Journal of Biophysical Chemistry
Vol.3 No.2(2012), Article ID:19451,5 pages DOI:10.4236/jbpc.2012.32023
Crystal structure of PqqB from Pseudomonas putida at 2.2 Å resolution
![]()
1Department of Molecular Biology, University of Salzburg, Salzburg, Austria; *Corresponding Author: fisher@ill.fr
2Helmholtz-Zentrum Berlin for Materials and Energy, Electron Storage Ring BESSY II, Berlin, Germany 3Institut Laue Langevin, Grenoble, France
Received 8 December 2011; revised 17 January 2012; accepted 15 March 2012
Keywords: PqqB; PQQ; Biosynthetic Pathway; X-Ray Crystallography
ABSTRACT
Pyrroloquinoline quinone (PQQ) is an important redox-active cofactor for many bacterial dehydrogenases. It’s biosynthetic pathway involves six or seven genes, one of which is pqqB. Former studies indicated that the protein encoded by pqqB, namely PqqB, functions as a PQQ transporter. Here we report the crystal structure of PqqB from Pseudomonas putida at 2.2 Å resolution together with functional studies to verify this theory.
1. INTRODUCTION
Many bacterial dehydrogenases (e.g. ethanol and glucose dehydrogenase) use Pyrroloquinoline quinone (PQQ) as a cofactor to oxidize their substrates (to ethanol and glucose respectively) [1]. PQQ has attracted considerable interest because of its presence in foods, its antioxidant properties, and its role as a growth-promoting factor [2, 3]. However its detailed physiological role still requires further investigation [4,5]. PQQ belongs to the orthoquinone redox cofactor family. Six or seven genes in the PQQ-operon [6] are required for its biosynthesis from the amino acids glutamate and tyrosine, which are encoded in the precursor peptide PqqA [7].
In an attempt to elucidate the biosynthetic pathway of PQQ the reported PQQ-transport-protein PqqB [8] was purified and crystallized. Previous studies [9] found that PqqB knock-out cells accumulate PQQ in the cytosol, but in lower yield than in wild type cells. This led to the conclusion that PqqB is not required for PQQ biosynthesis, but serves as a carrier that transports PQQ across the plasma-membrane to the periplasm [8,9]. Further evidence for this hypothesis is the fact that PqqC, the protein responsible for the penultimate step of PQQ biosynthesis, requires an acceptor for PQQ. This is because free PQQ readily reacts with oxygen to generate free radicals, and additionally the PqqC reaction is severely product inhibited [10,11]. Although the structure of PqqB has been previously deposited (PDB code 1TXO), no description of its structural features have been published. Here we report the detailed analysis of the X-ray structure of PqqB at 2.2A (higher resolution than previously deposited), along with the description of conserved residues and a putative binding site.
2. RESULTS
PqqB from Pseudomonas putida is a protein with 303 residues, a corresponding molecular mass of 33.3 kD, and a theoretical pI of 5.4. The protein was cloned, overexpressed in E. coli, purified, and crystallized. PqqB crystallizes in the tetragonal space group P 43212 having cell dimensions of a = b = 86.21 Å, c = 109.41 Å, with one molecule in the asymmetric unit. The structure was solved by molecular replacement using PDB entry 1XTO as the starting model. Statistics for data collection are shown in Table 1, and refinement and model quality are summarized in Table 2.
The structure reveals a half-moon shaped molecule with a metallo-hydrolase/oxidoreductase fold and approximate molecular dimension of 42 × 39 × 39 Å (see Figure 1). The model exhibits good geometry with one Ramachandran outlier, which is strongly supported by the electron density.
