American Journal of Plant Sciences
Vol.3 No.6(2012), Article ID:20046,6 pages DOI:10.4236/ajps.2012.36096
A New Fuzzless Seed Locus in an Upland Cotton (Gossypium hirsutum L.) Mutant
![]()
1Agricultural Research Service (ARS), United States Department of Agriculture (USDA), Stoneville, USA; 2Department of Plant and Soil Science, Texas Tech University, Lubbock, USA.
Email: efrem.bechere@ars.usda.gov
Received March 27th, 2012; revised April 20th, 2012; accepted April 30th, 2012
Keywords: Fuzzy; Fuzzless; Lint Percent; Mutagenesis; Mutant; Naked Seed
ABSTRACT
Various fiber mutants of cotton have been reported since 1920. Two of the best characterized mutants are the naked seed loci, N1N1 and n2n2. Recently, a naked-tufted mutant called 9023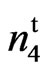 was developed from the cultivar SC 9023 through chemical mutagenesis. The mutant was tested to determine if it was allelic to either N1, or n2 or was a unique mutant in a new uncharacterized locus. In 1999, one M3 plant from SC 9023 with tufted partially naked seed coat (fuzzless) was identified. In 2004, 2006, and 2007 (Lubbock, TX), 2007 (College Station, TX), and 2011 (Stoneville, MS), the homozygous naked seed mutant was evaluated with the non-mutated wild type parent (SC 9023) in replicated trials for agronomic and fiber traits. Crosses between the mutant and the wild type was made at Stoneville, MS in 2009. The F2 of this cross segregated in a 3 fuzzy: 1 fuzzless ratio indicating that the fuzzless trait in the mutant is controlled by a recessive locus. Allelism tests with N1N1, n2n2 and n3n3, lint percent, and fiber trait data indicated that the new locus in the mutant differs from the previously characterized fuzzless seed alleles in that it does not appear to decrease lint percent. We have putatively designated this gene
was developed from the cultivar SC 9023 through chemical mutagenesis. The mutant was tested to determine if it was allelic to either N1, or n2 or was a unique mutant in a new uncharacterized locus. In 1999, one M3 plant from SC 9023 with tufted partially naked seed coat (fuzzless) was identified. In 2004, 2006, and 2007 (Lubbock, TX), 2007 (College Station, TX), and 2011 (Stoneville, MS), the homozygous naked seed mutant was evaluated with the non-mutated wild type parent (SC 9023) in replicated trials for agronomic and fiber traits. Crosses between the mutant and the wild type was made at Stoneville, MS in 2009. The F2 of this cross segregated in a 3 fuzzy: 1 fuzzless ratio indicating that the fuzzless trait in the mutant is controlled by a recessive locus. Allelism tests with N1N1, n2n2 and n3n3, lint percent, and fiber trait data indicated that the new locus in the mutant differs from the previously characterized fuzzless seed alleles in that it does not appear to decrease lint percent. We have putatively designated this gene .
.
1. Introduction
Normal cottonseed is covered with lint and fuzz. Lint is a textile fiber whereas fuzz is a short fiber that is difficult to gin from the seeds. However, spontaneous fiber mutants of cotton have been reported since 1920 (Kohel, 1973 [1] and Endrizzi et al., 1984 [2]). Cotton mutants have been useful to cotton researchers in various ways. They are used to detect and locate QTL for lint yield (An, C. et al., 2010) [3], fiber quality (Paterson et al., 2010) [4], seed traits (Song and Zhang, 2007) [5], and response to biotic and abiotic stresses (Shen et al., 2006) [6] to linkage groups on chromosomes in cotton. Turley and Ferguson (1996) [7] demonstrated that mutants can be used to determine differences in gene/protein regulation during development of ovular trichomes. Mutants are also a powerful resource for studying gene functions (Rong et al., 2005) [8]. Recent studies have also shown that naked seed fiber mutants gin faster and use less energy when compared to other conventional or transgenic cultivars. They also have reduced number of seed coat neps, nep size and short fiber contents (Bechere et al., 2011) [9].
Some of the first mutants were fuzzless but linted (Du et al., 2011) [10]. Two of thebest characterized of these are the naked seed loci, N1N1 and n2n2. N1N1 is dominant whereas n2n2 is recessive. The naked seed mutant (n2) was initially characterized and assigned to chromosome 26 using aneuploidy stocks (Endrizzi and Ray, 1991) [11]. A second mutant (N1) predicted to be homoeologous to n2 was mapped to chromosome 12 (Endrizzi and Ray, 1991 [11] and Samora et al., 1994 [12]). Turley and Kloth (2002) [13] developed a fiberless line, MD 17 from the cross of accession 143 (n2n2) and accession 243 (N1N1) and indicated that at least two loci (N1 and n2) interacted to produce this fiberless seed. Other fiberless mutants which have been reported in the literature include MU-5, a fiberless, lintless mutant from India (Nadarajan, N., and S.R. Sree Rangasamy, 1988) [14], SMA-4, a genetic stock containing a recessive mutation (ha) that confers fiberless seed, and an epistatic recessive mutation (fz) that produce lintless (i.e. fuzz fibers only) seed in the absence of homozygosity for ha (Beasley and Egli, 1977) [15], Fb1, an incompletely dominant fiberless mutation exhibiting no lint or fuzz fibers (Kearney and Harrison, 1927) [16], SL1-7-1, an inbred line with three loci conditioning the expression of the fiberless phenotype (Turley and Kloth, 2008) [17], XZ142w, with a fuzzless trait controlled by two gene loci (Zhang and Pan, 1991) [18], and L40 where the fuzz around the micropyle is controlled by two non-allelic major genes (Musaev and Abzalov, 1972) [19].
