International Journal of Astronomy and Astrophysics
Vol.3 No.3(2013), Article ID:36519,12 pages DOI:10.4236/ijaa.2013.33035
Analysis of the Effect of Space Radiations on the Nematode, Caenorhabditis elegans, through the Simulated Space Radiation
1Korea Aerospace Research Institute (KARI), Daejeon, Republic of Korea
2Korea Advanced Institute of Science and Technology, Daejeon, Republic of Korea
Email: *Ksyeon@kari.re.kr
Copyright © 2013 Soyeon Yi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received June 4, 2013; revised July 6, 2013; accepted July 14, 2013
Keywords: Astronaut; International Space Station; Space Experiment; Space Radiation; C. elegans; Microarray
ABSTRACT
In this study, we simulated space flight of the nematode, Caenorhabditis elegans, on the ground and examined how it is affected by space radiation and G-forces. We simulated G-forces during launch in a gravity acceleration laboratory device in order to identify and isolate the effects of the G-forces. Following this, we irradiated C. elegans with accelerated protons (MC-50 Cyclotron) and gamma rays (iR 222 machine) at the same physical dose. We calculated the expected radiation dose according to Reitz [1] and simulation programs (NASA AP8MIN [2], NASA AE8MAX [2], and CREAM86 [3]) for 1 month (dose rate: 6 × 10−3 Gy; 2.8 × 10−2 Gy), 6 months (dose rate: 36 × 10−3 Gy; 16.8 × 10−2 Gy), and 2 years (dose rate: 144 × 10−3 Gy; 67.2 × 10−2 Gy) of space flight. There have been several trials that aimed to take C. elegans into orbit on US space shuttle missions including a mission on the shuttle Columbia. In this study, we simulated longer duration space flights and performed a whole-genome microarray analysis to observe phenotype variations whereas most such experiments were carried out during short duration space flights and focused on mutations and genotypic variations. We expect that the results of this study will be useful to predict the effects of long-term exposure of space radiation on living organisms.
1. Introduction
There are many risks to humans when they explore space, and many researchers have studied the effects of space radiations. Radiations exert several effects on living organisms, such as mutations, apoptosis, death, and behavioral changes [4]; however, the specific mechanisms underlying these effects are still unclear.
In this study, we analyzed the effects of space radiation and G-forces on the nematode Caenorhabditis elegans a ground simulation of the long duration space flight. The nematode, C. elegans, has a variety of features that make it an attractive model for space experiments. In particular, C. elegans is easy to culture and is a self-fertilizing organism. Above all else, many of the developmental and biochemical pathways of C. elegans are conserved in humans [5]. Some space flight experiments involving live C. elegans have been carried out in space. On STS-42, C. elegans was examined in terms of how it is affected by space radiation. An 8-fold increase in the mutation rate was observed with large chromosomal rearrangements being found at a higher frequency in space-flown worms compared to ground controls [6,7]. On STS-76, increased mutations induced by low earth orbit space radiation were observed and were shown to be the direct consequence of space radiation. A 3.3-fold increase in the mutation rate in that region alone and the mutations were occurred due to large-scale alterations in their chromosome structure. In addition, G-forces exerted only a marginal effect [6]. On STS-95, all of the species died, therefore, no results were obtained. On STS-107, the Columbia space shuttle crashed and only 3 canisters with living C. elegans were found [8]. ICE-First has new organism hardware, CeMM, so Biologist wants to check new hardware performance. And they confirmed CeMM validates. The effects on nematodes that were grown for 3 - 4 generations in CeMM media and in space for 10 d were analyzed. In this experiment, space-faring animals underwent normal development, apoptosis [9], and DNA repair [10], but experienced altered muscle development [11]. Because it can sustain a high degree of radiation damage, C. elegans can be used as a potential biological dosimeter [12]. There are many changes in genes that are associated with ageing, oxidative stress, and muscle growth were observed, using C. elegans habitat hardware [13], because of long term flight. Our study examines the influence of space radiation on C. elegans for long-duration space flight, and the effects of G-force during launch on C. elegans.
