American Journal of Plant Sciences
Vol.3 No.4(2012), Article ID:18721,9 pages DOI:10.4236/ajps.2012.34063
Application of Marker Assisted Backcrossing to Introgress the Submergence Tolerance QTL SUB1 into the Vietnam Elite Rice Variety-AS996
![]()
1Agricultural Genetics Institute, Tu Liem, Vietnam; 2Cuu Long River Delta Rice Research Institute, Tan Thanh, Vietnam; 3International Rice Research Institute, Metro Manila, Philippine.
Email: *cucchi04@gmail.com, LHHAM@agi.ac.vn
Received December 20th, 2011; revised January 19th, 2012; accepted February 20th, 2012
Keywords: AS996-SUB1; Climate Change; Marker-Assisted Backcrossing (MABC); Rice; Submergence
ABSTRACT
The result of the study contributes to enhancing and sustaining future livelihoods and food security in Vietnam vis-a-vis climate change. An innovative strategy based on marker-assisted backcrossing (MABC) was used to transform popular rice variety AS996 into the one can tolerate submergence while maintaining its original characteristics preferred by farmers and consumers. The submergence tolerance QTL SUB1 counts for up to 70% of the submergence tolerant and provides a marked improvement of submergence tolerance in all genetic backgrounds and environments tested so far. Parental diversity was carried out with 460 markers. Of which, 53 polymorphic markers were used for assessment on BC1F1, BC2F1 and BC3F1 generations. The best BC1F1 plant was P422 with highest recipient allele was 87.5%, while the additional chosen plants were P412, P428, P215 and P39 (81% - 84%). All these plants were used to develop BC2F1 generation. The six BC2F1 plants were used to develop BC3F1 and BC2F2 were the plants number P422-11 and P422-14 having 93.75% recipient alleles and P422-12, P422-3, P39-17, P39-25 having 92.25% recipient alleles. Total of 445 BC3F1 plants were confirmed the introgresion of SUB1 using ART5 and SC3. After three generations of backcrossing, application of MABC resulted in the best BC3F1 individual P422-14-177 with 100% of recipient alleles based on the number of 53 markers used with only the introgression size of SUB1 was 0.3 Mb between ART5 and SC3. Phenotyping was carried out on BC3F1 and BC2F2 of the selected lines. The survival ratio of these selected lines and IR64SUB1 were the same. It convinced the successfully introgress SUB1 into AS996 rice variety. The breeding line BC4F1 having 100% genetic background of donor variety is ready for develop new submergence tolerant rice variety ASS996-SUB1 to cope with climate change.
1. Introduction
Vietnam is among the five countries hardest hit by climate change. The frequency of natural disasters like typhoon, flooding etc… are increasing annually. Submergence caused by flash-flooding is one of the most serious impacts to the agricultural productions of the country. In Vietnam, rice is the number one agricultural product, not only for second largest exporter but also accounts for 90% of national food production. Of the 4.5 million ha rice area, more than one million ha is affected by flooding, remains for at least one to two weeks in many parts of the country. Depending on the timing of flood with respect to growth stage, shorter duration and shallow flashfloods can result in less than 10% production losses while deeper and stagnant water with two weeks’ duration and >100 cm in depth can cause damage ranging from 40% to 77% [1]. Traditional varieties adapted to the submergence prone environments are low yielding due to their low tillering ability, long droopy leaves, susceptibility to lodging and poor grain quality. Development of submergence tolerant varieties is generally considered as the most effective entry point for improving productivity of rice varieties damaged from typhoon and flash flood, and it is also the cheapest option for farmers. Mackill (2006) proposed that adoption of a completely new variety could take considerable time, whereas the chances of rapid adoption of popular varieties converted through marker assisted backcrossing (MABC) were relatively higher [2].
MABC is a precise and effective method to introgress a single locus controlling a trait of interest while retaining the essential characteristics of the recurrent parent [3-5]. MABC has three main advantages over conventional backcrossing. This approach has been used with great success for ‘enhancing’ rice varieties for traits such as bacterial blight resistance gene XA21 [6], the waxy locus for grain quality and submergence tolerance SUB1 [7,8].
At the International Rice Research Institute (IRRI), a project was undertaken to transfer SUB1, a major QTL on chromosome 9 explaining almost 70% of the phenoltypic variance [9,10], into at least ten widely-grown rice cultivars in Asia. Continue with the success of the researchers in IRRI, the objective of the present study was to introgress SUB1 gene into widely accepted AS996, while maintaining all its characteristics unchanges, would assure increased production from submergence prone areas of the country.
2. Materials and Methods
2.1. Materials
The rice variety IR64 SUB1—derived submergence tolerant breeding line was used as the donor of SUB1. This variety was developed that can yield 4 ton/ha under water depth from 0.8 - 1 meter for 20 - 25 days in Vietnam conditions [11].
The recipient rice variety AS996 was widely grown in South of Vietnam. AS996 was derived from the cross IR64/Oryzarufipugon, official released in 2002. The yield potential of this variety is 8 ton/ha under optimum management. Already having most of the background advantages of IR64 variety, converting AS996 into AS996- SUB1 was a great choice.
2.2. Methods
• Breeding scheme (Figure 1)
• Data analysis The molecular weights of the different alleles were measured using Alpha Ease Fc 5.0 software. The marker data was analyzed using the software Graphical Genotyper (GGT 2.0) [12]. The homozygous recipient allele, homozygous dominant allele and heterozygous allele were scored as “A”, “B” and “H”. The percent markers homozygous for recipient parent (%A) and the percent recipient alleles including heterozygous plants (%R) were calculated.
