Open Journal of Medical Microbiology
Vol. 3 No. 1 (2013) , Article ID: 28694 , 7 pages DOI:10.4236/ojmm.2013.31013
Bovine Leukemia Virus Gene Segment Detected in Human Breast Tissue
1Environmental and Applied Science University, UDCA, Bogotá, Colombia
2Microbiology Departament, Pontificia Universidad Javeriana, Bogotá, Colombia
3Pathology Departament, Pontificia Universidad Javeriana, Bogotá, Colombia
Email: *mfgutier@javeriana.edu.co
Received November 20, 2012; revised December 25, 2012; accepted January 11, 2013
Keywords: Breast cancer; Bovine Leukemia Virus (BLV)
ABSTRACT
Background: Bovine Leukemia Virus (BLV) is known by infections in bovine cattle and produce, in 30% of infected animals, persistent lymphocytosis significantly impacts the beef industry. It has been proposed that this virus could be transmitted to humans and be present in cases of breast cancer. Aim: to determine the presence of 380 bp of gag gene segment of BLV in paraffin-embedded breast tissue. Study Design: Control-case study. Methodology: 106 tissue samples were collected. 53 were cancer positive samples and 53 were negative samples for this pathology. After dewaxing tissues, DNA was extracted, amplified and sequenced. Phylogenetic analysis was done in order to verify BLV gene segment, presence and origin. Results: 43 samples were positive (40.5%) for BLV segment. In the case group this segment was found in 35.8% of the samples and in the control group, BLV presented in 45.2% of the samples. Phylogenetic analysis confirmed BLV presence and had shown a high homology between amplified gene sequences obtained from human breast tissues and those coming from bovine cattle with leukosis reported by GenBank. Conclusion: The presence of BLV genes in humans and its location in breast tissue can be confirmed, however, it should be clarified as a possible promoter of malignancy processes on this tissue.
1. Introduction
Bovine Leukemia Virus (BLV) is known worldwide by infecting bovine cattle affecting in a significant way the bovine industry. Its prevalence varies between countries, and with reports of 60% throughout the world [1]: 28% in Japan [2], 33% in Argentina [3] and 45.8% in Colombia [4]. A recent study in Japan showed that there is more BLV in dairy cattle than in beef cattle [5]. It presents on infected animals and it is associated with a persistent lymphocytosis characterized by the nonmalignant polyclonal expansion of B-cells. From these animals, 5% of them develops a leukemia or a lymphoma [6,7]. Besides of infecting cattle, some other authors have proposed the possibility of transmission to other species, including humans [8].
This virus is member of the genus Deltaretrovirus, in the retroviridae family, the BLV genome has two identical RNA positive single strains, and it carries the reverse transcriptase enzyme, and also other retroviruses such as Murine Leukemia Virus (MMLV), Simian T-cell Leukemia Virus (STLV) and Human T-cell Lymphotropic Virus (HTLV 1), it has been related with some forms of cancer. Its molecular structure, is composed of three principal genes: env, gag and pol which encode the viral envelope, structural proteins and the polymerase respectively; and two other genes of a smaller size, rex and tax, in which the last one is associated with viral regulation. [9,10]
The mechanism by which this virus is associated with cancer in cattle is not clear at all. Some time ago, it was proposed that BLV did not have oncogenes, it was not found a place of insertion into the cell genome and was proposed that the provirus did not have a replicative cycle in tumor cells [6]. Furthermore, Murakami et al., showed that this provirus is not inserted in a particular chromosome, but it does in introns located far away from transcription factors starters [7]. Otherwise, Takahashi states that Tax protein cooperates with a viral oncoprotein to generate transformation in a mouse cell line, suggesting that in addition of having a regulatory function in the viral replication, it could induce leukemia in bovine cattle infected with BLV [9].
Breast cancer is one of the most frequent pathologies in human beings. In United States in 2009, 211,731 women were diagnosed with breast cancer and 40,676 women died of this disease and in Colombia each year are diagnosed nearly 7000 women with this pathology and are reported 2500 deaths [11,12]. Accepting the assumption that cancer etiology is multifactorial [13], and with some important points such as that, some viruses have been related with several kinds of cancer, that it has been found antibodies against BLV in people with breast cancer, that already exists in some tests for detecting other retroviruses (MMTV) in breast cancer tissue [14,15] and taking into account that in a previous study developed by our research group was reported the presence of gp51 protein in breast cancer tissue[16], the aim of this study was to continue showing the presence of the virus in the indicated tissue. To accomplish this objective, it determined the presence of a BLV gene segment, specifically looking for a 380 bp segment of gag gene.