Historically, fuzzless seed phenotypes have been strongly associated with both low lint yield and low lint percent. This had somehow diminished the interest of breeders in these phenotypes despite their positive attributes like lower neps, short fiber content and better ginning efficiency. The two loci N1 and n2 were reported to inhibit fuzz fiber development and had considerable negative effect on lint production (Ware et al., 1947 [20], Rong et al., 2005 [8], and Turley et al., 2007 [21]).
The objectives of this study were to evaluate the agronomic and fiber quality performance of a new fuzzless mutant of upland cotton and determine if the new mutant is allelic to either N1 or n2 or is a unique mutant at a new uncharacterized locus.
2. Materials and Method
2.1. Developing the Mutant
In 1997, a commercial variety of cotton SC 9023 (PVP # 9500237) was treated with 2.45% volume by volume ethyl methane sulfonate (EMS). The seeds were imbibed in aerated distilled water for 16 hours and rinsed with distilled water and treated with EMS for 2 hours. The seeds were thoroughly rinsed with distilled water again and hand planted in the field immediately. During 1997 (M1 generation) and 1998 (M2 generation), one boll per plant was harvested in bulk to form the next generation and to reduce the mutation load. In 1999, one M3 plant from SC 9023 with tufted partially naked seed coat (fuzzless) was identified. From 2000 to 2003, individual plant selections from this mutant were made at Lubbock, TX to stabilize this trait. A stable, homozygous line was identified and named 9023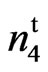 (Figure 1).
(Figure 1).
2.2. Evaluation of the Mutant Line for Agronomic and Fiber Traits
In 2004, 2006, and 2007 (at Lubbock, TX), 2007 (at College Station, TX), and 2011 (at Stoneville, MS), the homozygous naked seed mutant was evaluated with the non-mutated wild type parent (SC 9023). Materials were planted in 12.2 m single rows in randomized complete block design with 1.0 m between rows in 4 replications. Fertilizers, herbicides, fungicides and insecticides were applied on an “as needed” basis at each location. Fifty randomly selected bolls were hand-picked from each entry and the cotton was ginned on a 10-saw laboratory gin stand (Continental Eagle, Prattville, AL). Data were collected on lint yield, lint percent, fibers/seed, fiber density, HVI and AFIS quality traits, and yarn and spinning performance. Fibers were analyzed for HVI (High Volume Instrument) at the Fiber and Biopolymer Research Institute, Texas Tech University and Star Lab Inc., Knoxville, TN. Analyses for AFIS (Advanced Fiber Infor-

Figure 1. Phenotypes for seeds of n2n2, N1N1, SC 9023 (wild type) and 9023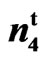 (mutant).
(mutant).
mation System) was conducted at the Fiber and Biopolymer Research Institute and at Stoneville, MS. Yarn spinning and performance studies were conducted at the Fiber and Biopolymer Research Institute.
To calculate fibers per seed and fiber density, acid delinted seeds were scanned for surface area with WinSeedle scanner (http://www.regent.qc.ca/products/needle/ NEEDLE/html). The mean length by number and fineness data from AFIS were then used to estimate the number of fibers per seed by dividing by the mean surface area to obtain the number of fibers/mm2 (Eric Hequet, personal communication). Lint percent was calculated by dividing the mass lint ginned by the mass of total weight of lint and seed (seed cotton) and expressed as a percentage of the mass of seed cotton. The SAS software package (SAS Institute Inc., SAS Circle, Carry, NC) was used to analyze all data.
2.3. Crosses for Inheritance Study and Allelism Test
Four lines, SC 9023 (wild type), its mutant 9023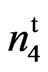 , accession 143 (n2n2n3n3) (PI 528543), accession 243 (N1N1) (PI 528610) alongwith the resulting F1, F2, and BC1 test crosses were grown at Stoneville, MS during 2009, 2010 and 2011 growing seasons. SC 9023 is an obsolete High Plains cultivar developed by Seedco Corporation. 9023
, accession 143 (n2n2n3n3) (PI 528543), accession 243 (N1N1) (PI 528610) alongwith the resulting F1, F2, and BC1 test crosses were grown at Stoneville, MS during 2009, 2010 and 2011 growing seasons. SC 9023 is an obsolete High Plains cultivar developed by Seedco Corporation. 9023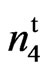 is a mutant developed from SC 9023 through chemical mutagenesis (Bechere et al., 2009) [22]. Accessions 143 and 243 were obtained from Rick Turley at USDA, Stoneville, MS and from the National Collection of Cotton Germplasm (Percival, 1987) [23]. Seeds from 143 and 243 were grown in the field at Stoneville, MS, verified for phenotype and seed increased. Accession 143 is the Mexican fuzzless seed UA 3-3 and has the recessive fuzzless seed alleles n2 and n3. Accession 243 is the Ballard fuzzless seed line and has the dominant fuzzless seed allele N1 (Kearney and Harrison, 1927 [16]; Endrizzi et al., 1984 [2]; Percy and Kohel, 1999 [24]; Turley and Kloth, 2002 [17]). Crosses of the mutant and wild type (269 F2 plants) and their reciprocal (106 F2 plants) were made to study the inheritance of the naked tufted mutant. Allelism tests were made between accession 143 and the mutant (682 F2 plants) and accession 243 and the mutant (265 F2 plants). Tests of homogeneity were conducted between values for different populations before the data were combined.
is a mutant developed from SC 9023 through chemical mutagenesis (Bechere et al., 2009) [22]. Accessions 143 and 243 were obtained from Rick Turley at USDA, Stoneville, MS and from the National Collection of Cotton Germplasm (Percival, 1987) [23]. Seeds from 143 and 243 were grown in the field at Stoneville, MS, verified for phenotype and seed increased. Accession 143 is the Mexican fuzzless seed UA 3-3 and has the recessive fuzzless seed alleles n2 and n3. Accession 243 is the Ballard fuzzless seed line and has the dominant fuzzless seed allele N1 (Kearney and Harrison, 1927 [16]; Endrizzi et al., 1984 [2]; Percy and Kohel, 1999 [24]; Turley and Kloth, 2002 [17]). Crosses of the mutant and wild type (269 F2 plants) and their reciprocal (106 F2 plants) were made to study the inheritance of the naked tufted mutant. Allelism tests were made between accession 143 and the mutant (682 F2 plants) and accession 243 and the mutant (265 F2 plants). Tests of homogeneity were conducted between values for different populations before the data were combined.