• Screening of precision introgression lines for submergence tolerance Submergence screening was performed in the greenhouse at Agricultural Genetics Institute, Hanoi-Vietnam following standard protocols [13]. Seeds of the six PILs including AS996-SUB1 of the BC2F2 generation with parents and susceptible check IR42 were germinated in rows in 35 cm/20 cm trays with three replications. Fourteen-day-old seedlings were submerged for 14 days. The
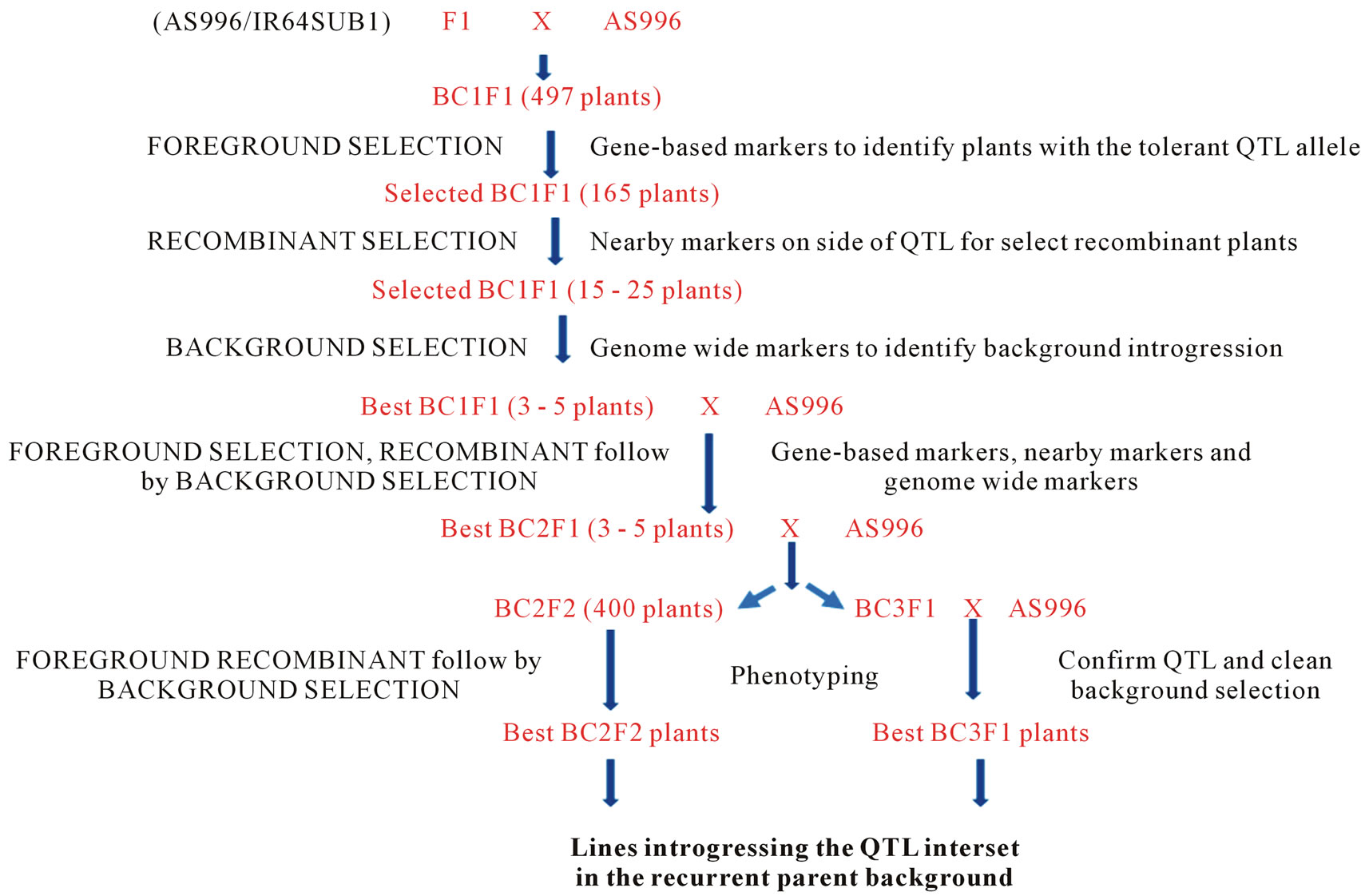
Figure 1. Breeding strategy for MABC method.
survival percentage and elongation ratio of plants were taken 21 days after de-submergence.
3. Results and Discussion
3.1. Parental SSR Polymorphism Screening
Total of 460 markers covering all the 12 chromosomes were used for the parental polymorphism screening. Of which, 53 polymorphic markers (Table 1) were used for assessment on every BC1F1, BC2F1 and BC3F1 generations.
The ratio of polymorphic marrkers were rather low, only 11.3% in this study. Figure 2 was the parental screening of polymorphic markers to use in this study.
3.2. Genotyping BC1F1 Generation
3.2.1. Foreground Selection
Of the 497 BC1F1 plants, 165 plants were heterozygous for the marker ART5 (6.3 Mb) and SC3 (6.6 Mb) tightly linked to SUB1 (Figure 3). The 165 plants with the “H” score for the tightly-linked marker were subjected to recombinant selection with flanking markers.
3.2.2. Recombinant Selection
Recombinant selection was carried out on 165 plants that were heterozygous for the marker used for foreground selection in the BC1F1 generation. Fourteen recombinants were obtained, five of them were double recombinant and the other nine single recombinants which minimized the size of the introgression size at the both distalproximal ends.
3.2.3. Background Selection
Among 460 SSR primers surveyed, 26 markers were used for selection initially on BC1F1. The maximum number of recombinant and background markers used was 8 for chromosome 9. The microsatellite markers that revealed fixed (homozygous) alleles at non-target loci in one generation were not screened in the next backcross generation. The segregants with fixed donor alleles were discarded from the selection.
From the result above, the recipient allele in 14 recombinant plants ranging from 62.5% to 87.5%. The best plant was P422, while the additional chosen plants were P412, P428, P215 and P39 with highest recipient allele was 87.5% in plant P422 (Table 2).
