Advances in Biological Chemistry
Vol. 2 No. 3 (2012) , Article ID: 21816 , 11 pages DOI:10.4236/abc.2012.23037
The polybasic region of cytohesin-2 determines paxillin binding specificity to mediate cell migration
![]()
1Department of Pharmacology, National Research Institute for Child Health and Development, Tokyo, Japan
2Department of Molecular and Functional Genomics, Shimane University, Matsue, Japan
3Immuno-Biological Laboratories (IBL) Co. Ltd., Fujioka, Japan
4The Japan Health Sciences Foundation (JHSF), Tokyo, Japan
Email: #jyamauchi@nch.go.jp
Received 25 April 2012; revised 27 May 2012; accepted 7 June 2012
Keywords: Cytohesin-2; Polybasic Region; Paxillin; Migration
ABSTRACT
The intracellular signaling pathways through ADPribosylation factors (Arfs) of the small GTPase family control cell morphological changes by regulating membrane components and/or cytoskeletal protein dynamics. We previously reported that cytohesin-2 (CYTH2), an Arf-guanine-nucleotide exchange factor (GEF), binds to the cytoskeletal scaffold protein paxillin through C-terminal region of CYTH2 and promotes the migration of mouse 3T3-L1 fibroblasts. In mammals, CYTH family GEFs are composed of four subfamilies. Among them, CYTH2 and CYTH3 are widely expressed in tissues and it remains to be clarified to determine whether they have specific biochemical and cellular functions or are redundant. Here, we show that the C-terminal short polybasic region of CYTH2 is necessary and sufficient for binding to paxillin to mediate cell migration. Although 3T3-L1 cells primarily express CYTH2 and CYTH3 of four CYTH family members, neither knockdown of CYTH3 by the specific siRNA nor expression of its C-terminal region inhibits migration. Importantly, replacing the C-terminal region of CYTH3 with that of CYTH2 adds the ability of paxillin-binding and mediating migration to CYTH3. Conversely, replacing the Cterminal region of CYTH2 with that of CYTH3 leads to loss of these abilities of CYTH2. These results reveal that paxillin is a unique binding partner with CYTH2 in migrating cells, presenting the first CYTH family GEF’s region that is involved in the selectivity of the binding protein.
1. INTRODUCTION
Cell migration is a crucial process for early development and is also seen in pathological states. It is established that the intracellular signaling mechanism responsible for cell morphological changes, such as migration and morphological differentiation, are controlled by small GTPases of the Rho family [1-4]. Recent evidence demonstrates that in addition to Rho GTPases, Arf branches of small GTPases are involved in cell morphological changes [5-8]. The mammalian Arf proteins are composed of three groups: class I (Arf1 and Arf3, and/or Arf2), class II (Arf4 and Arf5), and class III (Arf6) [5-8]. Like Rho GTPases, they act as molecular switches; they are active when bound to GTP and they are inactive when bound to GDP. In particular, Arf1 and Arf6 are well characterized to interact with the downstream effectors, including lipid-modifying enzymes such as phosphatidylinositol-4-phosphate 5-kinases (PIP5Ks) and phospholipase D isozymes (PLDs), and coat proteins, which coordinately contribute to cell morphological changes through modification of membrane lipid components and/or modulation of actin cytoskeletal dynamics [5-8].
The guanine-nucleotide-binding states of Arfs are primarily controlled by two types of regulatory proteins [1]. Guanine-nucleotide exchange factors (GEFs) catalyze the intrinsically slow exchange reaction of GDP with free cytoplasmic GTP to generate the active Arfs. GTPaseactivating proteins (GAPs) accelerate the very low, intrinsic GTPase activity to inactivate Arfs. This former reaction is a very important, rate-limiting step, since GEFs define the timing and strength of the Arf activation by integrating upstream, intracellular signals [7,8].
In mammals, there are least 15 Arf-GEFs, of which the type with the lowest molecular weight belongs to the cytohesin/Arf nucleotide binding site opener (ARNO) subfamily and is composed of four homologous molecules [7,8]. Cytohesin-2 (CYTH2, also called ARNO or pleckstrin homology [PH], Sec7, and coiled-coil [CC] domains 2 [PSCD2]) and CYTH3 (also called general receptor for phosphoinositides 1 (GRP1)/ARNO3/PSCD3) are widely expressed in tissues, whereas CYTH1 (also called B2-1 or PSCD1) and CYTH4 (also called PSCD4) display a specific expression in lymphocytes and related tissues [7,8]. CYTH family GEFs are primarily the GEFs for Arf1 and Arf6 [7-9].
We previously reported that CYTH2 regulates the migration of mouse 3T3-L1 fibroblasts through the direct interaction with paxillin, which is a cytoskeletal scaffold protein [10,11]. All CYTH family GEFs have the same domain structure, which contains the N-terminal CC domain, the catalytic Sec7 domain, and the PH domain plus the C-terminal basic amino acid extension [9]. The binding between CYTH2 and paxillin was through the region containing the PH domain in tandem with the C-terminal polybasic region of CYTH2 and the Leu-rich LIM2 domain of paxillin [11]. Here, we describe the small CYTH2’s region, which determines the specificity of binding to paxillin to mediate cell migration. The C-terminal polybasic region (14 amino acids) of CYTH2 is necessary and sufficient for binding to paxillin to regulate cell migration. This finding is unusual, as the polybasic amino acid sequence at the C-terminus participates in a well-known protein-phospholipid interaction [12] rather than in a protein-protein interaction. These findings not only add new information to the list of amino acid sequences linking signal transduction molecules to cytoskeletal proteins but also provide insight into whether and how each CYTH family protein plays a specific role in cells.