PqqB contains a cavity about 25 Å in length and 10 Å in diameter, which is lined by a number of strictly conserved residues (H93, H269 and Y169). The end of this cavity contains a metal ion tetrahedrally coordinated by three cysteines (C19, C21, and C24) and one asparagine (N272) (see Figure 2), all of which are strictly conserved. A search of the “metal coordination sites in proteins server” [12] showed that the coordination of this ion in PqqB is unique. The usual zinc coordination is tetrahedral with 3 cysteines and one histidine. The usual iron
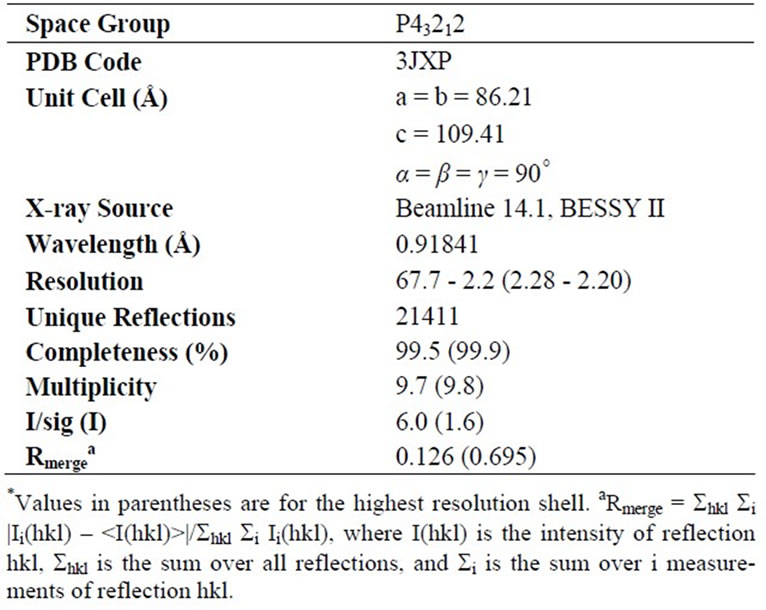
Table 1. Data statistics.
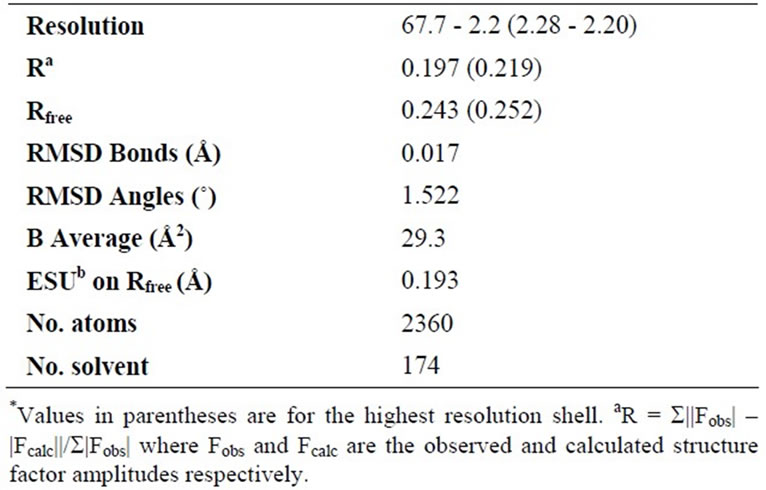
Table 2. Refinement parameters and statistics.
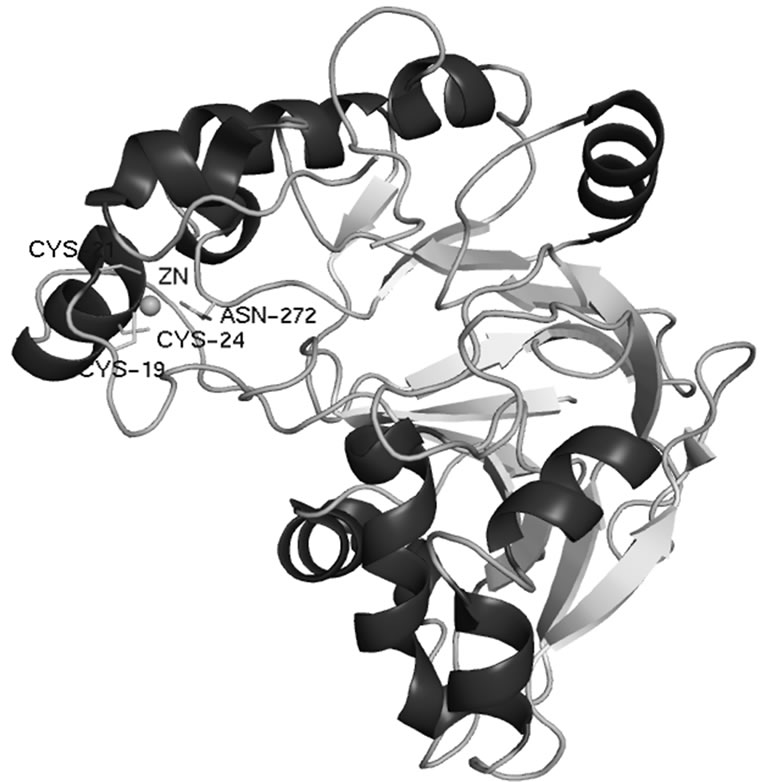
Figure 1. Crystal structure of PqqB from Pseudomonas putida (PDB code 3JXP) shown in ribbon representation.