The fuzzy/fuzzless phenotypes were scored as described by Ware et al. (1940) [25] and Ware et al. (1947) [20] with the fuzzy seed corresponding to classes 1 to 11 and fuzzless seed corresponding to classes 12 to 16 (Figure 2). Chi-squares were calculated to determine the best fit for all genetic models tested.
3. Results and Discussion
Lint yield and lint percent data for the wild type, the naked seed mutant, accession 143 (n2n2n3n3), accession 243 (N1N1) are summarized in Table 1. Overall, the lint yield of the 9023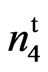 mutant was slightly lower than that of the wild type and check varieties at all locations. However, the 9023
mutant was slightly lower than that of the wild type and check varieties at all locations. However, the 9023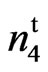 mutant significantly yielded higher than the other fuzzless seed mutants, n2n2n3n3 and N1N1. The lint percent of the 9023
mutant significantly yielded higher than the other fuzzless seed mutants, n2n2n3n3 and N1N1. The lint percent of the 9023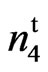 mutant was not significantly lower than the lint percent of the wild type except at the Stoneville location. The lint percent of the mutant averaged over locations and years was 35%. Accession 143 (n2n2n3n3) had 23% lint and accession 243 (N1N1) had only 7% lint. Turley et al. (2007) [21] reported lint percent ranging from 0.7 to 23.6 % for different N1 line and 24.4% for the n2 line. They proposed that only the genotype n1n1N2N2N3N3 would generate a normal lint percent of 40.5%. One of the reasons cotton breeders were reluctant to use the naked seed trait in their breeding programs was the low lint percent associated with this characteristic.
mutant was not significantly lower than the lint percent of the wild type except at the Stoneville location. The lint percent of the mutant averaged over locations and years was 35%. Accession 143 (n2n2n3n3) had 23% lint and accession 243 (N1N1) had only 7% lint. Turley et al. (2007) [21] reported lint percent ranging from 0.7 to 23.6 % for different N1 line and 24.4% for the n2 line. They proposed that only the genotype n1n1N2N2N3N3 would generate a normal lint percent of 40.5%. One of the reasons cotton breeders were reluctant to use the naked seed trait in their breeding programs was the low lint percent associated with this characteristic.