2. Materials and Methods
2.1. Preparation of C. elegans
Caenorhabditis elegans was cultured according to standard methods [14]. Worms were cultured on nematode growth medium (NGM) agar plates that were spread with Escherichia coli K-12 strain MG1655 as a food source. The worms were grown at 20˚C. Three hundred young adult worms were used for each experiment.
2.2. Calculation of Space Radiation
We used 2 methods to calculate the space radiation dose. First, based on a paper of the G. Reitz group [1], the space radiation dose on the International Space Station (ISS) was estimated to be 151 μGy to 231 μGy per day, and then we assumed an average dose rate of 200 μGy per day. Second, we simulated the space radiation with the programs NASA AP8MIN [2], NASA AE8MAX [2], and CREAM86 [3]. Through this method, we estimated the dose rate to be 100 Rad for 3 y. We simulated oversampling radiation using assumptions for aluminum thickness. In addition, we assumed the duration of a space flight such as a shuttle mission of 30 d, a Soyuz mission of 180 d (6 months), and a Mars mission of 730 d (2 years), and used the estimated radiation dose rates noted in Table 1.
We used 2 types of radiation: accelerated protons and gamma rays. Following irradiation, we compared the common expressed genes for the protons and gamma rays. In addition, we analyzed the effects of G-forces and space radiation, which was composed of protons and gamma rays, on commonly expressed genes.
First, we carried out irradiation using 3 types of accelerated protons, 144 × 10−3 Gy, 16.8 × 10−2 Gy, and 33.6 × 10−2 Gy, on C. elegans using an MC-50 Cyclotron. The MC-50 Cyclotron could not adjust to a tiny dose rate so
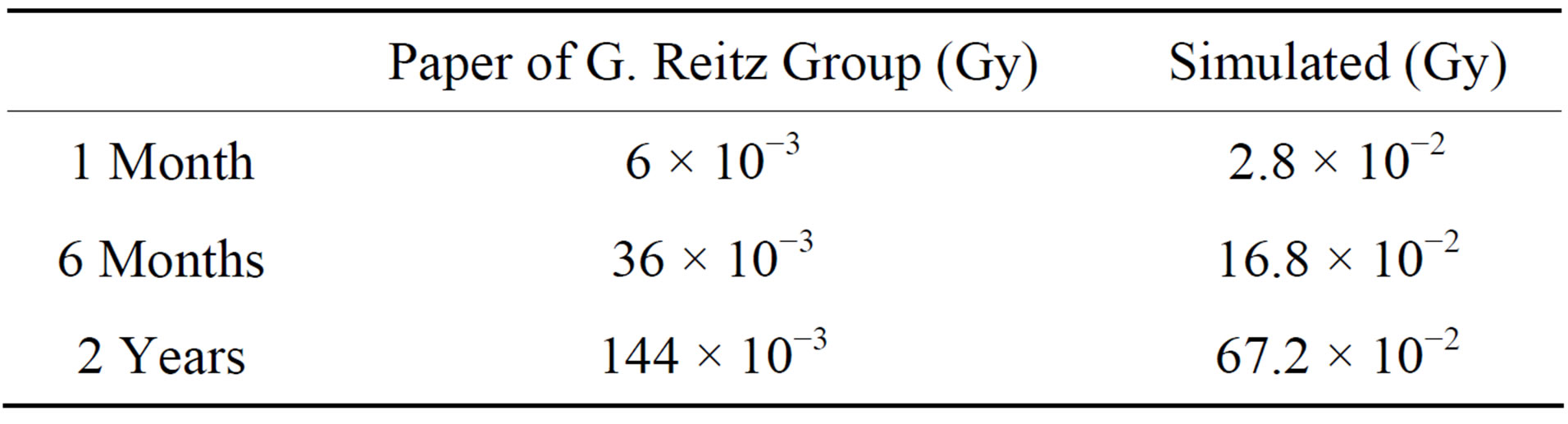
Table 1. Space radiation dose.
we only experimented with 3 types of radiation. Second, we carried out irradiation using 6 types of gamma rays 6 × 10−3 Gy, 36 × 10−3 Gy, 144 × 10−3 Gy, 2.8 × 10−2 Gy, 16.8 × 10−2 Gy, and 67.2 × 10−2 Gy, on C. elegans using an iR 222 machine. We carried out experiments on C. elegans by varying the exposure distance at the same time. Figure 1 shows our experiment setup and Table 2 shows the gamma rays produced by iR 222 machine. After exposure to radiation, C. elegans was collected, frozen in liquid nitrogen, and kept at −70˚C until the RNA extraction procedure was carried out.