2. Materials and Methods
2.1. Study Design
The study was done by using retrospective case-control design, in which human breast tissue segments embedded in paraffin and stored at the Pathology Laboratory in the San Ignacio Hospital (Bogota, Colombia) since 2006 were analyzed. The study included a total of 106 samples obtained from women aged between 18 and 80 years, this was a case group made up by samples with presumptive cancer diagnosis and confirmed cancer by pathological study; while in the control group were consider samples excluding cancer. As inclusion criterion, it was taken into account samples with enough cellularity to be able to extract the DNA. All selected samples were confirmed by pathological diagnosis and the patient’s age was confirmed as well; these data were analyzed as risk variables. The samples number was calculated based on the comparison of two proportions formula, considering a 90% of confidence level.
Statistical analysis determined that significancy was done by a bivariate analysis, in which Odds ratios (OR) were obtained and confidence interval (CI) with an error of 5%.
2.2. Paraffin-Embedded Breast Tissue DNA Extraction
Before DNA extraction, paraffin was removed. Briefly, tissue cuts of approximately 0.02 g and were placed in 500 mL of Xylol and centrifuged at 10,000 rpm per 7 minutes, to remove excess of paraffin and xylol. Pellet washes were initially performed only with xylol and then with 70% ethanol. When tissues were completely out of paraffin, they were resuspended in 50 mL of HPLC water and TNE buffer (17 mM Tris HCl pH 8; 50 mM NaCl; 7 mM EDTA).
DNA extraction was performed by “Salting Out” method. Lysis buffer (10 mM Tris HCl pH 8.0; 0.5% SDS; 5 mM EDTA) was added to samples and 10 mL of proteinase K (N.C. 95H014715 C.C. SIGMA) (20 mg/ mL), leaving in incubation and shaking at 55˚C overnight. After incubation was done, 500 mL of 8M ammonium acetate and 1mM EDTA were added, samples were centrifuged, then cold isopropanol was added and again centrifuged at 10000 rpm per 15 min. DNA pellet was washed in 70% ethanol, centrifuged, and supernatant was discarded. DNA was dried out at 50˚C per 15 min and then was resuspended in 30 mL of TE 1X.
2.3. BLV Gag Gene Fragment Amplification Isolated from Breast Tissue
The segment of 380 bp was amplified. It is located within the gag gene of VLB and it is considered highly conserved gene. This segment has been reported by Dus Santos et al, who proved to be a useful segment in terms of sensitivity and specificity for this type of study. Moreover, this segment is used in export processes to determine and certify VLB free bovine semen [17]
The 380 bp fragment was amplified using a PCR InvitrogenÒ kit and a pair of primers synthesized by InvitrogenÒ (19327211), Primer Forward (PF; 1068-1088 AAC ACTACGACTTGCAATCC) and Primer Reverse (PR; 1434-1453 GGTTCCTTAGGACTCCGTCG), obtained from standardized protocol for detection of BLV in bull semen samples. PCR conditions were the same as previously reported [17]. As a positive control, a bovine DNA from lymphocytes was used, with clinical and serological diagnosis of BLV.
To evaluate the DNA efficiency obtained through Salting Out method, a PCR test was designed to amplify a 600 bp fragment of the human constitutive gene encoding the glyceraldehyde phosphate dehydrogenase (GAPDH) enzyme. PCR InvitrogenÒ kit (C.N. 10966- 020) was used to amplify the desire fragment.