Number of fibers per seed and fiber density were lower in the 9023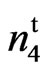 mutant than in the wild type which explains the low lint yield observed in the mutant line. However, when compared to accessions 143 and 243, the mutant had significantly much higher fibers/seed and fiber density (Table 2).
mutant than in the wild type which explains the low lint yield observed in the mutant line. However, when compared to accessions 143 and 243, the mutant had significantly much higher fibers/seed and fiber density (Table 2).
The naked seed mutant 9023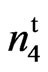 had lower short fiber content and seed coat neps than the wild type SC 9023. Accessions 143 and 243 had much lower short fiber content and seed coat neps than both the wild type and the mutant (Table 3). This, however, might be a function of the much lower fibers/seed in accessions 143 and 243 (Table 2). HVI data from Stoneville, MS indicated that the mutant had comparable fiber length and fiber strength with the wild type but had significantly longer fiber than accessions 143 (n2n2n3n3) and 243 (N1N1) and significantly stronger fiber than N1N1. Yarn quality data was obtained only from Lubbock, TX in 2007. The mutant had higher count strength product, similar tenacity and lower thin and thick places (Table 3). The count strength
had lower short fiber content and seed coat neps than the wild type SC 9023. Accessions 143 and 243 had much lower short fiber content and seed coat neps than both the wild type and the mutant (Table 3). This, however, might be a function of the much lower fibers/seed in accessions 143 and 243 (Table 2). HVI data from Stoneville, MS indicated that the mutant had comparable fiber length and fiber strength with the wild type but had significantly longer fiber than accessions 143 (n2n2n3n3) and 243 (N1N1) and significantly stronger fiber than N1N1. Yarn quality data was obtained only from Lubbock, TX in 2007. The mutant had higher count strength product, similar tenacity and lower thin and thick places (Table 3). The count strength
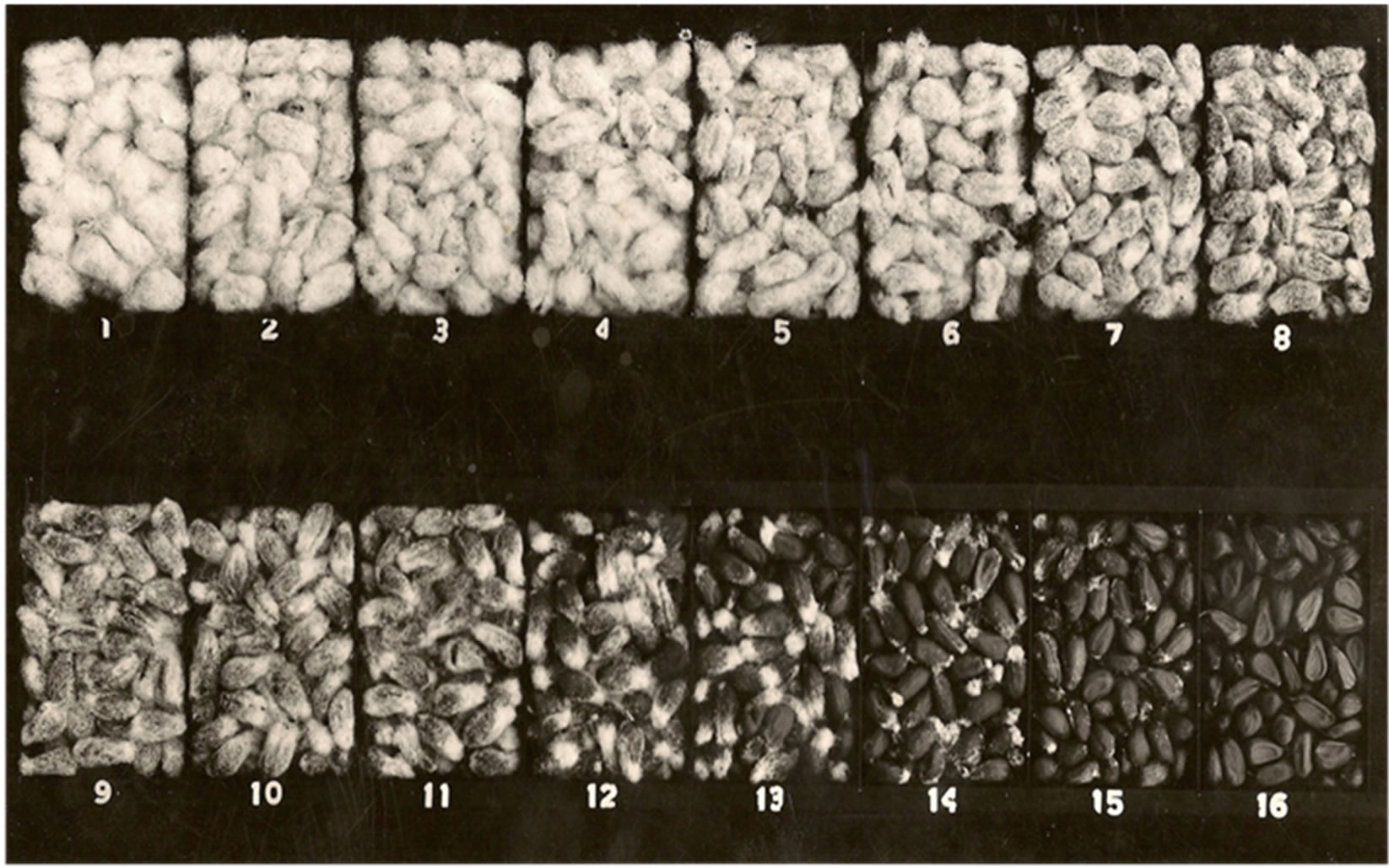
Figure 2. Grades of seed with fuzz in upland cotton (anonymous).
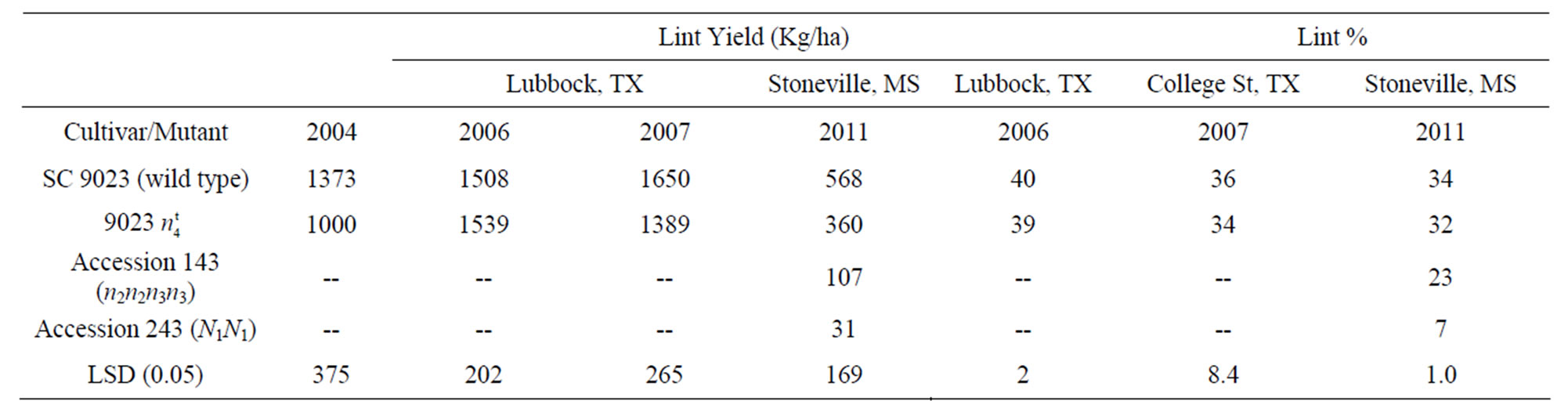
Table 1. Lint yield and lint % for 9023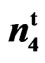 , its wild type SC 9023, and accessions 143 and 243 across locations and years.
, its wild type SC 9023, and accessions 143 and 243 across locations and years.