2.3. G-Force Experiment
We performed experiments on C. elegans according to private communication Soyuz User’s manual-Issue3 (2002), as shown in Figure 2, by using the acceleration equipment shown in Figure 3.
2.4. Microarray Analysis
RNA was extracted by following the Trizol method of the Trizol Molecular Research Center. For control and test RNA, the synthesis of target cRNA probes and hybridization were carried out using Agilent’s Low RNA Input Linear Amplification kit (Agilent Technology, USA) according to the manufacturer’s instructions. Briefly, 1 μg of total RNA and T7 promoter primer were mixed and


Figure 1. MC-50 cyclotron (left), iR 222 machine (right).
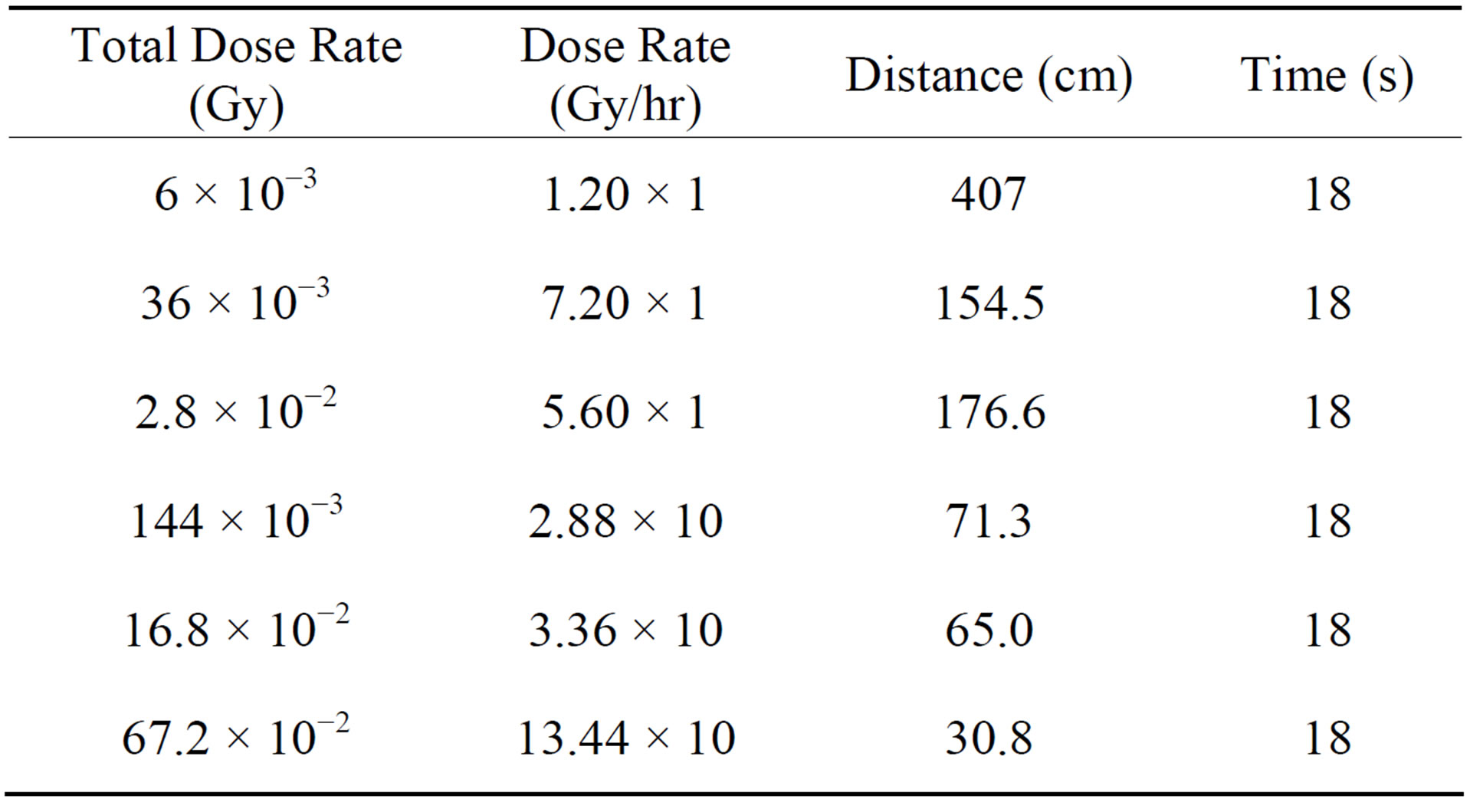
Table 2. Gamma radiations produced by the iR 222 machine.