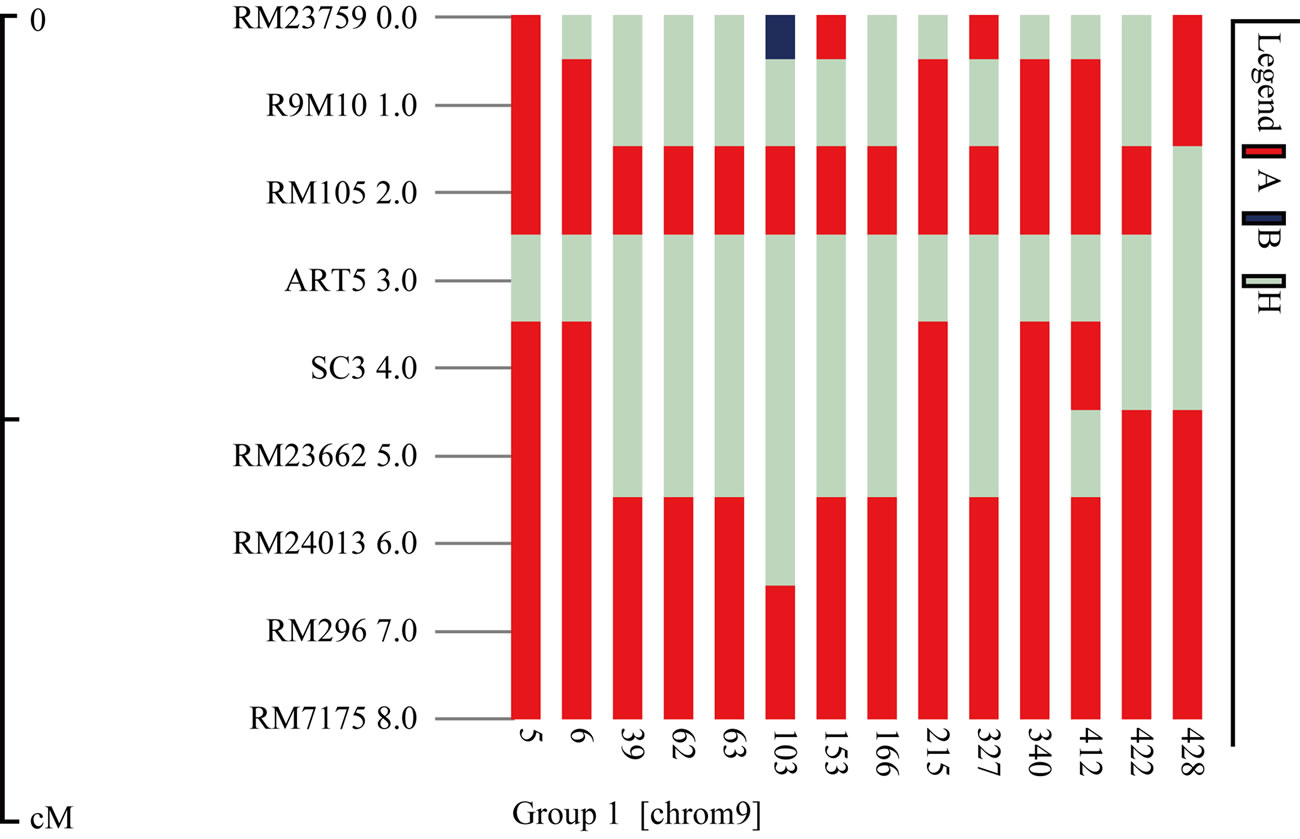
Analysing the data of 14 recombinant selected individuals in GGT 2.0 software was shown in Figures 4(a) and 4(b). All plants P422, P412, P428, P215 and P239 were used to develop BC2F1 generation.
3.3. Genotyping BC2F1 Generation
In 506 plants of BC2F1 generation, 245 plants were confirmed the introgresion of SUB1 using ART5 and SC3
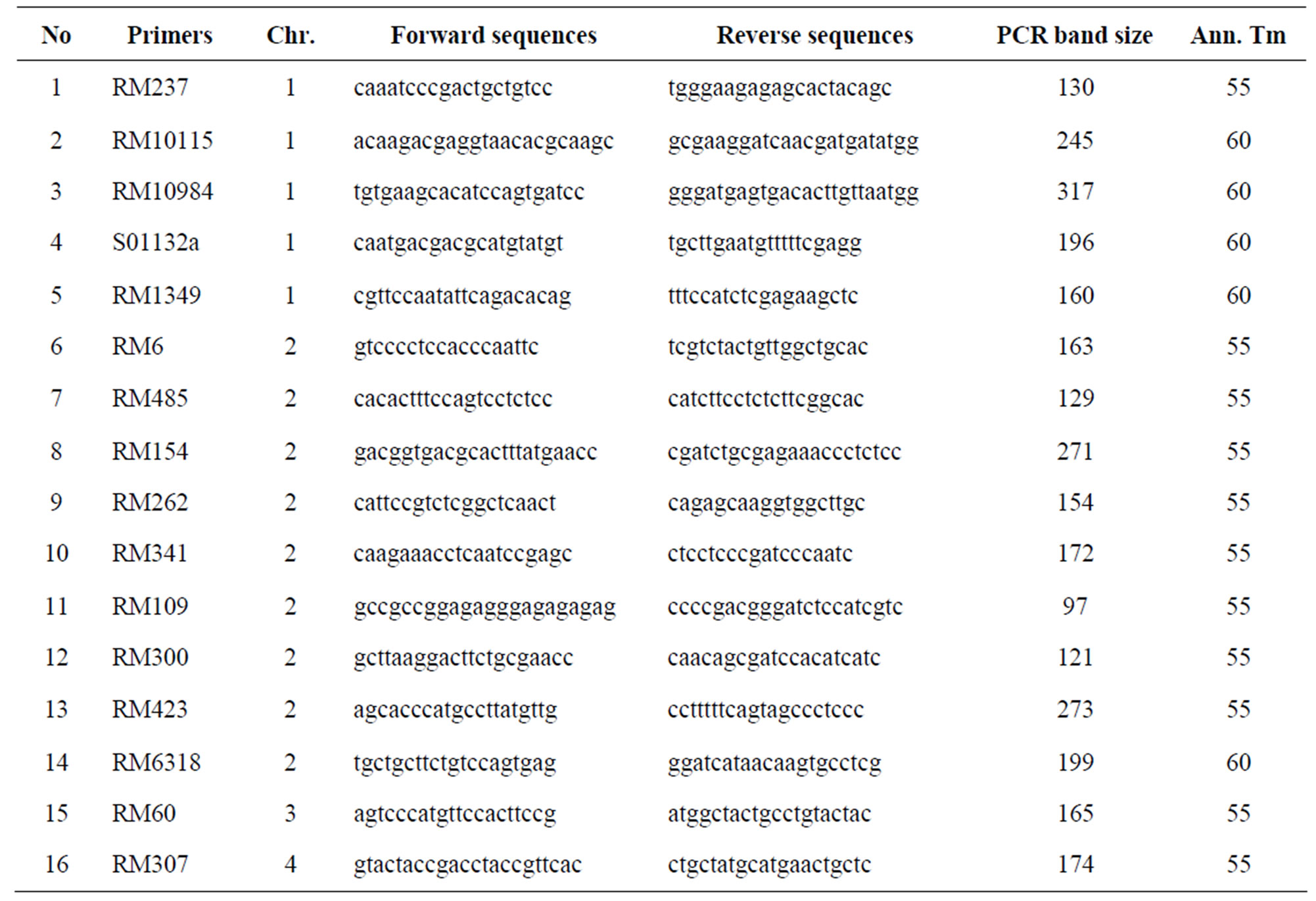
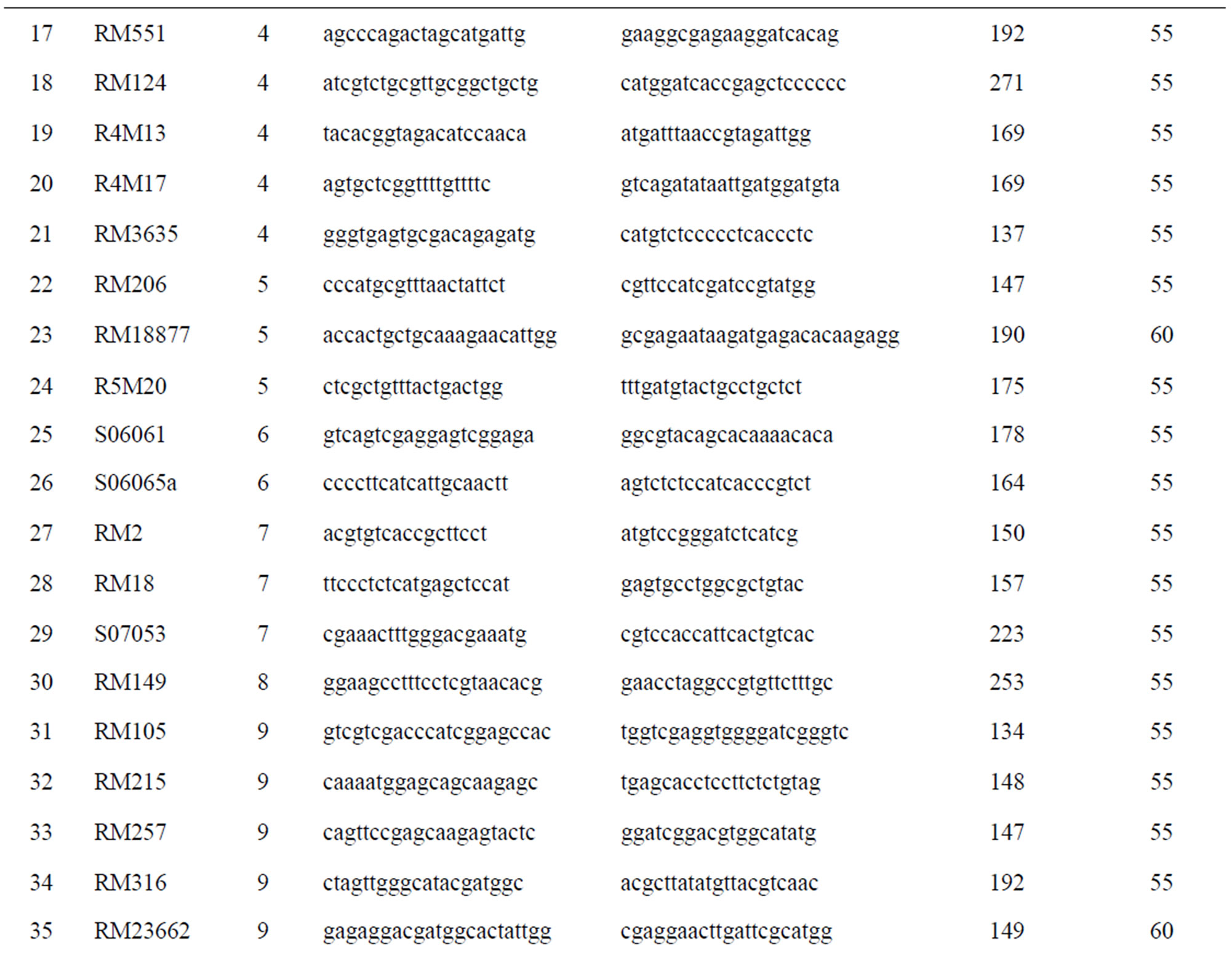