2.4. Phylogenetic Analysis
BLV positive samples obtained from PCR amplifications and with enough DNA were sent to Macrogen Inc for sequencing. Sequences were edited using CLC sequence viewer 6.0 program obtaining consensus sequences by ambisense alignments of each of them. Comparisons with previously reported sequences by GenBank were done by local alignments using the program BLAST for the National Center for Biotechnology Information, looking for similarities with BLV sequences registered in GenBank (see figure 1). Finally, global alignments were done using MEGA 5 program, in which were included BLV sequences obtained in the present study, GenBank
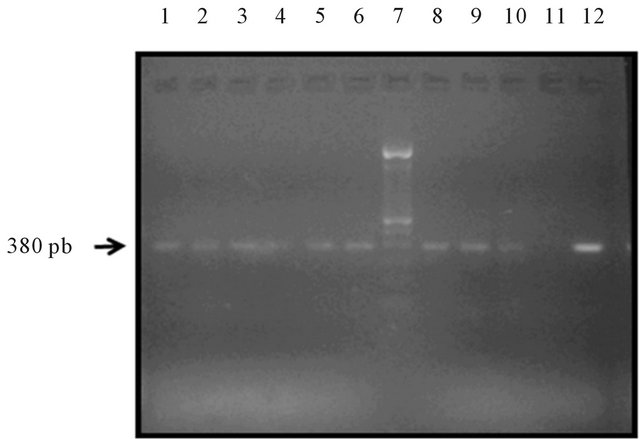
Figure 1. Electrophoresis of VLB segment. Lanes 1, 2, 3, 4, 5, 6, 8, 9 and 10 belonged to samples. They show a band of 380 bp (according to the weight marker located in lane 7) Lane 11 corresponds to the negative control and positive control lane 12.
BLV sequences and other human and animal retroviruses sequences such as MMLV. With these alignments was done a phylogram using Kimura 2 Parameters—K2P method.
3. Results and Discussion
3.1. BLV Gag Gene Fragment Detection in Paraffin-Embedded Breast Tissue Samples
From the total 106 samples collected, 53 belonged to women with breast cancer and other 53 were negative samples to malignancy. Cellular DNA extracted from the paraffin blocks determined the DNA concentration. In the study were only included samples with DNA concentrations between 5 and 4500 ng/L. Regarding DNA purity, averages obtained in the 260/280 index and 260/230 index were 1.92 and 1.9 respectively. To probe DNA quality and absence of PCR inhibitors, GAPDH gene was amplified to all analyzed samples. With these results, it was confirmed DNA quality after dewax process. Once purity and quality were verified, samples were able for BLV 380 bp fragment detection (Figure 1). 19 of 53 positive breast tissue cancer samples (35%) and 24 of 53 negative breast tissue cancer samples (45%) were positive to the amplification of 380 bp gag segment (Table 1). As a positive control was used DNA obtained from blood leucocytes of a bovine with clinical and serological diagnosis of Bovine Leukemia, and as negative controls, DNA used was obtained from healthy bovine paraffin-embedded tissue (gently supplied by the Pathology Lab at U.D.C.A), healthy bovine blood leucocytes and healthy women leucocytes.3.2. Socio-Demographic Characteristics
From all processed samples, 43 were positive for BLV
Table 1. BLV presence in breast tissue with and without cance.

(40.5%) of whom in the case group BLV presence was detected in 35% of samples (19/53) and in the control group was detected in 45% (24/53) p = 0.32. The average age of patients with breast cancer was 46.4 years and for women without breast cancer was 52.2 years.
Respecting to pathological diagnosis, samples were grouped according to their pathology, data shown in table 2 were for malignancies diagnosis and table 3 were for non-malignancies diagnosis.
3.3. Bivariate Analysis
To analyze the risk related with age and type of pathology associated with the presence of BLV and cancer, samples were distributed into 4 ages groups (see table 4), where it was found the presence and absence of BLV and cancer according to each age group.
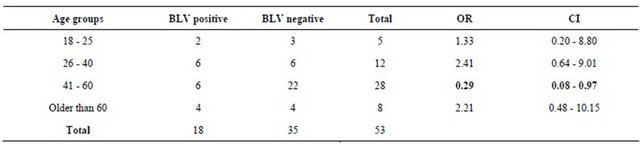
There were no significant differences related with the presence of BLV and breast cancer, due to the value obtained of the OR that was 0.62 and the CI was of 0.28 - 1.35. For the age variable, bivariate analysis was done comparing each age group with the other three groups. In table 4, it can be seen that the group of women between 41 and 60 years with breast cancer has the biggest risk of been infected with BLV compared with the other groups (OR: 0.29; CI: 0.08 - 0.97).
This analysis was performed likewise to non-pathological breast tissue, looking for associations between age groups and BLV presence, as well as association with the most prevalent pathologies (ductal carcinoma and fibroadenoma) and the BLV presence but no significant results were found in any of the evaluated conditions.