2. MATERIALS AND METHODS
2.1. Antibodies
The antibodies used were as follows: mouse monoclonal anti-CYTH2 (1:100, Sigma-Aldrich, St. Louis, MO, USA; 1:50, Santa Cruz, CA, USA), mouse monoclonal antiCYTH3 (1:50, Santa Cruz), mouse monoclonal anti-actin (1:1000, BD Biosciences Pharmingen, Franklin Lake, NJ, USA; 1:1000, Invitrogen, Carlsbad, CA, USA), mouse monoclonal anti-GFP (1:1000, MBL, Nagoya, Japan), rabbit polyclonal anti-RFP (1:1000, Evrogen, Moscow, Russia), mouse monoclonal anti-FLAG (1:1000, SigmaAldrich), and horseradish peroxidase or fluorescenceconjugated secondary antibody (1:10,000, GE Healthcare, Fairfield, CT, USA; 1:10,000, Nakalai, Kyoto, Japan).
2.2. Plasmids
The p3×FLAG (obtained from Sigma-Aldrich)-mouse CYTH1, p3×FLAG-mouse CYTH3, pEGFP (obtained from Takara, Shiga, Japan)-mouse paxillin, and pRFP (pTurboRFP, obtained from Evrogen, Moscow, Russia)mouse paxillin plasmids were constructed as previously described [13,14]. The human CYTH2, mouse CYTH2, and mouse CYTH4 cDNAs were purchased from NBRC (Chiba, Japan). The human or mouse CYTH2 cDNA was ligated into p3×FLAG or pMEI5-EGFP (Refs. 13, 14; obtained from Takara). The isolated C-terminal polybasic regions of human CYTH2 (C2, amino acids 386 - 400) and mouse CYTH3 (C3, amino acids 391 - 399) were inserted into the pEGFP vector. The CYTH2-C3 construct replacing the C-terminal region of human CYTH2 (amino acids 386 - 400) with that of mouse CYTH3 (amino acids 391 - 399) and the CYTH3-C2 construct replacing the C-terminal region of mouse CYTH3 with that of human CYTH2 were produced by the overlapping PCR method and ligated into the pMEI5-EGFP vector. These plasmid constructs used in this study are schematically shown in Figure S1. All nucleotide sequences were confirmed by Fasmac sequencing service (Kanagawa, Japan).
2.3. RT-PCR
Total RNA was extracted from 3T3-L1 cells using a Trizol (Invitrogen) reagent. The cDNAs were prepared from 1 mg of total RNA with Superscript III (Invitrogen) according to the manufacturer’s instructions. PCR amplification was performed with ExTaq polymerase (Takara) in 30 cycles, each cycle consisting of denaturation at 94˚C for 1 min, annealing at 58.5˚C to 61.5˚C (depending on the primer pair’s Tm value) for 1 min, and extension at 72˚C for 1 min. The primers used were as follows: 5’-ATGGA GGACGATGACAGCTATGTC-3’ (sense) and 5’-TCAG TGTCTCTTTGTGGAGGAGAC-3’ (antisense) for mouse cytohesin-1; 5’-ATGGAGGACGGTGTCTACGAG-3’ (sense) and 5’-TCAG-GGTTGTTCTTGCTTCTTCTTCAC- 3’ (antisense) for mouse cytohesin-2; 5’-ATGGACGAA GGCGGTG-GCGGTG-3’ (sense) and 5’-CTATTTATTG GCAATCCTCCTTTTCCTCGTGGCCAAC-3’ (antisense) for mouse cytohesin-3; and 5’-ATGGATGTGTGTCAC ACAGATCCAG-3’ (sense) and 5’-CTACTTGCCGAC AATCTTCTTTTTCCGA-3’ (antisense) for mouse cytohesin-4. The control primers for mouse b-actin were 5’- ATGGATGACGATATCGCTGCGCTC-3’ (sense) and 5’- CTAGAAGCATTTGCGGTGCACGATG-3’ (antisense).
2.4. siRNA Oligonucleotides
The 21-nucleotide siRNA duplexes were synthesized by EGT (Toyama, Japan). The target sequences were as follows: 5’-AAGAGCTAAGTGAAGTATGA-3’ specific for mouse CYTH2 and 5’-AAGAAAAAAGGAACTTATT GA-3’ specific for mouse CYTH3. The target sequence of the control P. pyralis luciferase siRNA was 5’-AAGC CATTCTATCCTCTAGAG-3’, which does not have significant homology to any mammalian gene sequences.

Figure S1. Schematic structures of the plasmids used in this study. In these panels, h indicates human and m indicates mouse.
2.5. Cell Cultures
Mouse 3T3-L1 fibroblasts were kindly provided by Drs. M. Imagawa and M. Nishizuka (Nagoya City University). 3T3-L1 fibroblasts and human embryonic kidney 293T cells were routinely cultured at 37˚C in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin.
2.6. Plasmid and siRNA Transfection
For 3T3-L1 cells, the plasmid DNAs and/or siRNAs were transfected using the Lipofectamine 2000 reagent (Invitrogen) or the Nucleofector system (Lonza, Basel, Switzerland) with the Nucleofector reagent kit, according to each manufacturer’s protocol. The medium was replaced 4 and 24 hours after lipofection and electroporation, respectively. Cells were cultured for an additional 44 or 24 hours [11]. For 293T cells, DNAs were transfected using the CalPhos reagent (Takara), according to a manufacturer’s protocol.
2.7. Fluorescence Images
The fluorescent images were captured using an Eclipse TE-300 microscope system (Nikon, Kawasaki, Japan) and analyzed with AxioVision software (Carl Zeiss, Oberkochen, Germany) or captured using a DMI4000B microscope system (Leica, Heerbrugg, Switzerland) and analyzed with AF6000 software (Leica).
2.8. Confocal Images
Cells were fixed in 4% paraformaldehyde in PBS, blocked with 20% heat-inactivated FBS in PBS and 0.05% Tween- 20, incubated with a primary antibody in PBS and 0.05% Tween-20, and then treated with a fluorescence-labeled secondary antibody in PBS and 0.1% Tween-20. The coverslips were mounted with the Vectashield reagent (Vector Laboratories, Burlingame, CA, USA) onto slides for observation by confocal microscopy. The confocal images were collected with an IX81 confocal microscope system (Olympus, Tokyo, Japan) and analyzed with FluoView software (Olympus).