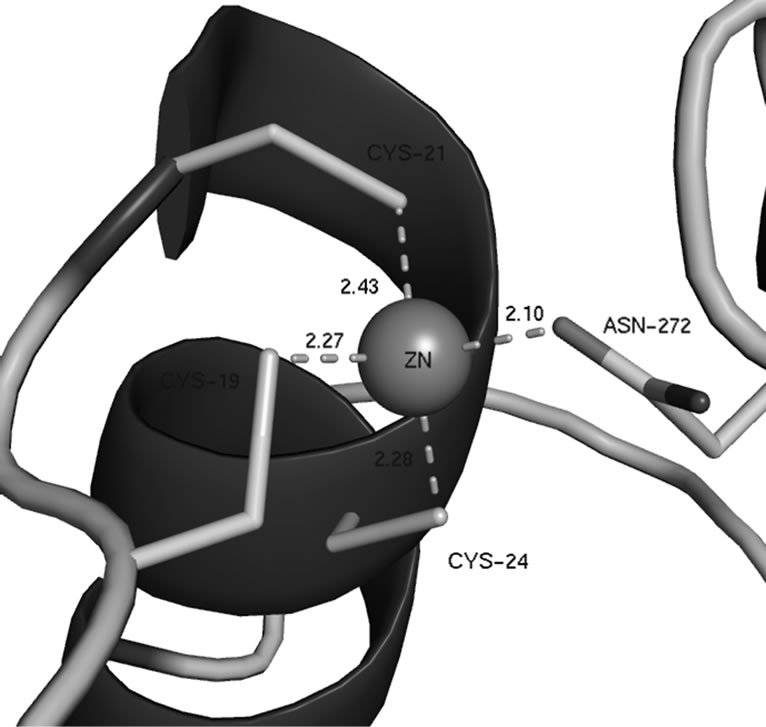
Figure 2. Close up of the metal coordination site in PqqB.
coordination is tetrahedral with 4 cysteines, although this is due mainly to the many Fe-S cluster containing proteins. In proteins containing non-heme mononuclear iron centres the preferred coordination is octahedral or trigonal bipyramidal, such as is the case for the 2 His-1 carboxylate facial triad family [13].
The electron density for the ion is consistent with both Zn2+ and Fe2+. The previously deposited structure (PDB code 1TXO) shows PqqB with Zn2+ bound. To determine which metal ion is present in the protein initially Inductive Coupled Plasma Mass Spectrometry (ICP-MS) experiments were performed. These experiments revealed that freshly purified PqqB reconstituted with Fe2+ (see methods section) has mostly iron (~80%), but also some zinc bound (~20%). Protein that was stored for several days showed mostly zinc bound. These results indicate that native PqqB actually contains Fe2+ which is replaced by Zn2+ over time. The Fe2+ ion is located at the end of the cavity and therefore this is proposed to be the active site with a putative substrate binding pocket that could easily harbour PQQ or a PQQ-precursor molecule (see Figure 3).
Structural and bioinformatical analysis (see methods section), reveal a significant similarity to Protein phnP (UniProt: P16692, sequence identity of 17%) and Ribonuclease Z (UniProt: P54548, sequence identity of 14%). It also showed a significant similarity to coenzyme F420H2 oxidase from Methanobacteriumthermoautotrophicum (UniProt: Q50497, sequence identity 10%) with the Metallo-beta-lactamase fold. F420H2 is however a diiron containing flavoenzyme [14] whereas PqqB contains only a mononuclear iron site. Therefore although their sequences may be comparable, the chemistry that they catalyse is not.