3.4. Phylogenetic Analysis
From all the samples confirmed for the 380 bp segment of BLV, only 26 could be sequenced and sent to GenBank (accession numbers from JQ480623 to JQ480649). These 26 sequences were used to build the phylogram, 13 coming from cases group and other 13 from control group; and 23 other sequences obtained from GenBank (NCBI) corresponding to the same 380 bp gene segment of the bovine origin BLV, HTLV, HIV, HBV and MMTV.
Table 2. Distribution of BLV in breast positive samples. Notice that most common cancer found was ductal carcinoma with 77.4%.
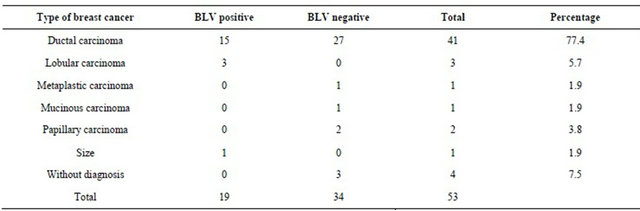
Table 3. Distribution of BLV in negative breast cancer tissue. Notice that most common disease was fibroadenoma with 56.6%.
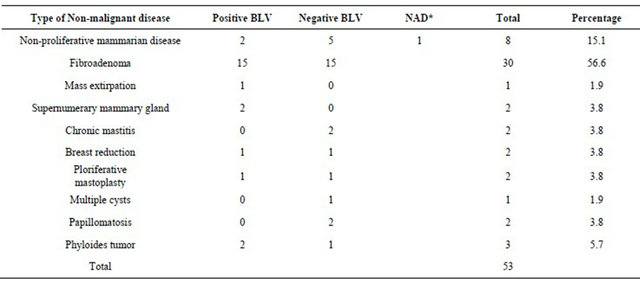
Table 4. Distribution of BLV in positive breast cancer tissue associated with women’ age group. Bivariate analysis taking as parameter the comparison of the different age groups.
All BLV sequences, both bovine previously reported in GenBank and those isolated from human breast tissue cancer showed an identity between 93% - 99%. This result was not the same when included other human virus sequences or other viral species.
Figure 2 shows the tree topology with 3 principal clusters, first one made up by the sequences isolated during the present study, which are closely interrelated with 9 BLV sequences coming from a bovine source. Bootstrap value was 99. The second cluster was made of HTLV1, HTLV2, HBV and MMTV segments; and the third one, clearly away from the other two arms that was

Figure 2. Phylogram built with a 380 bp segment from gag gene of bovine and human BLV, HTLV1-2, HBR and MMTV. The sequences isolated during the study are reported in GenBank with accession numbers from JQ486623 to JQ486649. In the phylogram appears the last 2 numbers of each registered sample in GenBank.
organized with HIV strains.
Each year, only in the United States are reported more than 170000 breast cancer cases, which is enough reason to look for the risk factors associated with origins and development of this pathology.
To confirm that the segment is present only in the BLV gag gen, alignments were performed with BLAST and homologies between 95 to 100% were found only with BLV genome There are few existing reports about BLV and other animal retroviruses present in humans. In 1995, Wang et al., reported the presence of MMLV segments in cases of breast cancer, after this, Hughes and coworkers proposed again this relation, despite having not found the virus by electronic microscopy but having detected the reverse transcriptase activity in some of the probed samples [17]. In 2002, Melana in Argentina reported gene segments of the same virus (MMLV) in 38% of breast cancer samples evaluated as well as in some cases in leukocytes [14]. These findings were then confirmed by Ford et al. Who found MMTV in the 40% of breast cancer tissues analyzed and in 2% of healthy breast tissues proposing an association between the virus and breast cancer [15]. Meanwhile, Michael Bock and Jonathan P. Stoye (2000), demonstrated endogenous retroviruses in germinal human cells genome and the possible association of them with the development of some cancers by the mutation done by the insertion of the viral genome in the human cells [18]. For the year 2003 Buering et al., reported antibodies against p24 protein of BLV in 74% of human sera [19] and in the same year, Jehoon Lee and coworkers presented an article in which they did not find BLV in leukemia nor human lymphomas even though it was argued that food workers, especially with meat, have a higher risk of developing a myeloid leukemia, Hodgkin’s lymphoma or lung cancer [20].