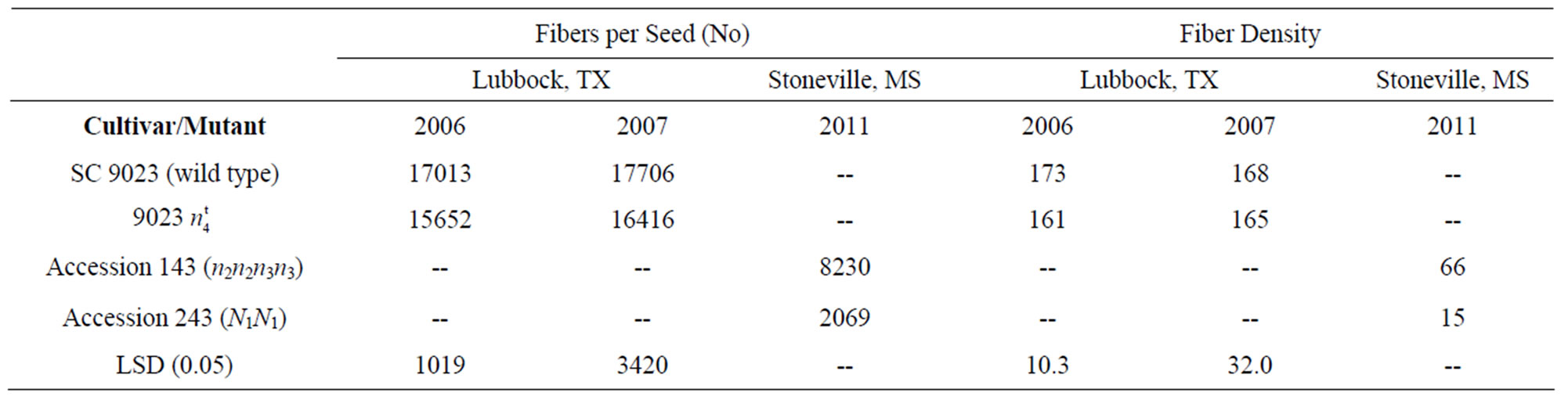
Table 2. Fibers per seed and fiber density for SC 9023, 9023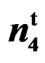 , accessions 143 and 243.
, accessions 143 and 243.
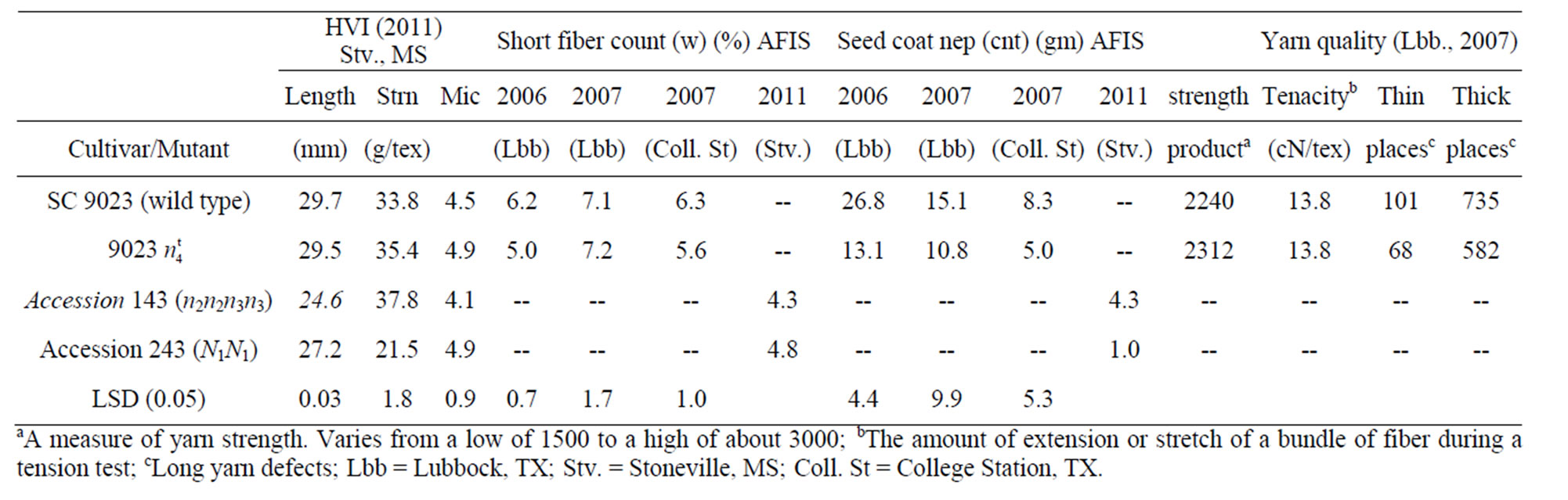
Table 3. HVI, AFIS, and yarn quality data for SC 9023, the mutant line, accessions 143 and 243.
product is a measure of yarn strength and varies from a low of 1500 to a high of about 3000. Tenacity is the amount of extension or stretch of a bundle of fiber during a tension test. Thin and thick places are long yarn defects.
A summary of the crosses, F1, F2, and BCF1 data with suggested genotypes for the parents is given in Table 4. The F2 progeny from the cross 9023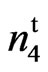 X SC 9023 (wild type) were used to determine the inheritance model for the mutant. All F1 plants from the cross (17 plants) and their reciprocals (25 plants) were all fuzzy. The observed phenotypic segregation ratios for the 264 second generation filial plants and their reciprocals (106 plants) are given in Table 4. The chi-square analyses from both the cross and reciprocals gave a good fit to a one locus model with a ratio of 3 fuzzy: 1 fuzzless with a χ2 = 0.7270, P = 0.3938 for the cross and χ2 = 0.0130, P = 0.9107 for the reciprocals. The fuzzless trait in the naked seed mutant appears to be controlled by a recessive gene. The BCF1 segregation data of 26 fuzzy and 27 fuzzless with a χ2 of 0.0190, P = 0.8907 fit a 1 fuzzy: 1 fuzzless genetic ratio, confirming the F2 result.
X SC 9023 (wild type) were used to determine the inheritance model for the mutant. All F1 plants from the cross (17 plants) and their reciprocals (25 plants) were all fuzzy. The observed phenotypic segregation ratios for the 264 second generation filial plants and their reciprocals (106 plants) are given in Table 4. The chi-square analyses from both the cross and reciprocals gave a good fit to a one locus model with a ratio of 3 fuzzy: 1 fuzzless with a χ2 = 0.7270, P = 0.3938 for the cross and χ2 = 0.0130, P = 0.9107 for the reciprocals. The fuzzless trait in the naked seed mutant appears to be controlled by a recessive gene. The BCF1 segregation data of 26 fuzzy and 27 fuzzless with a χ2 of 0.0190, P = 0.8907 fit a 1 fuzzy: 1 fuzzless genetic ratio, confirming the F2 result.