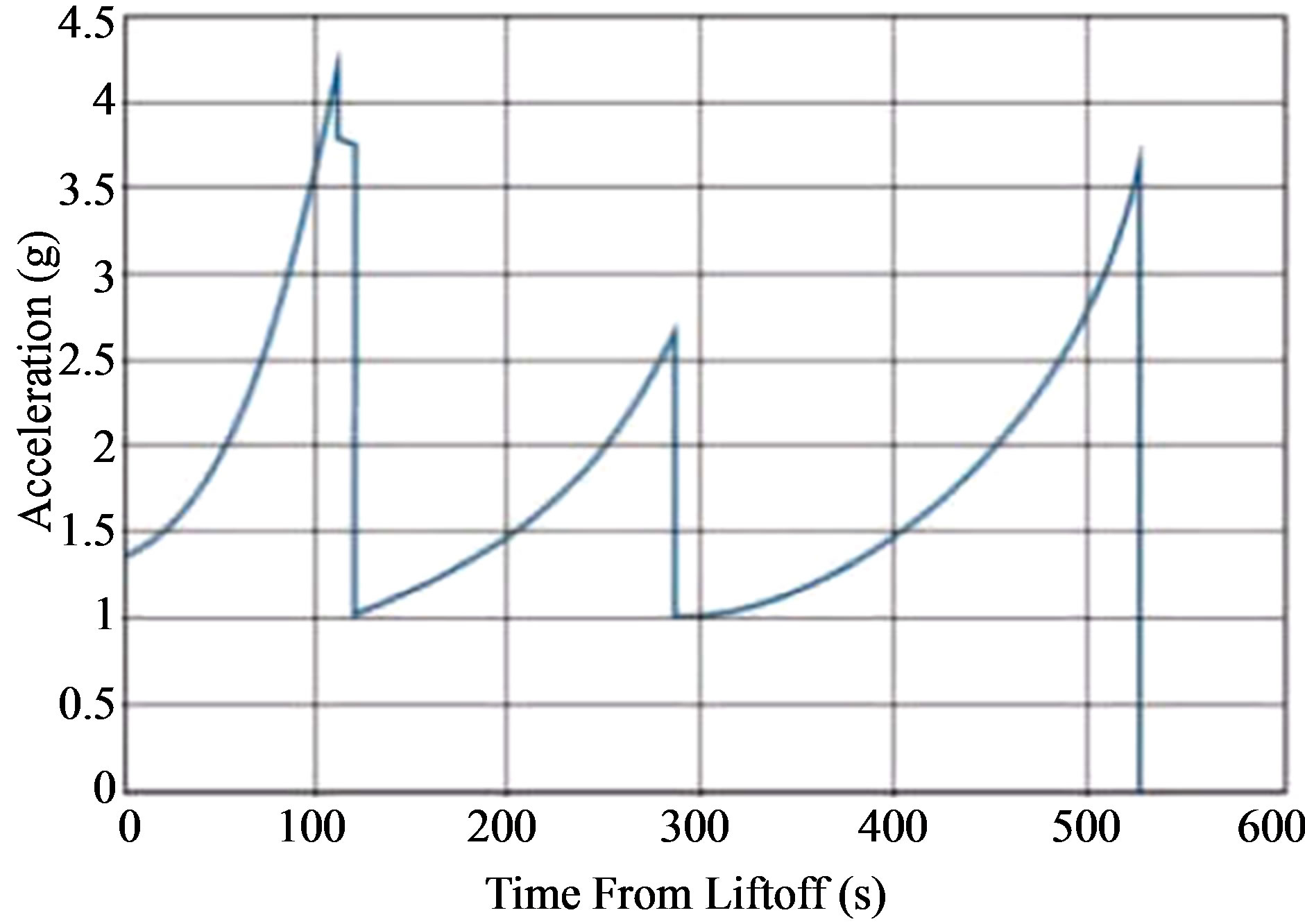
Figure 2. G-force profile for Soyuz launch.