2.9. Immunoblotting and Immunoprecipitation
Cells were lysed in lysis buffer (50 mM HEPES-NaOH, pH 7.5, 20 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethane sulfonylfluoride, 1 mg/ml leupeptin, 1 mM EDTA, 1 mM Na3VO4, 10 mM NaF, and 0.5% NP-40) [11]. After centrifugation, cell extracts were mixed with an antibody-absorbed protein G resin for immunoprecipitation. The immune-complexes or proteins in cell extracts were denatured and then subjected to SDS-PAGE. The electrophoretically separated proteins were transferred to PVDF membranes, blocked with the blocking reagent (Nakalai), and immunoblotted with a primary antibody and then with a peroxidase-conjugated secondary antibody. The bound antibodies were detected using the chemiluminescence reagent (Nakalai).
2.10. Wound Closure (Scratch-Induced) Migration Assay
3T3-L1 cells were incubated at 37˚C for 0 or 6 hours in the presence of 10 mM AraC [11]. The monolayers of confluent cells were scraped with the narrow end of a micropipette tip to generate a wound approximately 50 μm wound in width. Repair of the wound healing was quantified as the following: ratio of healing (%) = (width of wound at 0 hour minus width of wound at 0 or 6 hours/ width of wound at 0 hour) × 100.
2.11. Statistical Analysis
Values shown represent the mean ± SD from separate experiments. A one-way analysis of variance (ANOVA) was followed by a Fisher’s protected least significant difference (PLSD) test as a post hoc comparison (*, p < 0.01).
3. RESULTS
3.1. CYTH2 but Not CYTH3 Binds to Paxillin
We previously reported that CYTH2, through its C-terminal region containing the PH domain and the C-terminal polybasic region, binds to the cytoskeletal scaffold protein paxillin and mediates the migration of mouse 3T3- L1 fibroblasts [11]. RT-PCR analysis primarily detected CYTH2 and CYTH3 in 3T3-L1 cells, as seen in immunoblotting with the respective specific antibodies (Figure 1(a)). Thus, the 3T3-L1 cell line was a good model to study whether CYTH2 and CYTH3 have similar and/or totally different role in cells. Expression of CYTH1 was detected only in the RT-PCR analysis.
First, we asked if CYTH3 has the ability to bind to paxillin. We cotransfected the plasmids encoding paxillin with CYTH1, CYTH2, or CYTH3 in 293T cells and performed the coimmunoprecipitation from the cell lysate. Paxillin was coimmunoprecipitated with CYTH2 and weakly with CYTH1. In contrast, CYTH3 did not coimmunoprecipitate with paxillin (Figure 1(b)), presenting more specific involvement of CYTH2 in 3T3-L1 cell migration. The expression levels of transfected CYTH1, CYTH2, and CYTH3, as well as paxillin, were comparable.
To confirm that under these experimental conditions CYTH2 is required for migration, we transfected siRNA specific for mouse CYTH2 into 3T3-L1 cells and assessed the effect of knockdown of endogenous CYTH2 on scratch-induced migration. CYTH2 knockdown resulted in inhibition of migration. In contrast, transfection with siRNA for mouse CYTH3 did not have a significant inhibitory effect on migration (Figures 1(c) and (d)). Immunoblotting analysis with an antibody for CYTH2 or CYTH3 showed the specific knockdown for each siRNA (Figure 1(e)), as seen in the case of cell staining with the immunofluorescence method (Figures 2(A) and (B)). Knockdown of either CYTH2 or CYTH3 specifically reduced each expression at the cell periphery but not in the cytoplasm, including the perinuclear region. Thus, while CYTH2 and CYTH3 show a similar localization primarily at the cell periphery, CYTH2 specifically binds to paxillin and mediates cell migration.
3.2. The C-Terminal Polybasic Region of CYTH2 Is Important for Binding to Paxillin and Scratch-Induced Migration
Although paxillin binds to CYTH2’s C-terminus containing the PH domain and the polybasic region, the PH domain usually contributes to phospholipid-binding and determines their binding specificity [9]. Therefore, we focused on the study of whether the C-terminal polybasic region positioned in the outside of the PH domain determines paxillin-binding and the specificity. We made the
 (a)
(a)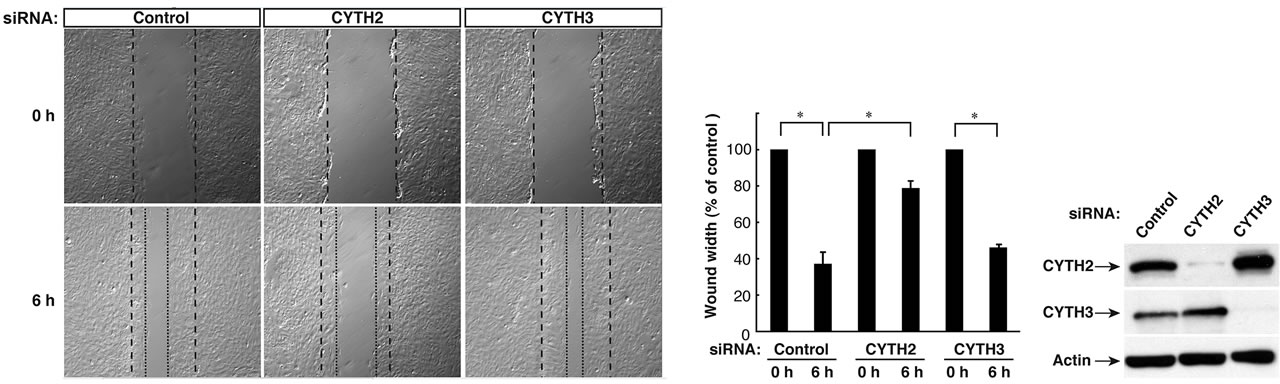 (b)
(b)
Figure 1. CYTH2, but not CYTH3, is specific for paxillin-binding and 3T3-L1 cell migration. (a) RT-PCR analyses for CYTH family GEFs show the primary transcription of CYTH2 and CYTH3 and the weak transcription of CYTH1 in 3T3-L1 cells. CYTH4 was also amplified using CYTH4 cDNA as the template. Expression levels of CYTH2 or CYTH3 were confirmed by means of immunoblotting with an anti-CYTH2 or anti-CYTH3 antibody, respectively; (b) 293T cells were cotransfected with the plasmid encoding FLAG-tagged CYTH1, CYTH2, or CYTH3 with GFP-tagged paxillin and lysed. CYTH1, CYTH2, or CYTH3 was immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-GFP antibody. The expression levels of CYTHs and paxillin were comparable; (c) 3T3-L1 cells were transfected with a control siRNA or a siRNA specific for mouse CYTH2 or mouse CYTH3. 3T3-L1 cell monolayers were wounded by scraping and observed microscopically at 0 and 6 hours. The position of the representative healing edge of migrating cells at 0 hours is indicated by a dashed line, and those at 6 hours are indicated by a dotted line; (d) The rate of healing was calculated based on the width of the wound at 6 hours; (e) 3T3-L1 cells transfected with a CYTH2 or CYTH3 siRNA were lysed and immunoblotted with an anti-CYTH2 or CYTH3 antibody. Data were evaluated with one-way ANOVA (*, p < 0.01).