Allelism tests were conducted to check if the gene causing the fuzzless mutant in 9023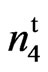 was allelic to N1, n2 or n3 or is a unique mutant at a new uncharacterized locus. The cross of n2n2n3n3 with 9023
was allelic to N1, n2 or n3 or is a unique mutant at a new uncharacterized locus. The cross of n2n2n3n3 with 9023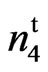 resulted in an F2 that segregated into 493 fuzzy and 189 fuzzless individuals fitting a 45:19 ratio with a 3 segregating loci model (χ2 = 1.2801, P = 0.4500). All twenty-two F1s scored fuzzless (Table 4). Turley and Kloth (2002) [13] reported that fuzzless seeds were obtained in n2n2 plants when a second recessive locus (n3) was present. According to their reports the n3 is required for the expression of the fuzzless phenotype in line 143 and fiberless phenoltype in line SL 1-7-1 (Turley and Kloth, 2008) [17]. The fourth locus coming from the mutant appears to be new and we designate this locus as
resulted in an F2 that segregated into 493 fuzzy and 189 fuzzless individuals fitting a 45:19 ratio with a 3 segregating loci model (χ2 = 1.2801, P = 0.4500). All twenty-two F1s scored fuzzless (Table 4). Turley and Kloth (2002) [13] reported that fuzzless seeds were obtained in n2n2 plants when a second recessive locus (n3) was present. According to their reports the n3 is required for the expression of the fuzzless phenotype in line 143 and fiberless phenoltype in line SL 1-7-1 (Turley and Kloth, 2008) [17]. The fourth locus coming from the mutant appears to be new and we designate this locus as 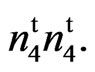 This locus appears to condition a partially naked and tufted seed phenotype in the mutant.The other allelism test conducted involved the cross N1N1 with 9023
This locus appears to condition a partially naked and tufted seed phenotype in the mutant.The other allelism test conducted involved the cross N1N1 with 9023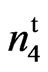 (Table 4). All 26 F1 plants were fuzzless. A total of 265 F2 progeny were scored and all werenaked-tufted, naked, or fuzzy. The independent assortment confirmed a 13 naked to 3 fuzzy ratio (dominant and recessive epistasis). These results differ from the crossing of N1N1 and n2n2 where 1 out of every 16 plants in the F2 progeny produced fiberless ovules. N1 and
(Table 4). All 26 F1 plants were fuzzless. A total of 265 F2 progeny were scored and all werenaked-tufted, naked, or fuzzy. The independent assortment confirmed a 13 naked to 3 fuzzy ratio (dominant and recessive epistasis). These results differ from the crossing of N1N1 and n2n2 where 1 out of every 16 plants in the F2 progeny produced fiberless ovules. N1 and 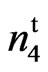 are therefore on separate loci and interact to produce the 9 naked, 4 naked and tufted and 3 fuzzy phenotypes. Endrizzi and Ray (1991) [11] crossed
are therefore on separate loci and interact to produce the 9 naked, 4 naked and tufted and 3 fuzzy phenotypes. Endrizzi and Ray (1991) [11] crossed 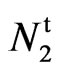 (Naked-tufted seed) with n2 (AG 208) and reported a 13 naked:
(Naked-tufted seed) with n2 (AG 208) and reported a 13 naked:
3 fuzzy independent assortment and concluded that 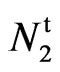 and n2 are alleles and are in linkage group IX. Based on the above results the following genotypes are proposed for the four lines involved in this study:
and n2 are alleles and are in linkage group IX. Based on the above results the following genotypes are proposed for the four lines involved in this study:
SC 9023 (Wild type and fuzzy) =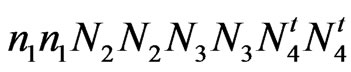 ;
;
9023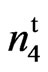 =
=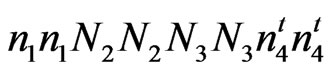 ;
;
accession 143 (fuzzless) =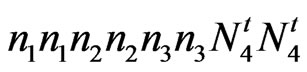 ;
;
accession 243 (fuzzless) =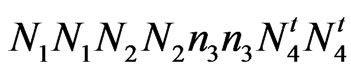 .
.
4. Conclusion
The 9023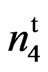 mutant is quite unique since it has a partially naked body with a small tuft attached to one end of the seed. The lint percent of the mutant is 39% higher than the lint percent of n2n2n3n3 and 57% higher than that of N1N1. The mutant had lower short fiber content, lower seed coat neps and better yarn quality than the wild type cultivar. A recent study (Bechere et al. (2011), has shown that this mutant gins faster and with less energy when compared to other conventional and transgenic cultivars. In summary, the agronomic, fiber trait, and phenotypic appearance of the mutant indicate that the locus in the mutant is not allelic to either N1, n2, or n3 and is a new mutant at a new uncharacterized locus. We designate this new locus as
mutant is quite unique since it has a partially naked body with a small tuft attached to one end of the seed. The lint percent of the mutant is 39% higher than the lint percent of n2n2n3n3 and 57% higher than that of N1N1. The mutant had lower short fiber content, lower seed coat neps and better yarn quality than the wild type cultivar. A recent study (Bechere et al. (2011), has shown that this mutant gins faster and with less energy when compared to other conventional and transgenic cultivars. In summary, the agronomic, fiber trait, and phenotypic appearance of the mutant indicate that the locus in the mutant is not allelic to either N1, n2, or n3 and is a new mutant at a new uncharacterized locus. We designate this new locus as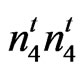 . The allelism tests in this study (Table 4) confirm this conclusion. This mutant can be included in cotton breeding programs to exploit its positive traits without the fear of adverse effect from low lint percent exhibited by N1, n2 and n3.