Figure 3. Acceleration equipment.
incubated at 65˚C for 10 min. The cDNA master mix (5 × First strand buffer, 0.1 M DTT, 10 mM dNTP mix, RNAe-Out, and MMLV-RT) was prepared and added to the reaction mixture. The samples were incubated at 40˚C for 2 h, and then dsDNA synthesis was terminated by incubating at 65˚C for 15 min. The transcription master mix was prepared according to the manufacturer’s protocol (4 × transcription buffer, 0.1 M DTT, NTP mix, 50% PEG, RNAe-Out, inorganic pyrophosphatase, T7- RNA polymerase, and Cyanine 3/5-CTP). Transcription of dsDNA was performed by adding the transcription master mix to the dsDNA reaction samples and incubating them at 40˚C for 2 h. The amplified and labeled cRNA was purified using a cRNA Cleanup Module (Agilent Technology) according to the manufacturer’s protocol. Labeled cRNA target was quantified using a ND- 1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). After checking the labeling efficiency, fragmentation of the cRNA was performed by adding 10 × blocking agent and 25 × fragmentation buffer and incubating the mixture at 60˚C for 30 min. The fragmented cRNA was re-suspended with 2 × hybridization buffer and directly pipetted onto an assembled Agilent’s C. elegans Oligo Microarray (44 K). The arrays were hybridized at 65˚C for 17 h using an Agilent Hybridization oven (Agilent Technology, USA). The hybridized microarrays were then washed according to the manufacturer’s protocol (Agilent Technology, USA).
2.5. Data Acquisition and Analysis
The hybridized images were scanned using Agilent’s DNA microarray scanner and quantified by using Feature Extraction Software (Agilent Technology, Palo Alto, CA). All data normalization and selection of fold-changed genes were performed using GeneSpringGX 7.3 (Agilent Technology, USA). The averages of the normalized ratios were calculated by dividing the average of the normalized signal channel intensity by the average of the normalized control channel intensity. Functional annotation of the genes was performed according to Gene OntologyTM Consortium [14] by GeneSpringGX 7.3. Gene classification was based on searches done by Bio-Carta [15], GenMAPP[16], and DAVID[17]. We identified the functions of genes based on WormBase.
3. Results and Discussion
3.1. Effects of Estimated Long-Term Exposure of Space Radiation in C. elegans
We analyzed the effect of estimated long-term exposure to space radiation on the global gene profiles of C. elegans. To gain insight into the biological processes associated with the regulated genes, we determined which GO annotation terms were over-represented. Further, we categorized these effects 3 groups: DNA repair, oxidative stress, and cell death. We analyzed all of the genes that were regulated upon proton and gamma ray, and set the fold cut-off value at 2.0. Later we compared the 2-fold up/down-regulated common genes between proton and gamma ray-irradiated C. elegans.
3.2. Changes in C. elegans Genes Due to the Effects to Protons Radiation
We examined 1 month, 6 months and 2 years radiation dose according to paper of Reitz and simulation program. However, there are few changes 1 month and 6 months radiation dose according to Reitz group and 1 month radiation dose according to simulation program. Therefore we only show 2 years radiation dose according to Reitz group and 6 months and 2 years radiation dose according to simulation program.