CYTH2 mutant (CYTH2-C3) by replacing its C-terminal 14 amino acid sequence with CYTH3’s C-terminal 9 amino acid sequence. Conversely, the CYTH3 mutant (CYTH3-C2) replacing the C-terminal sequence of CYTH3 with C-terminal sequence of CYTH2 was constructed (Figure 3(A)). We cotransfected the plasmid encoding each of these constructs with paxillin into 293T cells and performed coimmunoprecipita tion. As shown in Figure 3(B), CYTH3-C2, but not CYTH2-C3, was specifically coimmunoprecipitated with paxillin, indicating that C-terminal 14 amino acids of CYTH2 add the paxillin-binding ability to CYTH3. Under these experimental conditions, the expression levels of these two mutants were comparable.
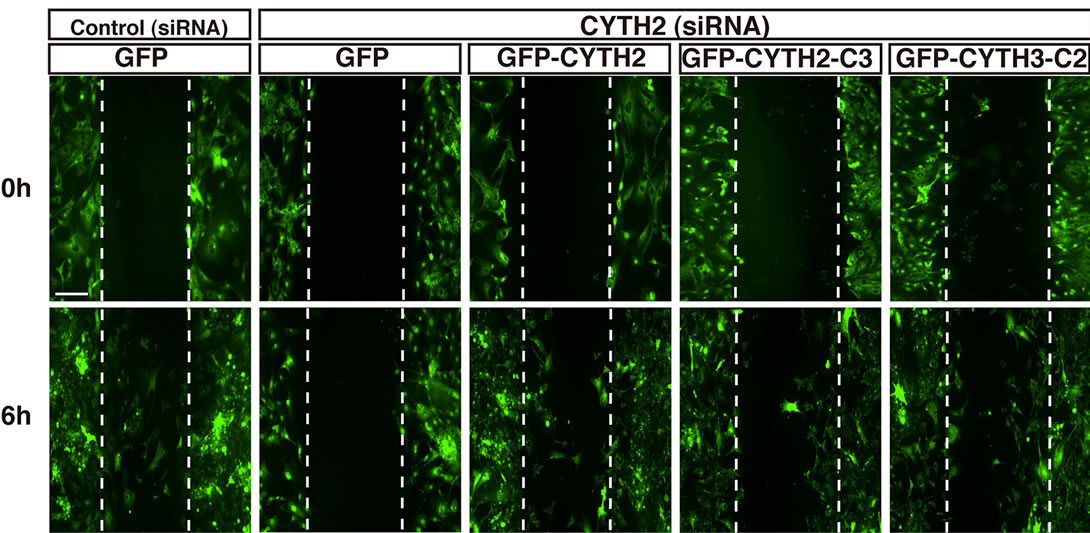
We further tested the effects of CYTH2-C3 and CYTH3- C2 on the migration of mouse 3T3-L1 cells. The CYTH2 or control luciferase siRNA was cotransfected with the plasmid encoding CYTH2, CYTH2-C3, or CYTH3-C2 into cells. Since the CYTH2 constructs that we used in these experiments were derived from human species, they were resistant to siRNA specific for mouse CYTH2 (Figure 4(c)). Expression of human CYTH2 in cells rescued CYTH2 siRNA inhibition of scratch-induced migration by 50% - 60% (Figures 4(a) and (b); in statistical data, lane 4 compared with lane 6 of Figure 4(b)). The similar result was observed in expression of CYTH3-C2, which recovered migration by 40% - 50% (lane 4 compared with lane 10 of Figure 4(b)). However, expression of CYTH2-C3 was not able to rescue CYTH2 siRNA inhibition of migration (lane 4 compared with lane 8 of Figure 4(b)). Taken together with the results from Figure 3, the C-terminal polybasic region of CYTH2 plays a key role in paxillin-binding and migration. These findings are very unique, as the polybasic amino acid sequence at the C-terminal position has well-known involvement in the protein-phospholipid interaction rather than the protein-protein interaction [12].