. The allelism tests in this study (Table 4) confirm this conclusion. This mutant can be included in cotton breeding programs to exploit its positive traits without the fear of adverse effect from low lint percent exhibited by N1, n2 and n3.
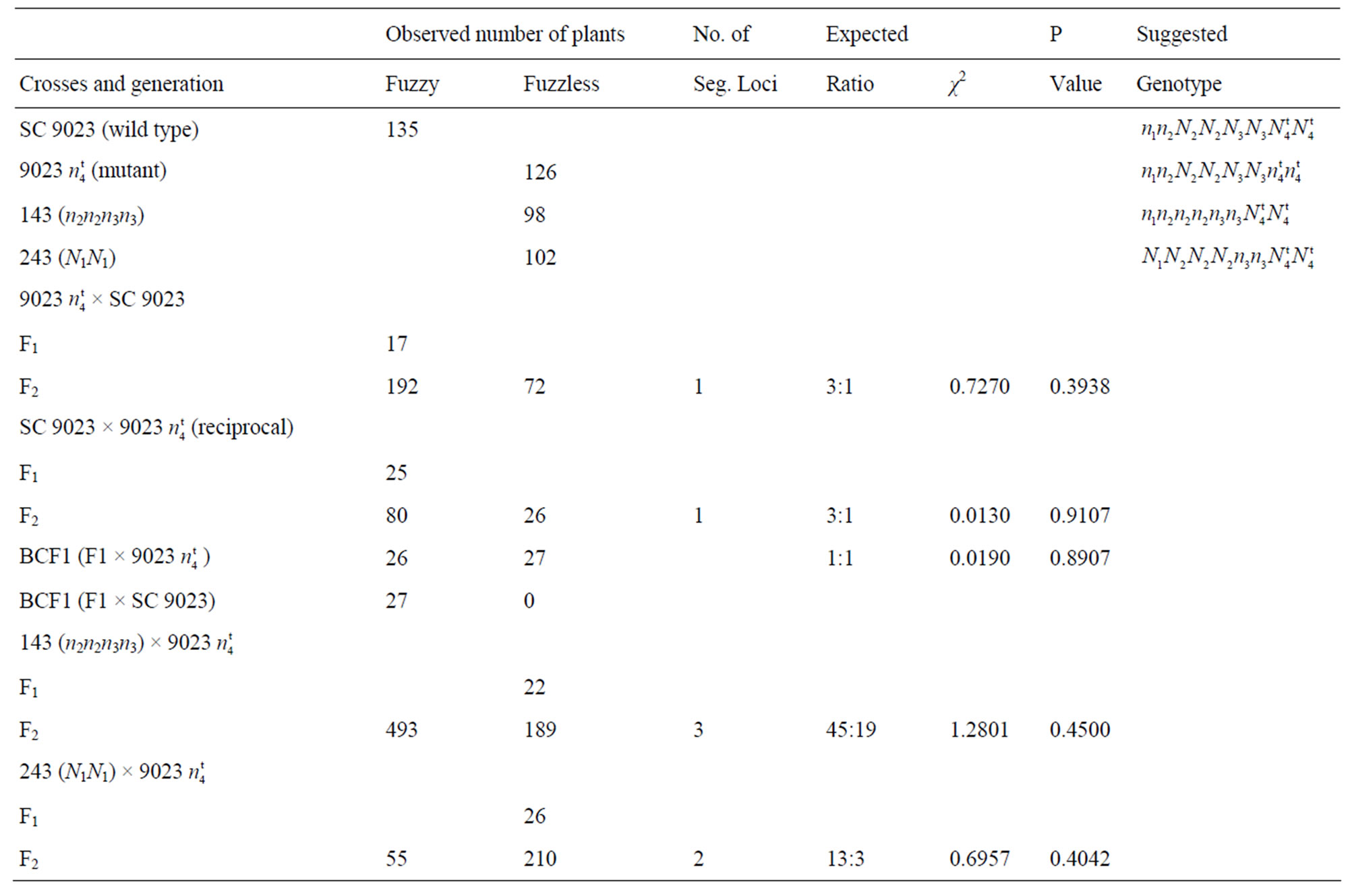
Table 4. Summary of Crosses and generations of SC 9023, 9023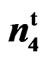 , accessions 143, and 243.
, accessions 143, and 243.
5. Acknowledgements
This research is part of the National Program 301 (Plant Genetic Resources, Genomics and Genetic Improvement) and funded by Cotton Incorporated and through ARS Project Number 6402-21000-033-00D.
6. Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
REFERENCES
- R. J. Kohel, “Genetic Nomenclature in Cotton,” Journal of Heredity, Vol. 64, No. 5, 1973, pp. 291-295.
- J. E. Endrizzi, E. L. Turcotte and R. J. Kohel, “Qualitative Genetics, Cytology, and Cytogenetics,” In: R. J. Kohel, and C. F. Lewis, Eds., Cotton, Agronomy Society of America, Madison, 1984, pp. 59-80.
- C. An, J. N. Jenkins, W. Jixiang, Y. Guo and J. C. McCarty, “Use of Fiberand Fuzz Mutants to Detect QTL for Yield Components, Seed, and Fiber Traits of Upland Cotton,” Euphytica, Vol. 172, No. 1, 2010, pp. 21-34. doi:10.1007/s10681-009-0009-2
- A. H. Paterson, Y. Saranga, M. Menz, C. X. Jiang and R. J. Wright, “QTL Analysis of Genotype × Environment Interactions Affecting Cotton Fiber Quality,” Theoretical and Applied Genetics, Vol. 106, No. 3, 2003, pp. 384-396.