3.2.1. DNA Repair
We analyzed the changes in the C. elegans genes that were induced by the effects to protons irradiation on as shown in Table 3 and found that 3 up-regulated genes are related to DNA repair functions. This category includes rad-23, msh-5, and rpt-4. rad-23 encodes a proteasomal ubiquitin receptor that is related to axon
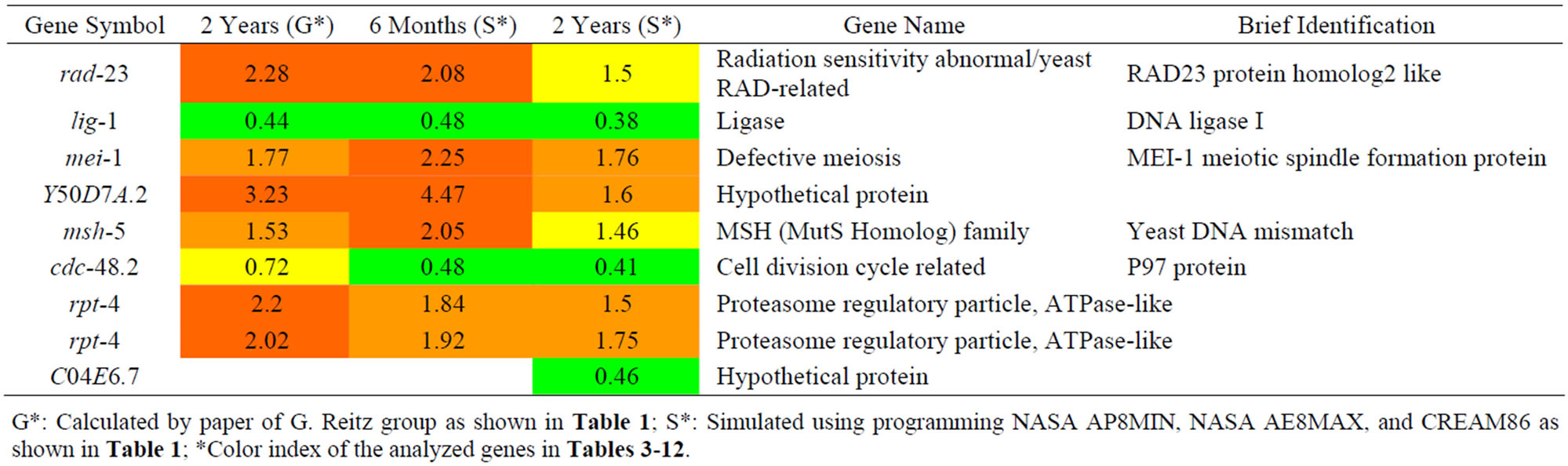
Table 3. Results of a microarray analysis that shows how genes influenced by proton irradiation that are related