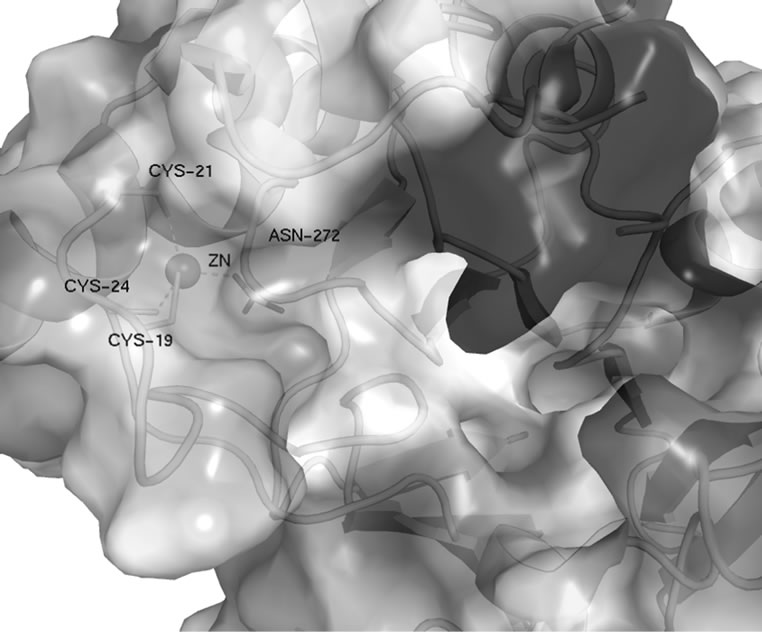
Figure 3. Putative substrate binding pocket with computed electrostatic surface.
The N-terminus of the protein is interesting as it contains a unique glycine rich region (7-GSAAGGG-13) which is a part of the putative substrate binding pocket. The flexibility of the four glycine residues could play a role as a hinge in conformational changes facilitating substrate recognition and binding.
In order to test the hypothesis of PqqB acting as a PQQ-transporter protein a number of binding studies were performed. These included fluorescence and UV/Vis spectrometry, as well as pull down assays (see methods section). The fluorescence scans showed that neither PqqB with Zn2+, nor PqqB reconstituted with Fe2+ bind PQQ, which would be indicated by a shift of the PQQ fluorescence to a shorter wavelength, as is observed for binding of PQQ to PqqC. The same results were obtained from UV/Vis spectra of samples of PqqB incubated with PQQ. To test the binding of PqqB to PqqC or the PqqC/PQQ complex His-tag pull down assays were performed with PqqB containing Zn2+ or Fe2+ as prey proteins. These experiments also showed that PqqB does not bind to either PqqC or the PqqC/PQQ complex. Finally the binding of PqqB to the PQQ precursor peptide PqqA was also tested. This experiment again showed no binding.
3. DISCUSSION
Previous studies [9] indicated that PqqB may be required for transport of PQQ to the periplasm, given the reduced amount of PQQ produced in cells lacking PqqB. The studies here however show that PqqB does not bind PQQ either alone or in complex with PqqC, nor does it bind to PqqA. Although our data indicates that PqqB does not transport PQQ under the conditions tested, these negative results do not exclude the possibility that PqqB could still be a transporter.
There is conflicting information in the literature regarding the function of PqqB, although regarded as a transporter protein [8], knock out cells lacking PqqB produced a similar intermediate to that found in cells lacking PqqC, however this intermediate cannot be converted to PQQ upon addition of PqqC [9]. This indicates that the intermediate produced from PqqB lacking cells is structurally different to the intermediate produced in PqqC lacking cells. However some PQQ is still produced in these PqqB lacking cells, indicating that the PQQ biosynthetic pathway is not completely hindered.
Recent studies on another protein from the PQQ operon, PqqD [15] seem to indicate that PqqD may actually be a PQQ transporter, as it features a novel saddle fold, capable of binding PQQ between two molecules forming a dimer. The function of PqqD was previously unidentified.
The precise function of PqqB still remains elusive, future studies will focus on the binding of PqqB to other gene products from the PQQ operon, as well as the binding of PqqB to PQQ when reconstituted with other metals.
4. MATERIALS AND METHODS
4.1. Bioinformatic Analysis
Bioinformatic analyses were carried out using the FFAS server (http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) [16], the Tempura server (http://www.ebi.ac.uk/thornton-srv/databases/tempura/) [17], the “metal coordination sites in proteins” server (http://tanna.bch.ed.ac.uk/) [12], and the Top-Match Server (http://topmatch.services.came.sbg.ac.at/) [18,19].
4.2. Inductive Coupled Plasma Mass Spectrometry
Two different samples of PqqB were prepared, an untreated one that was measured directly after purification, and a second where the metal was chelated with a 10-fold excess of EDTA after Size Exclusion Chromatography (SEC). Excess EDTA was removed by dialysis. The sample was then incubated with a 10-fold excess of Fe2+. Excess of iron was removed by dialysis. The two samples were analysed for zinc and iron content using an ICP-MS instrument (Elan 6100 DRC, Perkin Elmer) at the WACKER Analytic Lab in Burghausen, Germany.