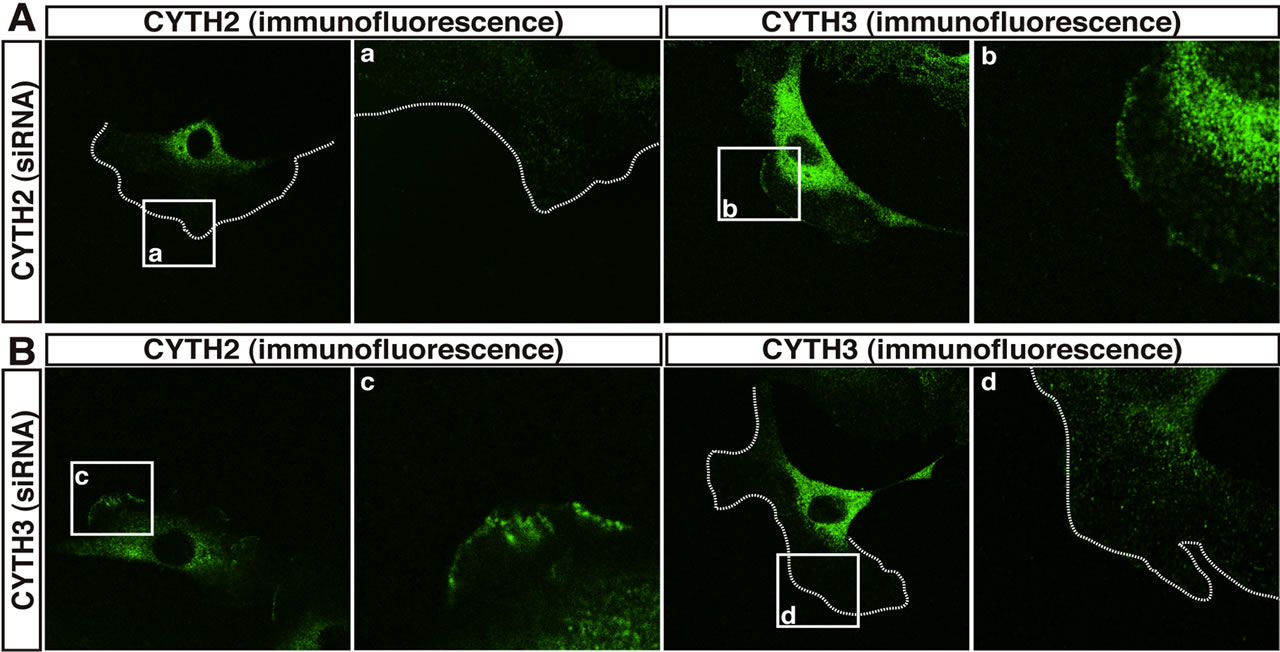
Figure 2. CYTH2 and CYTH3 are localized at the cell periphery of 3T3-L1 cells. (A, B) 3T3-L1 cells transfected with a CYTH2 or CYTH3 siRNA were stained with an anti-CYTH2 or CYTH3 antibody. The boxed areas indicated by small characters, a-d, are also shown in the magnification. CYTH2 and CYTH3 are stained in the cell peripheral region and the prinuclear region but each siRNA knockdown specifically deleted CYTH2 or CYTH3 staining in the cell peripheral region. The cell periphery was indicated by dotted lines.
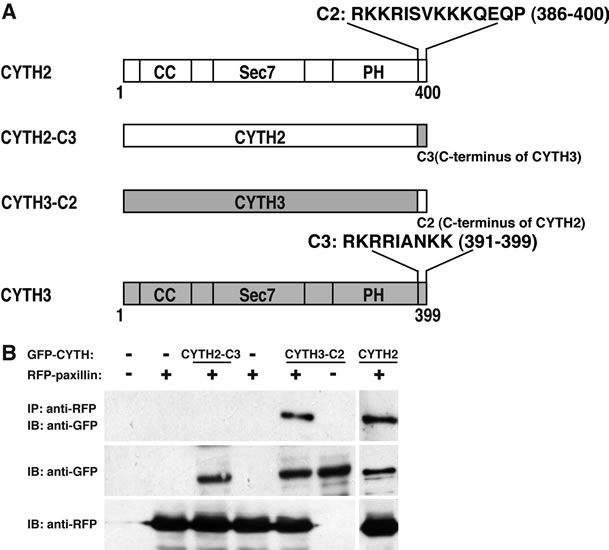
Figure 3. Replacing C-terminal polybasic region of CYTH3 with that of CYTH2 adds paxillin-binding ability to CYTH3. (A) A schematic diagram of the mutants replacing the C-terminal polybasic region of CYTH2 with that of CYTH3 (CYTH2-C3) and the C-terminal polybasic region of CYTH3 with that of CYTH2 (CYTH3-C2). CYTH2 and CYTH3 are very homologously composed of the coiled-coil (CC) region, the catalytic Sec7 domain, the pleckstrin homology (PH) domain, and the C-terminal extension; (B) The plasmid encoding GFP-tagged CYTH2-C3, CYTH3- C2, or CYTH2 were cotransfected with RFP-tagged paxillin, lysed, immunoprecipitated with an anti-RFP antibody, and immunoblotted with an anti-GFP antibody. The expresstion levels of CYTH mutants and paxillin were comparable.
3.3. The C-Terminal Region of CYTH2 Is Sufficient for Binding to Paxillin to Regulate Migration
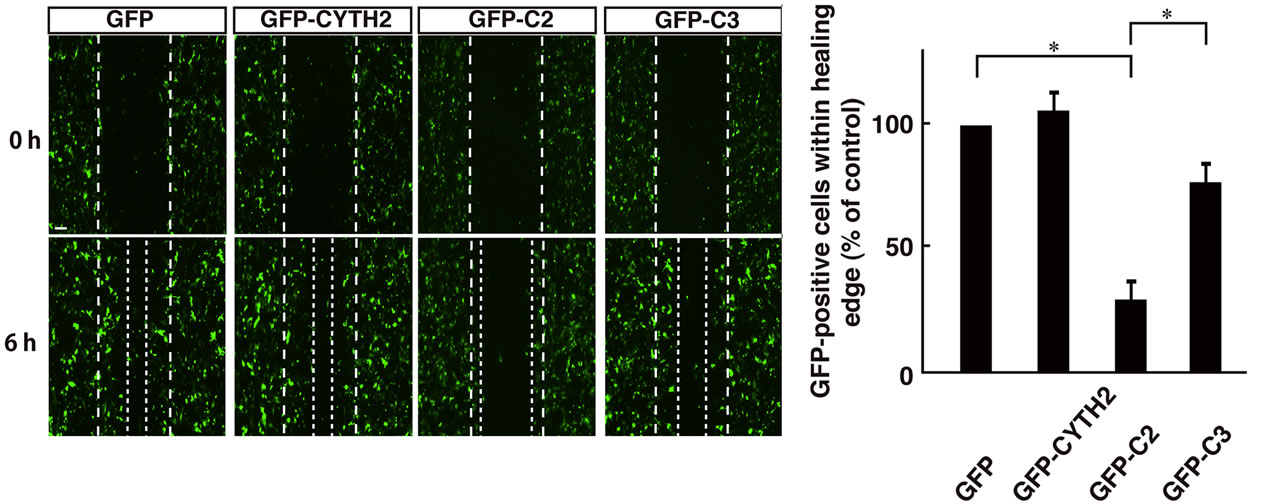
Thus, we asked if the C-terminal polybasic region of CYTH2 is sufficient for binding to paxillin. We cotransfected the plasmid encoding the isolated C-terminal region of CYTH2 or CYTH3 with paxillin into 293T cells. A coimmunoprecipitation shows that the isolated C-terminus of CYTH2, but not that of CYTH3, has the ability to form a complex with paxillin (Figures 5(A) and (B)). In addition, scratch-induced 3T3-L1 cell migration was inhibited by expression of the isolated C-terminus of CYTH2, but not that of CYTH3 (Figures 6(a) and (b)), indicating that the C-terminal region of CYTH2 unique-ly provides a platform to bind to paxillin to mediate migration. Although it is well known that the C-terminal polybasic region is involved in binding to phospholipids, the C-terminal region of CYTH2 did not bind to a phospholipid (Ref. 15; data not shown).
