) Ensembl variation resources. BMC Genomics, 11, 293. doi:10.1186/1471-2164-11-293
- Kinsella, R.J., et al. (2011) Ensembl BioMarts: A hub for data retrieval across taxonomic space Database (Oxford). doi:10.1093/database/bar030
- Dreszer, T.R., et al. (2012) The UCSC genome browser database: Extensions and updates 2011. Nucleic Acids Research, 40, 18-23. doi:10.1093/nar/gkr1055
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J. and Wheeler, D.L. (2006) Genbank. Nucleic Acids Research, 34, 16-20. doi:10.1093/nar/gkj157
- Ohyanagi, H., et al. (2006) The rice annotation project database (rap-db): Hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Research, 34, 741- 744. doi:10.1093/nar/gkj094
- Cochrane, G., et al. (2006) EMBL nucleotide sequence database: Developments in 2005. Nucleic Acids Research, 34, 10-15. doi:10.1093/nar/gkj130
- Goecks, J., Nekrutenko, A., Taylor, J. and Galaxy Team (2010) The Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology, 11, 86. doi:10.1186/gb-2010-11-8-r86
- Hsi-Yang, Fritz, M., Leinonen, R., Cochrane, G. and Birney, E. (2011) Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Research, 21, 734-40. doi:10.1101/gr.114819.110
- Schatz, M., Langmead, B. and Salzberg, S. (2010) Cloud computing and the DNA data race. Nature Biotechnology, 28, 691-693. doi:10.1038/nbt0710-691