Table 1. Polymorphic markers were used for screening the BC1F1, BC2F1 and BC3F1 generations.
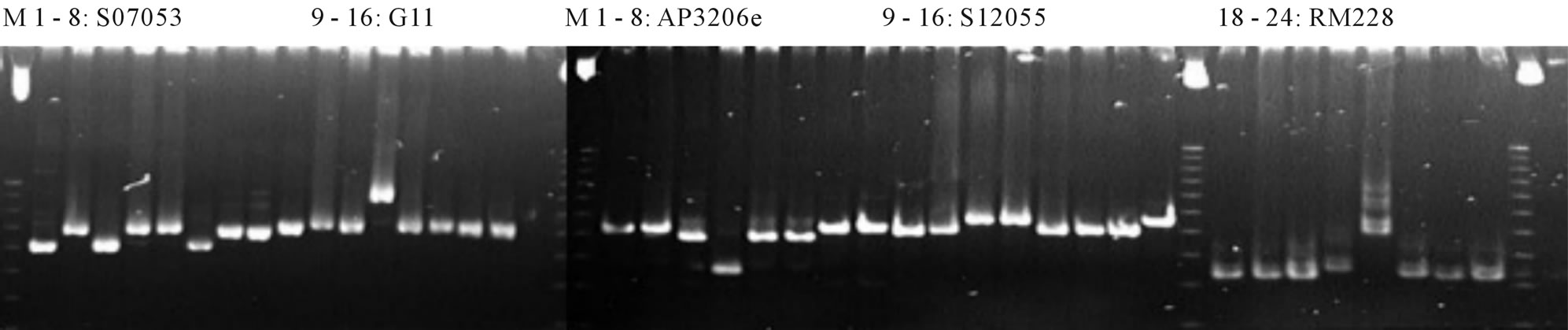
Figure 2. Parental screening on 6% polyacrylamide gel DNA: 1. Q5; 2. Q5; 3. OM5472; 4. FL478; 5. IR64SUB1; 6. AS996; 7. KDDB; 8. Bac thom. Primers: SO7053; G11; AP3206e; S12055; RM228.
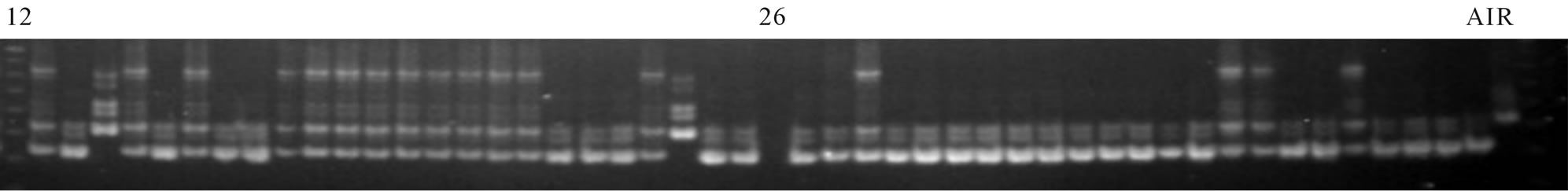
Figure 3. Screening of BC1F1 individuals (crossAS996/IR64SUB1) using marker SC3. Wells 1: 25bp marker, 2 - 25 and 27 - 48: BC1F1 individuals, Well 49: AS996, Well 50: IR64SUB1.