4. DISCUSSION
Of four CYTH family GEFs, CYTH2 and CYTH3 display wide expression in tissues and more than 90% of homology at the amino acid level [7-9]. Thus, it is important to determine whether CYTH2 and CYTH3 interact with a specific binding partner to play a specific role or they are redundant in cells. Herein, we demonstrate that CYTH2 specifically binds to the cytoskeletal scaffold
 (a)
(a) (b)
(b)
Figure 4. Mutant replacing C-terminal polybasic region of CYTH3 with that of CYTH2 rescues CYTH2 siRNA-mediated inhibition of scratch-induced migration. (a), (b) The plasmid encoding GFP-tagged, mouse CYTH2 siRNA-resistant CYTH2-C3, CYTH3-C2, or CYTH2 was cotransfected with a control or mouse CYTH2 siRNA into 3T3-L1 cells and cell migration was assayed; (c) The plasmid encoding GFP-tagged mouse CYTH2 construct (mouse CYTH2 siRNA-sensitive one) or mouse CYTH2 siRNA-resistant construct (CYTH2-C3 or CYTH3-C2) was cotransfected with a control or mouse CYTH2 siRNA (siCon or siCYTH2) into 3T3-L1 cells. The cell lysates were used for immunoblotting with an anti-GFP antibody. Data were evaluated with one-way ANOVA (*, p < 0.01).
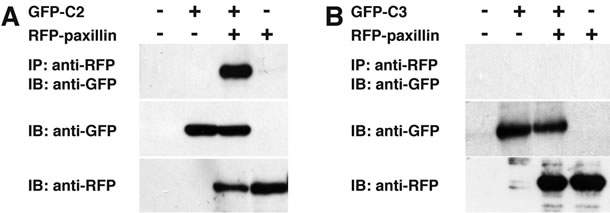
Figure 5. C-terminal region of CYTH2 is sufficient with paxillinbinding. (A, B) GFP-tagged C-terminus of CYTH2 or CYTH3 (C2 or C3) was cotransfected with RFP-tagged paxillin into 293T cells and cells were immunoprecipitated with an anti-RFP antibody and immunoblotted with an anti-GFP antibody. Expression of the transfected genes is also shown.
protein paxillin to mediate the migration of 3T3-L1 fibroblasts. The specificity is added by the C-terminal short polybasic region of CYTH2. This conclusion is supported by the result that replacing the C-terminal region of CYTH3 with that of CYTH2 adds the ability to bind to paxillin to mediate the migration to CYTH3 though it does not work the other way around. In addition, the C-terminal region of CYTH2 is necessary and sufficient for binding to paxillin to mediate the migration. These findings suggest that CYTH2 and CYTH3 play a role in cells and are not redundant, although they are similarly localized at the cell periphery of 3T3-L1 cells and do not exhibit an observable
 (a) (b)
(a) (b)
Figure 6. Expression of C-terminal polybasic region of CYTH2 inhibited 3T3-L1 cell migration (a) 3T3-L1 cells were transfected with the plasmid encoding GFP-tagged CYTH2, C2, or C3 and migration assay was performed. The position of the representative healing edge of migrating cells at 0 hours is indicated by a dashed line, and those at 6 hours are indicated by a dotted line; (b) The number of GFP-positive cells in the area between a dashed line and a dotted line was counted and shown in the relative value. Data were evaluated with one-way ANOVA (*, p < 0.01).
difference in cellular distribution. It is possible that CYTH2 and CYTH3 may be present in a similar but different signaling complex that is bridged by a scaffold protein including paxillin, at least in 3T3-L1 cells. To the best of our knowledge, paxillin is the first example of a protein that selectively binds to CYTH family GEFs.
Many CYTH family GEF-binding proteins are known, such as Grp1 signaling partner (GRSP) 1/mKIAA1013 [16], Grp1-associated scaffold protein (GRASP)/tamalin [17,18], cytohesin-associated scaffold protein (CASP)/Cybr/cytohesin-interacting protein (CYTIP) [19-21], interacting protein for cytohesin exchange factor (IPCEF) [22], and FERM domain-containing 4A (FRMD4A)/KIAA1294 [23]. The interaction with these proteins occurs through the CC domain and seems likely to bind to all CYTH family GEFs. Their interaction occurs through the CC domain. It has recently been identified that the connector enhancer of KSR1 (CNK1) [24] and the epidermal growth factor receptor (EGFR) [25] are binding partners with CYTH2. CNK1 links the insulin receptor to CYTH2/Arf6 to the effector PIP5Ks, resulting in recruiting the insulin receptor substrate 1 (IRS1) to the plasma membrane [24]. Thus, CNK1, acting through the CYTH2/Arf6/PIP5K unit, functions as a positive regulator of insulin receptor signaling. In some pathological states, CYTH2 also functions as an accessory protein of EGFR and helps to elevate EGFR transphosphorylation [25]. In this case, it is likely that the GEF activity of CYTH2 is not directly related to the transphosphorylation. Although it remains unknown which region of CYTH2 is involved in binding to CNK1 and EGFR, it would be interesting to determine whether CNK1 and EGFR are specific to CYTH2 or not and whether their signalings are involved in cell migration.