4.3. Expression and Purification of PqqB
The PqqB gene from Pseudomonas putida (GenBank ID: GQ913853) was cloned in expression vector pET 21d (Invitrogen, Carlsbad) and expressed in E. coli strain BL21 (DE3) star. Cells were induced with 0.5 mM IPTG (25˚C overnight) harvested and frozen at –20˚C. After cell lysis the crude extract was purified on a Ni-affinity column (Pharmacia). The fractions containing the PqqB protein were pooled and applied to a Superdex 75 gel filtration column (Pharmacia). The protein was concentrated using a centrifugal concentrator (Amicon) to about 10 mg/ml in a buffer containing 50 mM Tris pH 7.5, 150 mM NaCl and 5 mM DTT.
4.4. Fluorescence Analysis
Binding of PQQ to PqqB was measured fluorimetrically on a Tecan Infinite M200 microplate reader. The PqqB/PQQ complex should form by mixing equimolar amounts of protein and PQQ (Sigma, D7783) following incubation for 15 minutes at 25˚C. The absorbance spectrum of PQQ shows a characteristic maximum at 340 nm, corresponding to the absorbance maximum of the quinoline-quinone moiety. The fluorescence spectrum was recorded from 370 nm to 650 nm with an excitation wavelength at 340 nm.
4.5. Pull Down Assay
To test the binding to PqqC the bait protein PqqC (with His-Tag) was immobilized on a Ni-affinity column. Afterwards the prey protein PqqB (without His-Tag) was applied to the column. After an incubation for 30 minutes at room temperature the beads were washed with wash buffer (20 mM Tris pH 7.8, 300 mM NaCl, 5 mM Imidazole).
4.6. Crystallisation
Crystals were grown at 20˚C using the sitting drop vapor diffusion method in 2 µL droplets composed of one part protein solution and one part reservoir solution containing of 0.1 M TRIS hydrochloride pH 8.5, 0.2 M magnesium chloride hexahydrate, 15% w/v PEG 4000 (Hampton Pre-Crystallization Test, Reagent A2, Hampton Research). The crystals are of space group P 43212 and contain one molecule in the asymmetric unit. For data collection, these crystals were transferred into cryobuffer (crystallization buffer with 30% (v/v) sucrose) and flash-cooled in liquid nitrogen.
4.7. Data Collection, Structure Solution, and Refinement
Data were collected at BESSYII-MX BL14.1 (Berlin, Germany) [20]. Oscillation data were recorded in frames of 1˚ through a continuous angular range of 120˚. All data were processed with the HKL-2000 package [21]. The structure was solved using MOLREP [22] with PDB entry 1XTO as the starting model. The PqqB crystal structure was also solved by Forouhar et al. (PDB ID: 1XTO, unpublished) from the Northeast Structural Genomics Consortium, but we report the crystal structure at a significantly better resolution (2.2 Å). Crystallographic refinement was carried out using TLS refinement in REFMAC5.2 [23] and model modifications were made with the model building program Coot [24]. The PqqB structure is a monomer with 304 residues, 174 water molecules and one zinc ion. No electron density was observed for residue E241. Data collection parameters are show in Table 1 and detailed refinement statistics in Table 2. All figures were prepared with PYMOL [25].
4.8. Coordinate Deposition
Coordinates and structure factors for PqqB have been deposited in the Protein Data Bank (http://pdbbeta.rcsb.org/pdb/home/home.do) under accession code 3JXP.
5. ACKNOWLEDGEMENTS
We would like to thank Dr. Guido Kallinger and the analytic lab of Wacker Chemie, Burghausen for measuring the metal concentrations. We acknowledge the Helmholtz-Zentrum Berlin-Electron storage ring BESSY II for provision of synchrotron radiation at beamline 14.1, and would like to thank Dr. Uwe Müller for assistance. This work and SF were supported by FWF grant P22862 from the Austrian Science Fund.
REFERENCES
- Anthony, C. (2001) Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxidants & Redox Signaling, 3, 757-774. doi:10.1089/15230860152664966
- Stites, T.E., Mitchell, A.E. and Rucker, R.B. (2000) Physiological importance of quinoenzymes and the o-quinone family of cofactors. The Journal of Nutrition, 130, 719-727.