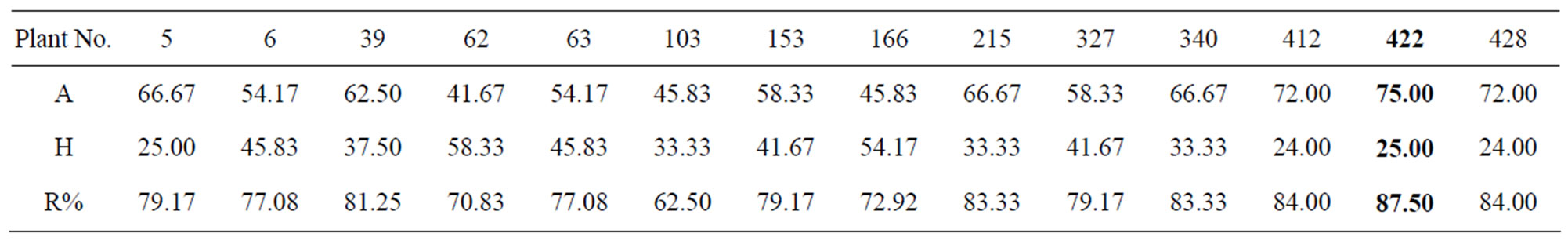
Table 2. The recippient allele of the fourteen BC1F1 recombinant plants.
(Figure 5). The recombinant selection was carried out using heterozygous flanking markers specific to the five BC2F1 populations produced from the five best plants of the BC1F1 generation.
Total of 17 double recombinant plants were used for background selection using 20 heterozygous markers. The maximum percentage of recipient alleles was 93.75% obtained in plant number P422-11 and P422-14. The minimum percentage of recipient alleles was 89.7%, which was obtained in plant number P39-80. Additionally plants were P422-12, P422-3, P39-17, P39-25 having the recipient alleles of 92.25%. Those six BC2F1 plants were used to develop BC3F1 generation.
The BC2F2 seed of the plants having SUB1 region also collected. Because of raining at the flowering time, so the ratio of unfilled grain were rather high (20% - 60% of the collected lines). Combine with conventional breeding, the breeding lines from BC2F3 and BC3F2 will be used for submergence tolerance screening on the field.
3.4. Genotyping BC3F1 Generation
Total of 445 BC3F1 plants were confirmed the introgresion of SUB1 using ART5 and SC3.
The recombinant selection was carried out on 124 plants using heterozygous flanking markers. After that, there were only 22 BC3F1 plants used for background selection with 19 markers (Figure 6). Two plants P422- 14-164 and P422-14-178 were receiving the equal recipient alleles up to 96.15% compare to AS996 original. The plant P422-14-177 was the best plants with 100% recipient alleles from AS996 with the total of 52 primers from BC3F2 seeds from those three plants will be used for screening for submergence tolerance as well as produce BC4F1 generation. Figure 7 was the analysing result from GGT2.0 software of the plant P422-14-177.
3.5. Submergence Screening
The selected BC2F2 and BC3F1 lines showed submergence tolerance just after de-submergence and after recovery against submergence stress of 21 days. The submergence tolerance scores of these BC2F2 lines were similar to those of the tolerant donor IR64SUB1. The result confirmed the introgression of SUB1 QTL in these lines. The situation of heterozygous SUB1 in BC3F1 resulted in lower survival scores than the homozygous SUB1 in BC2F2 lines.
4. Discussion
Plant adaptation to variable environments is reflected by
 (a)
(a)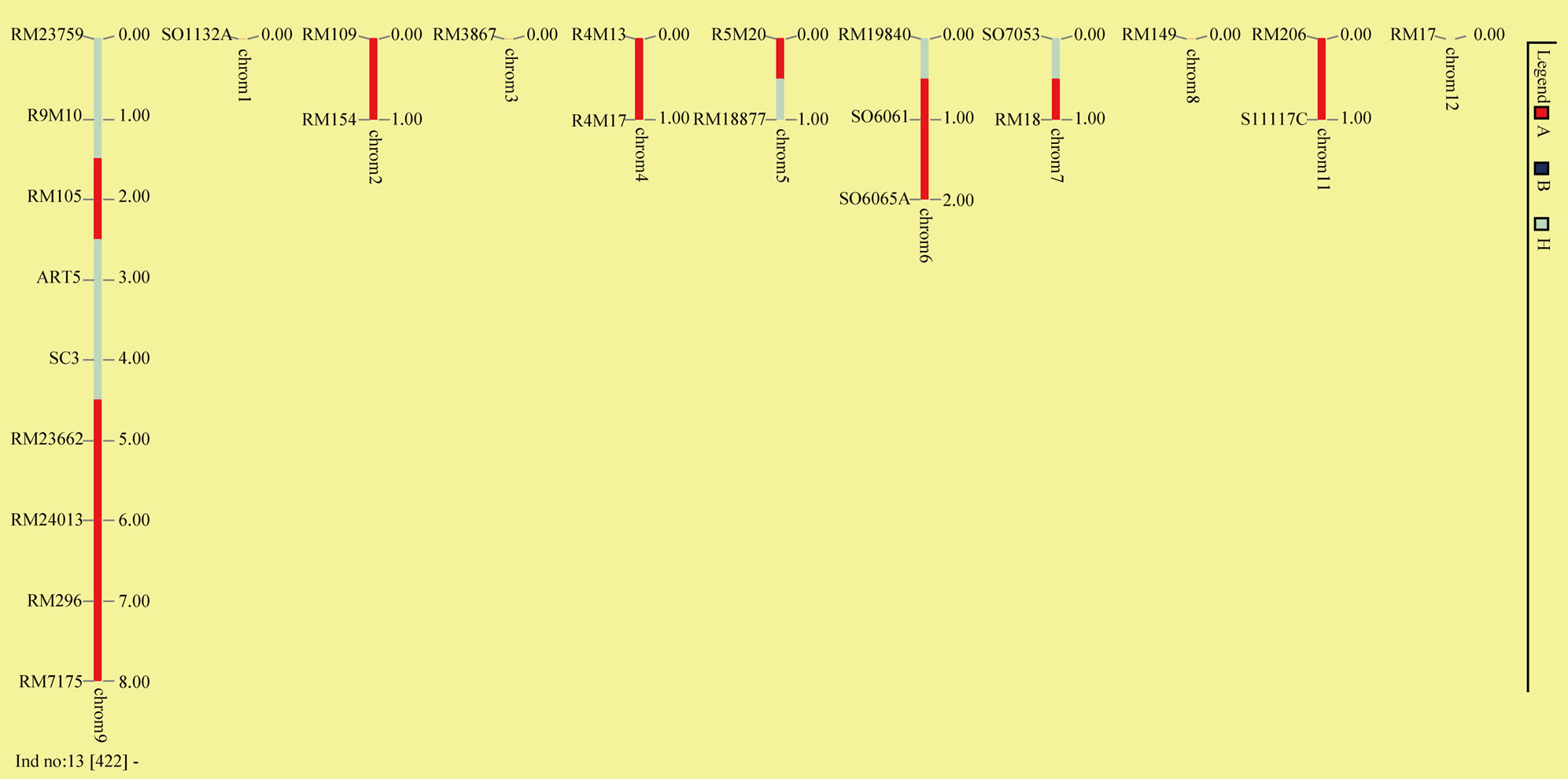 (b)
(b)
Figure 4. Graphic of fourteen recombinant plants (a) and plant BC1F1-P422 (b).