In addition to these binding partners with CYTH family GEFs, recent reports have added Arf-like protein, Arl4D, to the list [26,27]. Arl4D binds to the entire region containing the PH domain and the C-terminal region of CYTH family GEFs to regulate further downstream Arf6. Although CYTH2’s region to bind to Arl4D is overlapped with the paxillin-binding region, Arl4D binds equally to CYTH2 and CYTH3. Both the PH domain and the C-terminal region are required for the interaction The Arl4D/CYTH2/Arf6 signaling unit regulates the migration of epithelial cells. Thus, it is conceivable that Arl4D acts together with paxillin through CYTH2 and Arf6 to regulate the migration.
When beginning to study the role of the C-terminal polybasic region of CYTH2, we inferred that it could possess phospholipid-binding activity. The polybasic sequence at the C-terminal position provides lipid-binding activity with the signaling proteins [12]. The small GTPase member that contains a polybasic sequence at the C-terminal position binds to phosphoinositides such as phosphatidylinositol-3-phosphate (PIP3), which help to translocate small GTPases to the plasma membrane [12]. This is more important in some of the small GTPases that do not contain an amino acid sequence involved in the posttranslational modification such as farnesylation and geranylgeranylation. We previously reported that the C-terminal polybasic regions of sorting nexins (SNXs), belonging to the SNXPX group with one Phox (PX) domain flanked with very short Nand C-terminal regions, bind to phosphatidylinositol monophosphates including PIP3 and that they contribute to translocation to endosomes [15]. However, this rule did not apply to the case of CYTH2. Further studies on the comparison of polybasic amino acid sequences at the C-terminus in signaling proteins may clarify a common feature to determine whether one has the specificity for lipid-binding or the other does for paxillin-binding.
The only known difference between CYTH2 and CYTH3 in cells is that relating to their function of recycling endosomes with specific cargo [28]. CYTH2 is required for beta1 integrin recycling, whereas CYTH3 is not essential for it. In spreading human breast cancer MCF-7 cells, CYTH2 is localized on the edge and the interior of cells, but CYTH3 is specifically localized on the very peripheral edge. It is clear that CYTH2 differs from CYTH3 in paxillin-binding ability, but the difference in cellular functions of CYTH2 and CYTH3 may also depend on the differing on cell types and the contexts of signaling proteins.
In this study, we identify CYTH2’s amino acid sequence, which adds specificity for paxillin-binding. The specificity determines the involvement of CYTH2 in fibroblast migration. Further studies on the cellular role of the polybasic region in paxillin-binding and the identifycation of a possible additional protein binding to the polybasic region will enable us to understand whether and how each CYTH family GEF independently and/or cooperatively functions in various types of cells, as well as in vivo. Such studies may aid in the development of suitable drug-target-specific medicines for a disease such as type 2 diabetes, as SecinH3 of the first-phase CYTH inhibitor is known to cause insulin resistance [29,30].
5. ACKNOWLEDGEMENTS
We thank Drs. W. Furmanski and K. Spicer for reading this manuscript. We thank Drs. M. Imagawa and M. Nishizuka for providing 3T3-L1 cells and for their participation in helpful discussions. This work was supported by Grants-in-Aid for Scientific Research from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (22300123 and 23650200 to JY) and the Japanese Ministry of Health, Labor, and Welfare (KHA1001 and KHA1202 to JY), and by research grants from the Astellas Foundation (to JY), the Mochida Foundation (to JY), and the Takeda Foundation (to Y. M. and JY).
![]()
![]()
REFERENCES
- Bar-Sagi, D. and Hall, A. (2000) Ras and Rho GTPases: A family reunion. Cell, 103, 227-238. doi:10.1016/S0092-8674(00)00115-X
- Schmidt, A. and Hall, A. (2002) Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes & Development, 16, 1587-1609. doi:10.1101/gad.1003302
- Rossman, K.L., Der, C.J. and Sondek, J. (2005) GEFmeans go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nature Reviews Molecular Cell Biology, 6, 167-180. doi:10.1038/nrm1587
- Miyamoto, Y. and Yamauchi, J. (2010) The cellular signaling of Dock family in neural function. Cellular Signalling, 22, 175-182. doi:10.1016/j.cellsig.2009.09.036
- Donaldson, J.G. and Honda, A. (2005) Localization and function of Arf family GTPases. Biochemical Society Transactions, 33, 639-642.