Finding BLV in 40.5% of breast tissues was a relevant result. Now, having the presence of the virus in 35% (19/53) of positive breast cancer tissues and 45% (24/53) in negative tissues for this pathology, questioned whether the presence of BLV could be a protective factor for cancer developing, which was not confirmed in the statistical analysis, giving way to other questions such as which is the probability that over time, these tissues without actual malignancy but BLV positives, could begin a neoplastic process and develop a breast cancer; which is the process of adaptation for a bovine virus into human tissue; which adaptative mutations have undergone the virus to be able to integrate with the human genome, and once there, if virus behaves in an infectious state, can complete successfully its replicative cycle or stay dormant inside the host. When answers to all of these questions are solved, it is important to determine in which other tissues this virus could be found, how does it behave at the immunological system and which is its role as an etiological cancer agent.
Trying to give an answer of which was the way of access of a bovine virus into human, some reports have proposed that this could be through milk and colostrum due to the fact that some retroviruses have been found in it [5]. Grego and coworkers affirm that Jaagsiekte Sheep Retrovirus (JSRV), is transmitted through milk and colostrum and causes Ovine Pulmonary Adenocarcinoma (OPA) [21]. Other authors such as Motton et al. found this virus in breast epithelial cells, what made to ask themselves if the virus was really in the milk or just in glandular cells that produced it [22].
Molecular analysis in this work confirms BLV presence in human breast tissue, suggesting that the virus is able to get into human epithelial cells and begin a kind of processes to integrate into the cellular genome. This generates other concerns, for example if BLV could be a zoonotic agent and if infected epithelial breast cells have recognition receptors or molecules similar to BLVRcp1 or other membrane molecules implicated such as IgM, CD5+ and CD11b integrins as demonstrated experimentally in infected sheep studies [23].
Taking into account that in 35.8% of women with breast cancer was BLV present in the tissue, it could be supposed a transformant role of BLV in human breast tissue, what can be associated with one of the possibilities presented below: 1) tax genes expression, a protein that is implicated in transcriptional viral processes, where the increase in the expression of bcl-2/bax ratio is related with leukemia genesis in infected cows [9]. Several laboratory trials showed that Tax could transform cells in in vitro cultures and being oncogenic in animal models where cells can be transformed when Tax is combined with ha-ras oncogene to produce tumor cells in athymic mice and fibroblasts immortalization coming from rat embryo [24,25]; 2) Viral genome integration process close to highly active cellular genome segments associated with cell division [26]. This is how the Avian Sarcoma Leukosis Virus (ASLV) can integrate in many places of cellular genome, but when there is tumor in the host, the virus has been found upstream of the c-myc cellular gene, which codifies for the Myc proteins implicated in the cellular DNA synthesis control [26,27]. A similar situation happens with MMTV that locates in cellular genome places that induce cellular proliferation by insertional mutagenesis or transcriptional activation of close oncogenes [28,30]; 3) Blocking of tumor suppressors genes and apoptosis activation genes processes [14]. Mutations in the suppressor gene p53 have been reported in approximately 20% of women with breast cancer, and mutations of ras oncogene have been found in 30% in malignancy processes in humans [29].
With these three possibilities, it could be considered that BLV can be responsible of cellular transformation either if are benign, pre-malign or malign mutations; also is possible to consider BLV could be found in healthy breast tissue, as occurs with MMTV in 1.8% of human breast tissues studied by Ford et al. [15]. In these cases the virus could be present in latency without presenting changes in tissues, as also is presented with other human retroviruses such as HIV and HTLV-1, where the immunological system, cancer suppressor mechanisms, genetic and hormonal factors of the host, may be critical in the moment of maintaining cellular control and physiopathological processes despite of BLV infection.
4. Acknowledgements
We want to thank the Environmental and Applied Science University (Universidad de Ciencias Aplicadas) for the economical aid to fund this study, Dr. Jorge Almanza for his good ideas and Dr. Marcela Mercado for her guidance in data analysis.