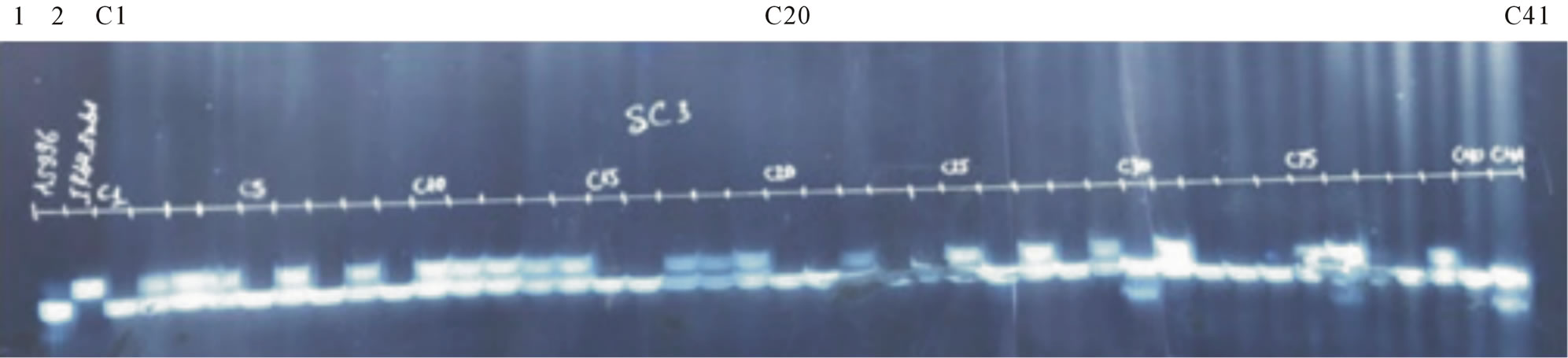
Figure 5. BC2F1 (AS996xIR64SUB1) individuals screening using primer SC3. Lane 1: AS996, Lane 2. IR64SUB1, C1-C41: BC2F1 individuals.
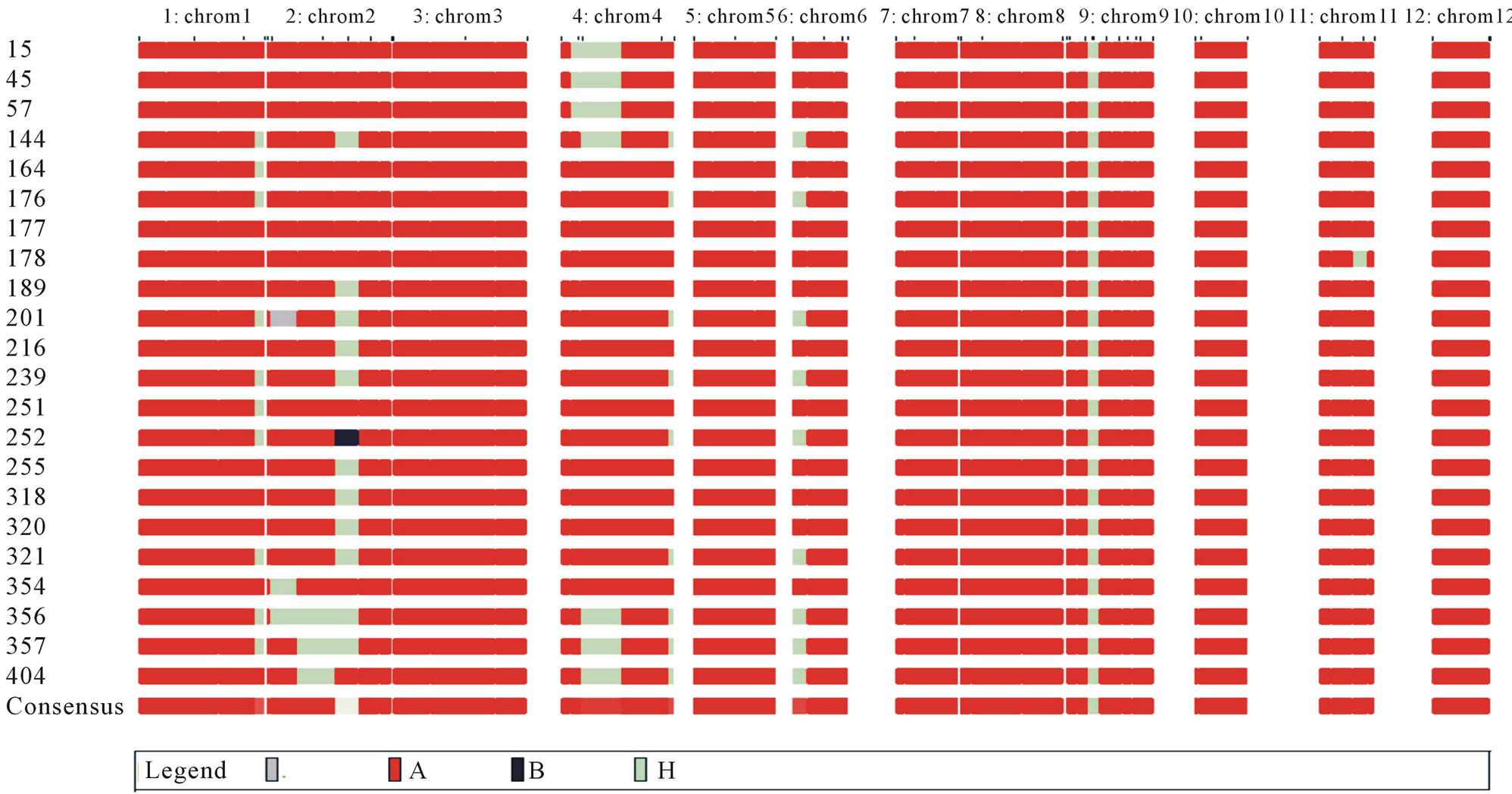
Figure 6. Data of 22 recombinant individuals of BC3F1 (AS996xIR64SUB1) were analysed using GGT3.2. Of which, individuals BC3F1 number 177 was the best candidate.