to DNA repair were up/down-regulated after one dose of simulated radiation at each dose rate.
G*: Calculated by paper of G. Reitz group as shown in Table 1; S*: Simulated using programming NASA AP8MIN, NASA AE8MAX, and CREAM86 as shown in Table 1; *Color index of the analyzed genes in Tables 3-12.
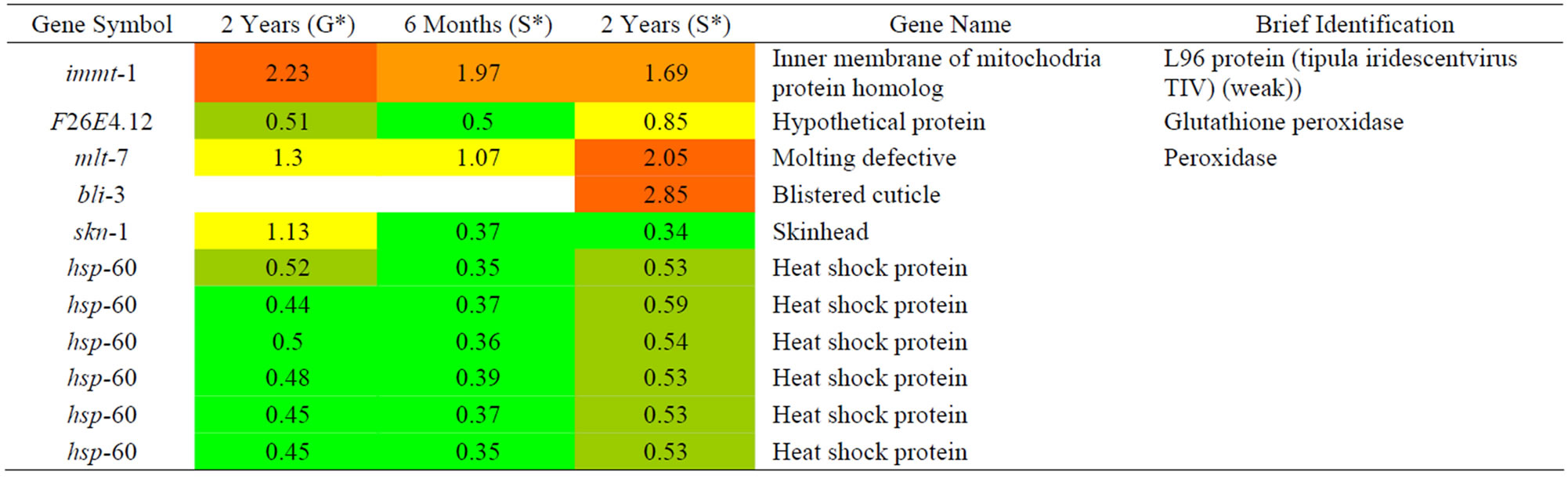
Table 4. Results of a microarray analysis that shows how genes that are related to oxidation repair were up/down-regulated after one dose of simulated gamma ray irradiation at each dose rate.