- X. L. Song and T. Z. Zhang, “Identification of Quantitative Trait Loci Controlling Seed Physical and Nutrient Trait in Cotton,” Seed Science Research, Vol. 17, No. 4, 2007, pp. 243-251. doi:10.1017/S0960258507834957
- X. Shen, G. V. Becelaere, P. Kumar, R. F. Davis, O. L. May and P. Chee, “QTL Mapping for Resistance to RootKnot Nematodes in the M-120 RNR Upland Cotton Line (Gossypiumhirsutum L.) of the Auburn 623 RNR Source,” Theoretical and Applied Genetics, Vol. 113, No. 8, 2006, pp. 1539-1549. doi:10.1007/s00122-006-0401-4
- R. B. Turley and D. L. Ferguson, “Changes in Ovule Proteins during Early Fiber Development in a Normal and Fiberless Lines of Cotton (Gossypium hirsutum L.),” Journal of Plant Physiology, Vol. 149, No. 6, 1996, pp. 695- 702. doi:10.1016/S0176-1617(96)80094-0
- J. R. Rong, G. J. Pierce, V. N. Waghmore, C. J. Rogers, A. De Sai, P. W. Chee, O. L. May, J. R. Gannaway, J. F. Wendel, T. A. Wilkins and A. H. Paterson, “Genetic Mapping and Comparative Analysis of Seven Mutants Related to Seed Fiber Development in Cotton,” Theoretical and Applied Genetics, Vol. 111, No. 6, 2005, pp. 1137-1146. doi:10.1007/s00122-005-0041-0
- E. Bechere, J. C. Boykin and W. R. Meredith, “Evaluation of Cotton Genotypes for Ginning Energy and Ginning Rate,” Journal of Cotton Science, Vol. 15, 2011, pp. 11-21.
- X. M. Du, J. J. Pan, R. H. Wang, T. Zh. Zhang and Y. Zh. Shi, “Genetic Analysis of Presence and Absence of Lint and Fuzz in Cotton,” Plant Breeding, Vol. 120, No. 6, 2001, pp. 519-522. doi:10.1046/j.1439-0523.2001.00643.x
- J. E. Endrizzi and D. T. Ray, “Monosomic and Monotelodisomic Analysis of 34 Mutant Loci in Cotton,” Journal of Heredity, Vol. 82, No. 1, 1991, pp. 53-57.
- P. J. Samora, D. M. Stelly and R. J. Kohel, “Localization and Mapping of the Le1 and Gl2 of Cotton (Gossypium hirsutum L.),” Journal of heredity, Vol. 85, No. 2, 1994, pp. 152-157.
- R. B. Turley and R. H. Kloth, “Identification of a Third Fuzzless Seed Locus in Upland Cotton (Gossypium hirsutum L.),” The Journal of Heredity, Vol. 93, No. 5, 2002, pp. 350-364. doi:10.1093/jhered/93.5.359
- N. Nadarajan and S. R. Rangasamy, “Inheritance of the Fuzzless-Lintless Character in Cotton (Gossypium hirsutum L.),” Theoretical and Applied Genetics, Vol. 75, No. 5, 1988, pp. 728-730. doi:10.1007/BF00265595
- C. A. Beasley and E. Egli, “Fiber Production in Vitro from a Conditional Fiberless Mutant of Cotton,” Developmental Biology, Vol. 57, No. 1, 1977, pp. 234-237. doi:10.1016/0012-1606(77)90371-2
- T. H. Kearney and R. J. Harrison, “Inheritance of Smooth Seed in Cotton,” Journal of Agricultural Research, Vol. 35, 1927, pp. 193-217.
- R. B. Turley and R. H. Kloth, “The Inheritance Model for the Fiberless Trait in Upland Cotton (Gossypium hirsutum L.) Line SL1-7-1: Variation on a Theme,” Euphytica, Vol. 164, No. 1, 2008, pp. 123-132. doi:10.1007/s10681-008-9670-0
- T. Z. Zhang and J. J. Pan, “Genetic Analysis of a Fuzzless-Lintless Mutant in Gossypium hirsutum L.,” Jiangsu Journal of Agricultural Science, Vol. 7, pp. 13-16.
- D. A. Musaev and M. M. Abzalov, “Some Questions Concerning the Inheritance of Fuzzy in Cotton Seeds (G. hirsutum L.),” Genetika, Vol. 8, 1972, pp. 7-16.
- J. O. Ware, L. I. Benedict and W. H. Rolfe, “A Recessive Naked-Seed Character in Upland Cotton,” Journal of Heredity, Vol. 38, No. 10, 1947, pp. 313-320.
- R. B. Turley, K. C. Vaughn and J. A. Scheffler, “Lint Development and Properties of Fifteen Fuzzless Seed Lines of Upland Cotton (Gossypium hirsutum L.),” Euphytica, Vol. 156, No. 1-2, 2007, pp. 57-65. doi:10.1007/s10681-006-9351-9
- E. Bechere, D. L. Auld and E. Hequet, “Development of ‘Naked-Tufted’ Seed Coat Mutant for Potential Use in Cotton Production,” Euphytica, Vol. 167, No. 3, 2009, pp. 333-339. doi:10.1007/s10681-009-9890-y
- A. E. Percival, “The National Collection of Gossypium Germplasm,” Southern Cooperative Series Bulletin 321, Department of Agricultural Communication, 1987.
- R. G. Percy and R. J. Kohel, “Qualitative Genetics,” In: C. W. Smith and J. J. Cothren, Eds., Cotton: Origin, History, Technology, and Production, John Wiley & Sons, New York, 1999, pp. 329-360.
- J. O. Ware, “Relation of Fuzz Pattern to Lint in an Upland Cotton Cross,” Journal of Heredity, Vol. 31, No. 11, 1940, pp. 489-498.