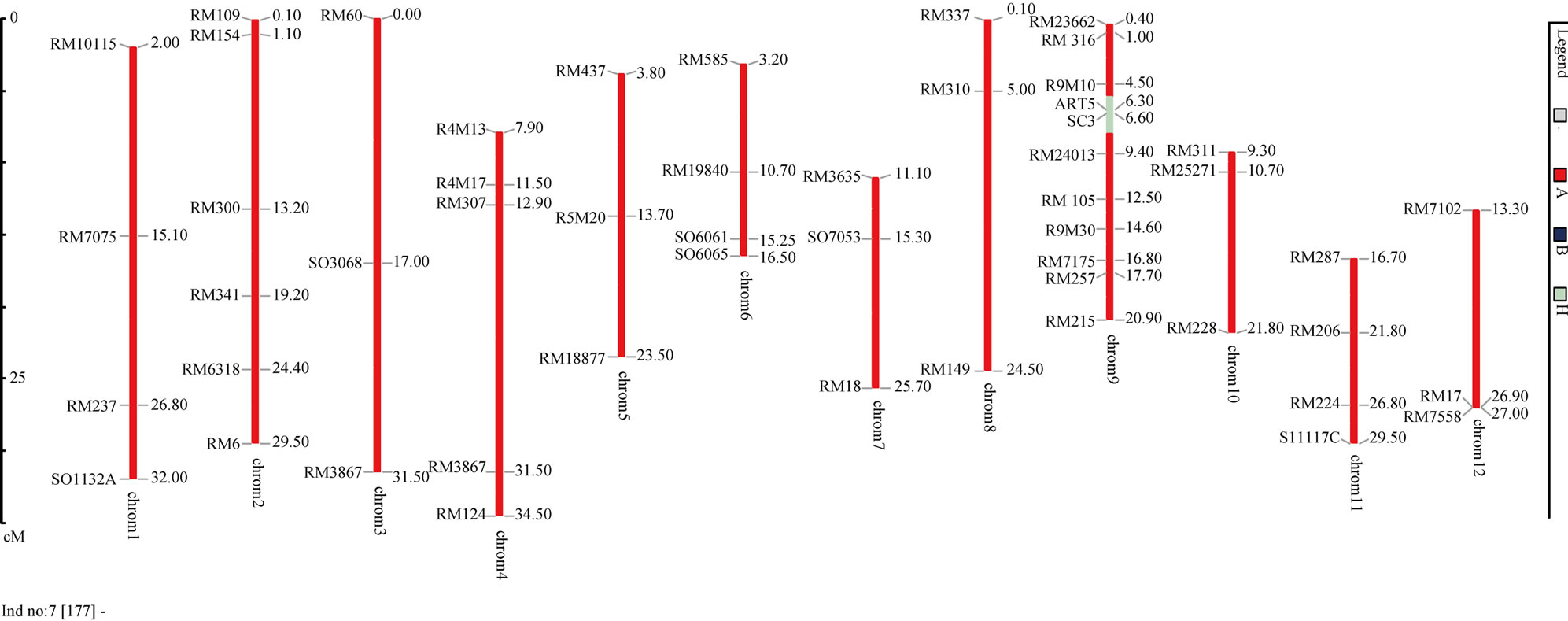
Figure 7. Plant P422-14-177 was the best plants with 100% recipient alleles from AS996. Data were analysed using GGT3.2.
an interrelated set of complex physiological and genetic strategies, we can obtain a deeper understanding of the underlying molecular mechanisms, which opens the way towards a more targeted breeding approach for higher stress tolerance in crop plants. The breakthrough that has made this approach possible was the introduction of easy-to-use DNA markers that brought QTL mapping into the mainstream, making it possible to efficiently map the genetic loci controlling complex traits. This was made possible through genetic linkage analysis, allowing the construction of linkage maps, and the identification of QTLs controlling particular traits based on statistical methods that help establish the association between molecular markers and phenotypic data. Currently, SSRs are predominantly being used to map and introgress agronomically important QTLs into popular varieties using MABC.
Because of the success of using the SUB1 QTL for molecular breeding, where a single allele at a major locus can provide submergence tolerance [7,8,14], the SUB1 QTL counts for up to 70% of the submergence tolerant and provides a marked improvement of submergence tolerance in all genetic backgrounds and environments tested so far. But also, after the MABC lines are completed using marker selection, their level of tolerance needs to be confirmed under greenhouse and field conditions before amplifying seed for large-scale testing and validation in farmers’ fields at target areas.
As discussed above, this approach is most effective when major QTLs are available for the traits of interest, and this has been the case for several abiotic stresses. Nevertheless, a similar approach could be feasible for smaller QTLs. Even a relatively small-effect QTL could have a significant impact if deployed over a large area. The MABC strategy is a relatively conservative approach aimed at getting incremental improvements in the best varieties, while plant breeders will continue to aim at the goal of new mega varieties through multiple approaches relying on conventional breeding and MAS.
Using MABC method for screening BC1F1 plants, we can know quite well about the map of the plant selected. From that result, determining the next step for selecting of the next generation is very importance.
From the result of selected BC1F1 plant, the maximum percentage of recipient alleles was 93.75%, which obtained in plant number P422-11 and P422-14 in BC2F1, following the P422-14-177 with 100% of recipient alleles based of number of 52 markers used with only the introgression size of SUB1 was 0.3 Mb between ART5 and SC3.