REFERENCES
- G. Moratorio, G. Obal, A. Dubra, A. Correa, S. Bianchi, A. Buschiazzo, J. Cristina and O. Pritsch, “Phylogenetic Analysis of Bovine Leukemia Viruses Isolated in South America Reveals Diversification in Seven Distinct Genotypes,” Archives of Virology, Vol. 155, No. 4, 2010, pp. 481-489. doi:10.1007/s00705-010-0606-3
- K. Murakami, S. Kobayashi, M. Konishi, K.-i. Kameyama, T. Yamamoto and T. Tsutsui, “The Recent Prevalence of Bovine Leukemia Virus (BLV) Infection among Japanese Cattle,” Veterinary Microbiology, Vol. 148, No. 1, 2011, pp. 84-88. doi:10.1016/j.vetmic.2010.08.001
- K. G. Trono, D. M. Pérez-Filgueira, S. Duffy, M. V. Borca and C. Carrillo, “Seroprevalence of Bovine Leukemia Virus in Dairy Cattle in Argentina: Comparison of Sensitivity and Specificity of Different Detection Methods,” Veterinary Microbiology, Vol. 83, No. 3, 2001, pp. 235- 248. doi:10.1016/S0378-1135(01)00420-5
- M. Rola and J. Kuzmak, “The Detection of Bovine Leukemia Virus Proviral DNA by PCR-ELISA,” Journal of Virologican Methods, Vol. 99, No. 1, 2002, pp. 33-40.
- K. Murakami, S. Kobayashi, M. Konishi, K. Kameyama, T. Yamamoto and T. Tsutsui, “The recent Prevalence of Bovine Leukemia Virus (BLV) Infection among Japanese Cattle,” Veterinary Microbiology, Vol. 146, No. 1, 2011, pp. 84-88. doi:10.1016/j.vetmic.2010.08.001
- N. Rice, R. Stephens, A. Burny and R. Gilden, “The Gag and Pol Genes of Bovine Leukemia Virus: Nucleotide Sequence and Analysis,” Virology, Vol. 142, No. 2, 1985, pp. 357-377. doi:10.1016/0042-6822(85)90344-7
- H. Murakami, T. Yamada, M. Suzuki, Y. Nakahara, S. Kazuhiko and H. Sentsui, “Bovine Leukemia Virus Integration Site Selection in Cattle That Develp Leukemia,” Virus Research, Vol. 156, No. 1-2, 2011, pp. 107-112. doi:10.1016/j.virusres.2011.01.004
- X. Zhao and G. C. Buehring, “Natural Genetic Variations in Bovine Leukemia Virus Envelope Gene: Possible Effects of Selection and Escape,” Virology, Vol. 366, No. 1, 2007, pp. 150-165. doi:10.1016/j.virol.2007.03.058
- M. Takahashi, S. Tajima, K. Okada, W. Davis and Y. Aida, “Involvement of BLV Virus in Induction and Inhibition of Apoptosis,” Microbes and Infection, Vol. 7, No. 1, 2005, pp. 19-28. doi:10.1016/j.micinf.2004.09.014
- M. Van den Heuvel, B. Jefferson and R. Jacobs, “Purified Bovine Plasma Blocking Factor Decreases Bovine Leukemia Virus p24 Expression While Increasing Protein Synthesis and Transcriptional Activity of Peripheral Blood Mononuclear Cells in Short-Term Culture,” The Canadian Journal of Veterinary Research, Vol. 69, No. 3, 2005, pp. 186-192.
- CDC, “Breast Cancer, Center for Disease Control and Prevention,” 2013. http://www.cdc.gov/spanish/cancer/breast/
- CDC, “Draft Comprehensive Care Guidelines (IEG) of Adult Cancer,” National Cancer Institute, Ministry of Social Protection, Colombia, 2001. http://www.minsalud.gov.co/salud/DropOffLibrary/Presentaci%C3%B3n%20GAI%20Instituto%20Nacional%20de%20Cancerolog%C23%ADa.pdf
- M. Andrujar, “Virus y Cáncer,” Biocancer, 2004. http://www.biocancer.com/Journal/247/virus-y-cancer>
- S. Melana, M. Picconi, C. Rossi, J. Mural, L. Alonio, A. Teyssie, J. Holland and B. Pogo, “Detección de Secuencias Homologas al Gen env del Virus del Tumor Mamario Murino (MMTV) en Cáncer de Mama de Pacientes Argentinas,” Medicina, Vol. 62, No. 4, 2002, pp. 323- 327.
- M. Juliarena, S. Gutierrez and C. Ceriani, “Determination of Proviral Load in Bovine Leukemia Virus Cattle with and without Lymphocytosis,” American Journal of Veterinary Research, Vol. 68, No. 11, 2007, p. 1220. doi:10.2460/ajvr.68.11.1220
- A. Ochoa-Cruz, A. Uribe and M. Gutierrez, “Zoonotic Behavior of the Bovine Leukemia Virus and Its Relationship with Breast Cancer,” Universitas Scientiarum, Vol. 11, No. 2, 2006, pp. 31-40.