- Kahn, R.A., Cherfils, J., Elias, M., Lovering, R.C., Munro, S. and Schurmann, A. (2006) Nomenclature for the human Arf family of GTP-binding proteins: ARF, ARL, and SAR proteins. Journal of Cell Biology, 172, 645-650. doi:10.1083/jcb.200512057
- D’Souza-Schorey, C. and Chavrier, P. (2006) ARF proteins: Roles in membrane traffic and beyond. Nature Reviews Molecular Cell Biology, 7, 347-358. doi:10.1038/nrm1910
- Casanova, J.E. (2007) Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic, 8, 1476-1485. doi:10.1111/j.1600-0854.2007.00634.x
- DiNitto, J.P., Delprato, A., Gabe Lee, M.T., Cronin, T.C., Huang, S., Guilherme, A., Czech, M.P. and Lambright, D.G. (2007) Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Molecular Cell, 28, 569-583. doi:10.1016/j.molcel.2007.09.017
- Brown, M.C. and Turner, C.E. (2004) Paxillin: Adapting to change. Physiological Reviews, 84, 1315-1339. doi:10.1152/physrev.00002.2004
- Torii, T., Miyamoto, Y., Sanbe, A., Nishimura, K., Yamauchi, J. and Tanoue, A. (2010) Cytohesin-2/ARNO, through its interaction with focal adhesion adaptor protein paxillin, regulates preadipocyte migration via the downstream activation of Arf6. Journal of Biological Chemistry, 285, 24270-24281. doi:10.1074/jbc.M110.125658
- Heo, W.D., Inoue, T., Park, W.S., Kim, M.L., Park, B.O., Wandless, T.J. and Meyer, T. (2006) PI(3,4,5)P3 and PI (4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science, 314, 1458-1461. doi:10.1126/science.1134389
- Yamauchi, J., Miyamoto, Y., Chan, J.R. and Tanoue, A. (2008) ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. Journal of Cell Biology, 181, 351-365. doi:10.1083/jcb.200709033
- Yamauchi, J., Miyamoto, Y., Torii, T., Mizutani, R., Nakamura, K., Sanbe, A., Koide, H., Kusakawa, S. and Tanoue, A. (2009) Valproic acid-inducible Arl4D and cytohesin-2/ARNO, acting through the downstream Arf6, regulate neurite outgrowth in N1E-115 cells. Experimental Cell Research, 315, 2043-2052. doi:10.1016/j.yexcr.2009.03.012
- Mizutani, R. Yamauchi, J., Kusakawa, S., Nakamura, K., Sanbe, A., Torii, T., Miyamoto, Y. and Tanoue, A. (2009) Sorting nexin 3, a protein upregulated by lithium, contains a novel phosphatidylinositol-binding sequence and mediates neurite outgrowth in N1E-115 cells. Cellular Signalling, 21, 1586-1594. doi:10.1016/j.cellsig.2009.06.005
- Klarlund, J.K., Holik, J., Chawla, A., Park, J.G., Buxton, J. and Czech, M.P. (2001) Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. Journal of Biological Chemistry, 276, 40065-40070. doi:10.1074/jbc.M105260200
- Nevrivy, D.J., Peterson, V.J., Avram, D., Ishmael, J.E., Hansen, S.G., Dowell, P., Hruby, D.E., Dawson, M.I. and Leid, M. (2000) Interaction of GRASP, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. Journal of Biological Chemistry, 275, 16827-16836. doi:10.1074/jbc.275.22.16827
- Kitano, J., Kimura, K., Yamazaki, Y., Soda, T., Shigemoto, R., Nakajima, Y. and Nakanishi, S. (2002) Tamalin, a PDZ domain-containing protein, links a protein complex formation of group 1 metabotropic glutamate receptors and the guanine nucleotide exchange factor cytohesins. Journal of Neuroscience, 22, 1280-1289.
- Mansour, M., Lee, S.Y. and Pohajdak, B. (2002) The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. Journal of Biological Chemistry, 277, 32302-32309. doi:10.1074/jbc.M202898200
- Boehm, T., Hofer, S., Winklehner, P., Kellersch, B., Geiger, C., Trockenbacher, A., Neyer, S., Fiegl, H., Ebner, S., Ivarsson, L., Schneider, R., Kremmer, E., Heufler, C. and Kolanus, W. (2003) Attenuation of cell adhesion in lymphocytes is regulated by CYTIP, a protein which mediates signal complex sequestration. EMBO Journal, 22, 1014-1024. doi:10.1093/emboj/cdg101
- Tang, P., Cheng, T.P., Agnello, D., Wu, C.Y., Hissong, B.D., Watford, W.T., Ahn, H.J., Galon, J., Moss, J., Vaughan, M., O’Shea, J.J. and Gadina, M. (2002) Cybr, a cytokine-inducible protein that binds cytohesin-1 and regulates its activity. Proceedings of the National Academy of Sciences USA, 99, 2625-2629. doi:10.1073/pnas.052712999
- Venkateswarlu, K. (2003) Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. Journal of Biological Chemistry, 278, 43460-43469. doi:10.1074/jbc.M304078200
- Ikenouchi, J. and Umeda, M. (2010) FRMD4A regulates epithelial polarity by connecting Arf6 activation with the PAR complex. Proceedings of the National Academy of Sciences USA, 107, 748-753. doi:10.1073/pnas.0908423107
- Lim, J., Zhou, M., Veenstra, T.D. and Morrison, D.K. (2010) The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes & Development, 24, 1496-1506. doi:10.1101/gad.1904610
- Bill, A., Schmitz, A., Albertoni, B., Song, J.N., Heukamp, L.C., Walrafen, D., Thorwirth, F., Verveer, P.J., Zimmer, S., Meffert, L., Schreiber, A., Chatterjee, S., Thomas, R.K., Ullrich, R.T., Lang, T. and Famulok, M. (2010) Cytohesins are cytoplasmic ErbB receptor activators. Cell, 143, 201-211. doi:10.1016/j.cell.2010.09.011
- Hofmann, I., Thompson, A., Sanderson, C.M. and Munro, S. (2007) The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Current Biology, 17, 711-716. doi:10.1016/j.cub.2007.03.007
- Li, C.C., Chiang, T.C., Wu, T.S., Pacheco-Rodriguez, G., Moss, J. and Lee, F.J. (2007) ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Molecular Biology of the Cell, 18, 4420-4437. doi:10.1091/mbc.E07-02-0149
- Oh, S. J. and Santy, L. C. (2010) Differential effects of cytohesins 2 and 3 on beta1 integrin recycling. Journal of Biological Chemistry, 285, 14610-14616. doi:10.1074/jbc.M109.043935
- Hafner, M., Schmitz, A., Grune, I., Srivatsan, S.G., Paul, B., Kolanus, W., Quast, T., Kremmer, E., Bauer, I. and Famulok, M. (2006) Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature, 444, 941-944. doi:10.1038/nature05415
- Fuss, B., Becker, T., Zinke, I. and Hoch, M. (2006) The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature, 444, 945-948. doi:10.1038/nature054
NOTES
*These authors contributed equally to this work.
#Corresponding author.