According to Hospital (2001) [4] flanking markers for recombinant selection should be chosen as closely linked as possible to the introgressed gene to minimize linkage drag. He suggested computing the minimal population sizes required to obtain double recombinants for such closely linked markers and to optimize the population size. The target of recombinant selection in BC1F1 generation was to obtain single recombinants at one side of the SUB1 QTL. These were segregants in which the size of the donor segment was minimized at one side of the QTL. In this study, with a large number of plants with eight flanking markers from the SUB1 region of chromosome 9 were used in BC1F1 generation. For the development of AS996-SUB1, RM24013 (9.4 Mb) was used as flanking marker at proximal end and RM105 (5 Mb) at distal end. Foreground selection was confirmed by the use of markers from the SUB1 genes ART5 (6.3 Mb) and SC3 (6.6 Mb) in all the plants. The used of two precise primers located in the SUB1 region absolutely resulted in minimized the introgression size of the SUB1 in AS996 variety. It also reduced the crossing over between the gene region, which decreased the false-positive results in foreground selection. The SUB1 introgression gene will be used for further assessment in the BC4F1 and BC3F2 generations.
5. Conclusion
In conclusion, the MABC approach described in the present study was successfully adopted to introgress SUB1 into AS996 and could be successfully utilized to introgress not only SUB1 in the other important varieties with a minimum introgression segment and within a short time frame. By limiting the size of the introgression, the chance of introducing donor genes that might change the essential characteristics of this popular variety was reduced. This will become increasingly important as other desirable genes are introduced into AS996-SUB1. It is expected that the newly developed SUB1 lines will be able to increase rice production in the submergence areas of Vietnam.
6. Acknowledgements
We would like to thank Danish Ministry of Foreign Affair and DANIDA Fellowship (Project code: 09-P01-VIE) for supporting this work. Thank was send to staff in Cuu Long River Delta Rice Research Institute for field work. We also thank the staff of the International Rice Research Institute (IRRI), Los Baños, Laguna, Philippines, for their colaboration in this research.
REFERENCES
- D. O. Manzanilla, T. R. Paris, G. V. Vergara, A. M. Ismail, S. Pandey, R. V. Labios, G. T. Tatlonghari, R. D. Acda, T. T. N. Chi, K. Duoangsila, I. Siliphouthone, M. O. A. Manikmas and D. J. Mackill, “Submergence Risks and Farmers’ Preferences: Implications for Breeding SUB1 Rice in Southeast Asia,” Agricultural Systems, Vol. 104, No. 4, 2011, pp. 353-347. doi:10.1016/j.agsy.2010.12.005
- D. J. Mackill, “Breeding for Resistance to Abiotic Stresses in Rice: The Value of Quantitative Trait Loci,” In: K. R. Lamkey and M. Lee, Eds., Plant Breeding: The Arnel R Hallauer International Symposium, Blackwell.
- B. C. Y. Collard and D. J. Mackill, “Marker-Assisted Selection: An Approach for Precision Plant Breeding in the 21st Century,” Philosophical Transactions of the Royal Society, Vol. 363, No. 1491, 2008, pp. 557-572. doi:10.1098/rstb.2007.2170
- F. Hospital, “Size of Donor Chromosome Segments around Introgressed Loci and Reduction of Linkage Drag in Markerassisted Backcross Programs,” Genetics, Vol. 158, No. 3, 2001, pp. 1363-1379.
- F. Hospital and A. Charcosset, “Marker-Assisted Introgression of Quantitative Trait Loci,” Genetics, Vol. 147, No. 3, 1997, pp. 1469-1485.
- S. Chen, X. H. Lin, C. G. Xu and Q. F. Zhang, “Improvement of Bacterial Blight Resistance of ‘Minghui 63’, an Eliteestorer Line of Hybrid Rice, by Molecular Marker-Assisted Selection,” Crop Science, Vol. 40, No. 1, 2000, pp. 239-244. doi:10.2135/cropsci2000.401239x
- C. N. Neeraja, R. Maghirang-Rodriguez, A. Pamplona, S. Heuer, B. C. Y. Collard, E. M. Septiningsih, G. Vergara, D. Sanchez, K. Xu, A. M. Ismail and D. J. Mackill, “A Marker-Assisted Backcross Approach for Developing Submergence-Tolerant Rice Cultivars,” Theoretical and Applied Genetics, Vol. 115, No. 6, 2007, pp. 767-776. doi:10.1007/s00122-007-0607-0
- E. M. Septiningsih, A. M. Pamplona, D. L. Sanchez, C. N. Neeraja, G. V. Vergara, S. Heuer, A. M. Ismail and D. J. Mackill, “Development of Submergence Tolerant Rice Cultivars: The SUB1 Locus and Beyond,” Annals of Botany, Vol. 103, No. 2, 2009, pp. 151-160. doi:10.1093/aob/mcn206
- K. Xu and D. J. Mackill, “A Major Locus for Submergence Tolerance Mapped on Rice Chromosome 9,” Molecular Breeding, Vol. 2, No. 3, 1996, pp. 219-224. doi:10.1007/BF00564199
- K. Xu, X. Xia, T. Fukao, P. Canlas, R. Maghirang-Rodriguez, S. Heuer, A. M. Ismail, J. Bailey-Serres, P. C. Ronald and D. J. Mackill, “SUB1A Is an Ethylene Response Factor-Like Gene that Confers Submergence Tolerance to Rice,” Nature, Vol. 442, No. 7103, 2006, pp. 705-708. doi:10.1038/nature04920
- N. T. Lang , T. T. Luy, T. T. Binh, T. D. Thuan, T. Van Canh, N. Van Bang, R. Labios, B. C. Buu, A. Ismail and D. Mackill, “Evaluation Methodology and Utilization Submergence Gene in Southern of Vietnam,” Omonrice, Vol. 17, 2010, pp. 41-53.
- R. Van Berloo, “GGT 2.0: Versatile Software for Visualization and Analysis of Genetic Data,” Journal of Heredity, Vol. 99, No. 2, 2008, pp. 232-236. doi:10.1093/jhered/esm109
- K. Xu, X. Xu, P. C. Ronald and D. J. Mackill, “A HighResolution Linkage Map in the Vicinity of the Rice Submergence Tolerance Locus SUB1,” Molecular and General Genetics, Vol. 263, No. 4, 2000, pp. 681-689. doi:10.1007/s004380051217
- M. J. Thomson, M. D. Ocampo, J. Egdane, M. A. Rahman, A. G. Sajise, D. L. Adorada, E. Tumimbang-Raiz, E. Blumwald, Z. I. Seraj, R. K. Singh, G. B. Gregorio and A. M. Ismail, “Characterizing the Saltol Quantitative Trait Locus for Salinity Tolerance in Rice,” Rice, Vol. 3, No. 2-3, 2010, pp. 148-160. doi:10.1007/s12284-010-9053-8
NOTES
*Corresponding author.