- Zhang, Y. and Rosenberg, P.A. (2002) The essential nutrient pyrroloquinoline quinone may act as a neuroprotectant by suppressing peroxynitrite formation. European Journal of Neuroscience, 16, 1015-1024. doi:10.1046/j.1460-9568.2002.02169.x
- Felton, L.M. and Anthony, C. (2005) Biochemistry: Role of PQQ as a mammalian enzyme cofactor? Nature, 433, E10-E10. doi:10.1038/nature03322
- Rucker, R., Storms, D., Sheets, A., Tchaparian, E. and Fascetti, A. (2005) Biochemistry: Is pyrroloquinoline quinone a vitamin? Nature, 433, E10-E11. doi:10.1038/nature03323
- Meulenberg, J.J.M., Sellink, E., Riegman, N.H. and Postma, P.W. (1992) Nucleotide sequence and structure of the klebsiella pneumoniae PQQ-operon. Molecular and General Genetics MGG, 232, 284-294.
- Goosen, N., Huinen, R.G. and Van de Putte, P. (1992) A 24-amino-acid polypeptide is essential for the biosynthesis of the coenzyme pyrrolo-quinoline-quinone. Journal of Bacteriology, 174, 1426-1427.
- Goosen, N., Horsman, H.P., Huinen, R.G. and Van de Putte, P. (1989) Acinetobacter calcoaceticus genes involved in biosynthesis of the coenzyme pyrrolo-quinoline-quinone: Nucleotide sequence and expression in escherichia coli k-12. Journal of Bacteriology, 171, 447-455.
- Velterop, J.S., et al. (1995) Synthesis of pyrroloquinoline quinone in-vivo and in-vitro and detection of an intermediate in the biosynthetic-pathway. Journal of Bacteriology, 177, 5088-5098.
- Aizenman, E., Hartnett, K., Zhong, C., Gallop, P. and Rosenberg, P. (1992) Interaction of the putative essential nutrient pyrroloquinoline quinone with the n-methyl-d-aspartate receptor redox modulatory site. The Journal of Neuroscience, 12, 2362-2369.
- Magnusson, O.T., et al. (2004) Quinone biogenesis: Structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proceedings of the National Academy of Sciences of the United States of America, 101, 7913-7918. doi:10.1073/pnas.0402640101
- Harding, M. (2004) The architecture of metal coordination groups in proteins. Acta Crystallographica Section D, 60, 849-859. doi:10.1107/S0907444904004081
- Hegg, E.L. and Jr, L.Q. (1997) The 2-his-1-carboxylate facial triad—An emerging structural motif in mononuclear non-heme iron(II) enzymes. European Journal of Biochemistry, 250, 625-629. doi:10.1111/j.1432-1033.1997.t01-1-00625.x
- Seedorf, H., et al. (2007) Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic archaea catalyzing the reduction of O2 to H2O. FEBS Journal, 274, 1588-1599. doi:10.1111/j.1742-4658.2007.05706.x
- Tsai, T.-Y., Yang, C.-Y., Shih, H.-L., Wang, A.H.J. and Chou, S.-H. (2009) Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins: Structure, Function, and Bioinformatics, 76, 1042-1048. doi:10.1002/prot.22461
- Jaroszewski, L., Rychlewski, L., Li, Z., Li, W. and Godzik, A. (2005) FFAS03: A server for profile—profile sequence alignments. Nucleic Acids Research, 33, W284-W288.
- Laskowski, R.A., Watson, J.D. and Thornton, J.M. (2005) Protein function prediction using local 3D templates. Journal of Molecular Biology, 351, 614-626. doi:10.1016/j.jmb.2005.05.067
- Sippl, M.J. (2008) On distance and similarity in fold space. Bioinformatics, 24, 872-873. doi:10.1093/bioinformatics/btn040
- Sippl, M.J. and Wiederstein, M. (2008) A note on difficult structure alignment problems. Bioinformatics, 24, 426-427. doi:10.1093/bioinformatics/btm622
- Mueller, U., et al. (2012) Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. Journal of Synchrotron Radiation, 19, 442-449.
- Otwinowski, Z. and Minor, W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology, 276, 307-326. doi:10.1016/S0076-6879(97)76066-X
- Vagin, A.A. and Teplyakov, A. (1997) Molrep: An automated program for molecular replacement. Journal of Applied Crystallography, 30, 1022-1025. doi:10.1107/S0021889897006766
- Murshudov, G.N., Vagin, A.A. and Dodson, E.J. (1997) Refinement of macromolecular structures by the maxi-mumlikelihood method. Acta Crystallographica Section D, 53, 240-255. doi:10.1107/S0907444996012255
- Emsley, P. and Cowtan, K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography, 60, 2126-2132. doi:10.1107/S0907444904019158
- DeLano, W.L. (2002) The PyMol molecular graphics system. http://www.pymol.org
NOTES
#Equal Contributors.