- G. Hughes, G. McKerr, J. Allen and Y. Barnett, “Are Retroviruses Involved in the Aetiology of Human Breast Cancer?” Cancer Letters, Vol. 103, No. 2, 1996, pp. 219- 225. doi:10.1016/0304-3835(96)04217-6
- J. Lee, Y. Kim, C. Suk and D. Hyun, “Investigation of the Bovine Leukemia Virus Proviral DNA in Human Leukemias and Lung Cancers in Korea,” Journal of Korean Medical Science, Vol. 20, No. 4, 2005, pp. 603-606. doi:10.3346/jkms.2005.20.4.603
- G. Buehring, S. Philpott and K. Y. Choi, “Humans Have Antidodies Reactive with Bovine Leukemia Virus,” AIDS Research and Human Retrovirus, Vol. 19, No. 12, 2003, pp. 1105-1113.
- X. Zhao and G. Buehring, “Natural Genetic Variations in Bovine Leukemia Virus Envelope Gene: Possible Effects of Selection and Escape,” Virology, Vol. 366, No. 1, 2007, pp. 150-165. doi:10.1016/j.virol.2007.03.058
- S. Dube, L. Abbott, D. Dube, G. Dolcini, S. Gutierrez, C. Ceriani, M. Juliarena, J. Ferrer, R. Perzova and B. Poiesz, “The Complete Genomic Sequence of an in Vivo Low Replicating BLV Strain,” Virology Journal, Vol. 6, 2009. doi:10.1186/1743-422X-6-120
- D. Motton and G. Buehring, “Bovine Leukemia Virus Alters Growth Properties and Casein Syntesis in Mammary Epithelial Cells,” Journal of Dairy Science, Vol. 86, No. 9, 2003, pp. 2826-2838. doi:10.3168/jds.S0022-0302(03)73880-6
- E. Chamizo, “Enzootic Bovine Leucosis,” REDVET, Vol. 6, No. 7, 2005, pp. 1-20.
- A. Florins, N. Gillet, M. Boux, P. Kerkhofs, R. Kettmann and L. Willems, “Even Attenuated Bovine Leukemia Virus Proviruses can Be Pathogenic in Sheep,” Journal of Virology, Vol. 81, No. 18, 2007, pp. 10195-10200. doi:10.1128/JVI.01058-07
- N. Gillet, A. Florins, M. Boxus, C. Burteau, A. Nigro, F. Vandermeers, H. Balon, A. Bouzar, J. Defoiche, A. Burny, M. Reichert, R. Kettmann and L. Willems, “Mechanisms of Leukemogenesis Induced by Bovine Leukemia Virus: Prospects for Novel Anti-Retroviral Therapies in Human,” Retrovirology, Vol. 4, 2007, pp. 1-32. doi:10.1186/1742-4690-4-18
- B. Fulton, M. Portella and K. Radke, “Dissemination of Bovine Leukemia Virus-Infected Cells from a Newly Infected Sheep Lymph Node,” Journal of Virology, Vol. 80 No. 16, 2006, pp. 7873-7884. doi:10.1128/JVI.00529-06
- P. Klener, M. Szynal, Y. Cleuter, M. Merimi, H. Duvillier, F. Lallemand, C. Bagnis, P. Griebel, C. Sotiriou, A. Burny, P. Martiat and A. Van der Broeke, “Insights into Genes Expression Changes Impacting B-Cell Transformation: Cross-Species Microarray Analysis of Bovine Leukemia Virus Tax-Responsive Genes in Ovine B Cells,” Journal of Virology, Vol. 80, No. 4, 2006, pp. 1922-1938. doi:10.1128/JVI.80.4.1922-1938.2006
- M. Gallego, “Manejo del Problema Reproductivo en Ganado de leche,” Editorial. U.D.C.A., Bogotá, 2009, pp. 125-130.
- R. Javier and J. Butel, “The Hostory of Tumor Virology,” Cancer Research, Vol. 68, No. 19, 2008, pp. 7693-7706. doi:10.1158/0008-5472.CAN-08-3301
Abbreviations
BLV: Bovine leukemia Virus.
NOTES
*Corresponding author.