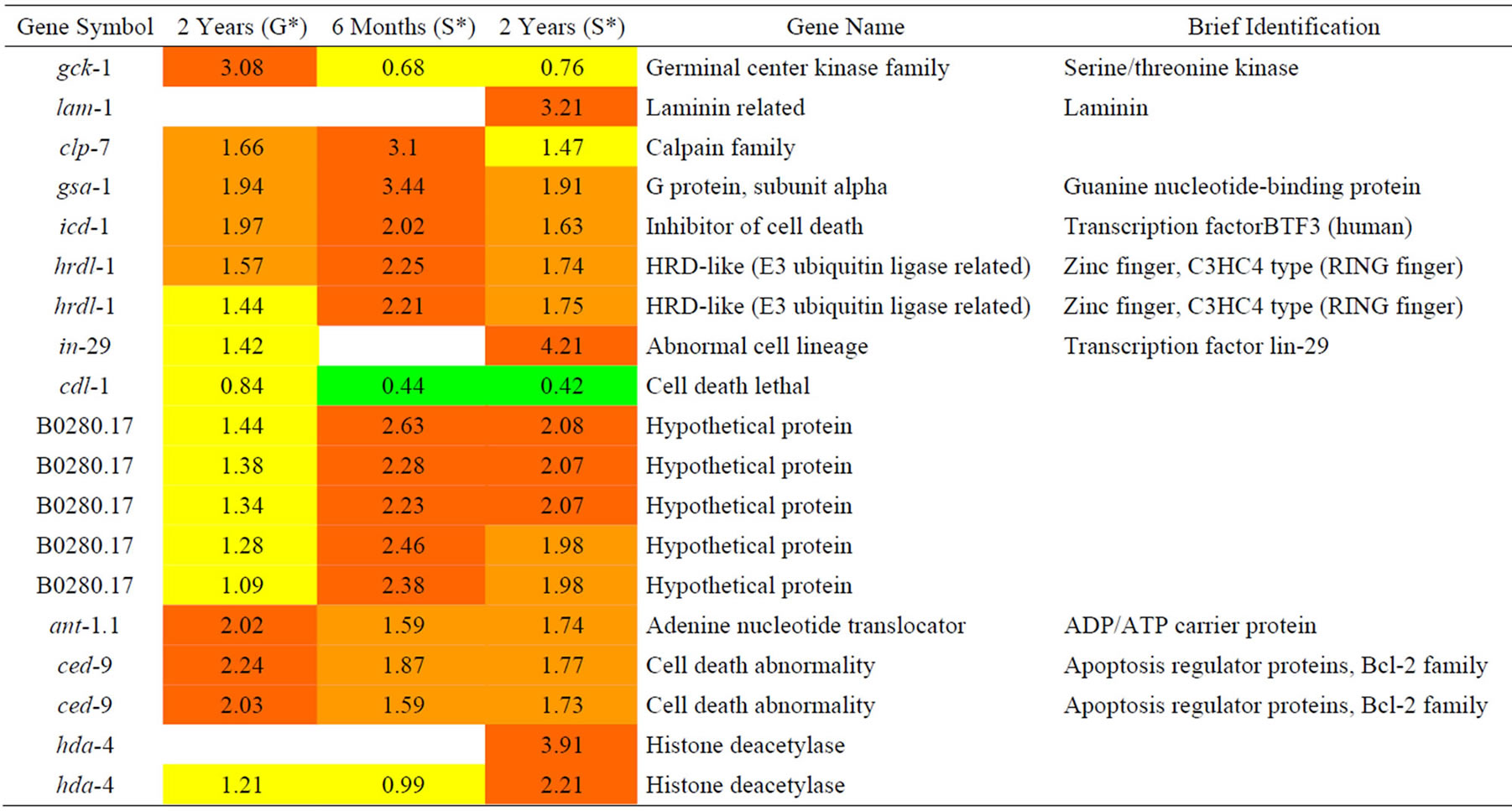
Table 5. Results of a microarray analysis that shows how genes that are related to cell death were up/down-regulated after one dose of simulated gamma ray irradiation at each dose rate.
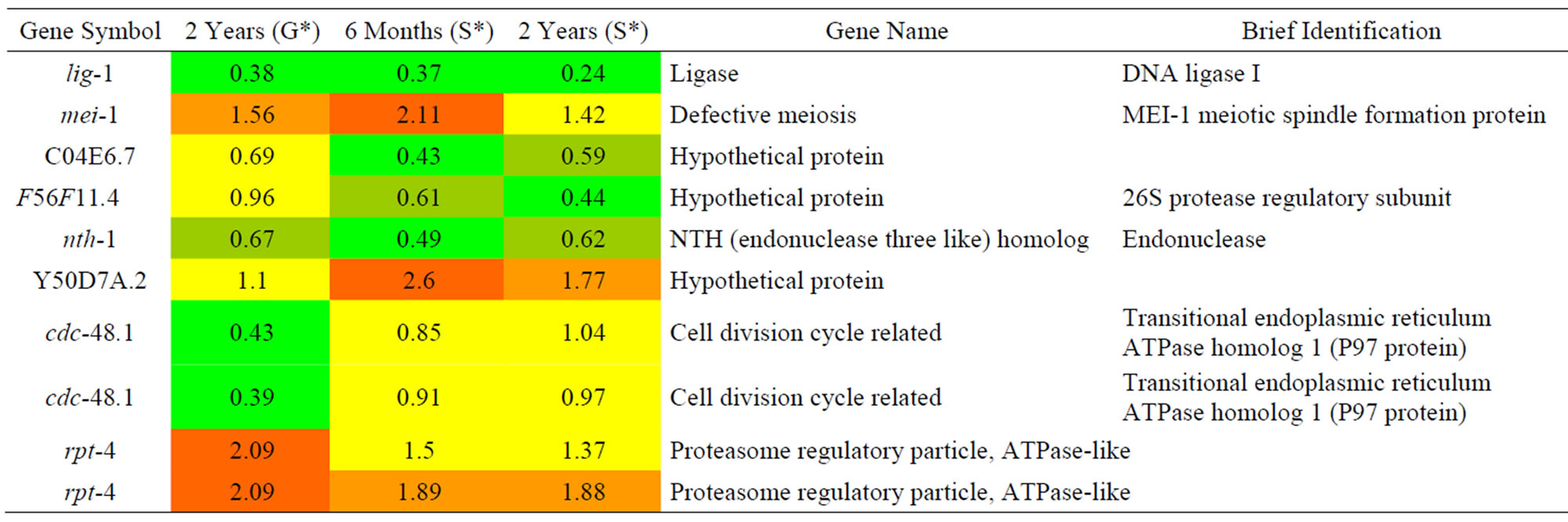
Table 6. Results of a microarray analysis that shows how genes that are related to DNA repair were up/down-regulated after one dose of simulated gamma ray irradiation at each dose rate.
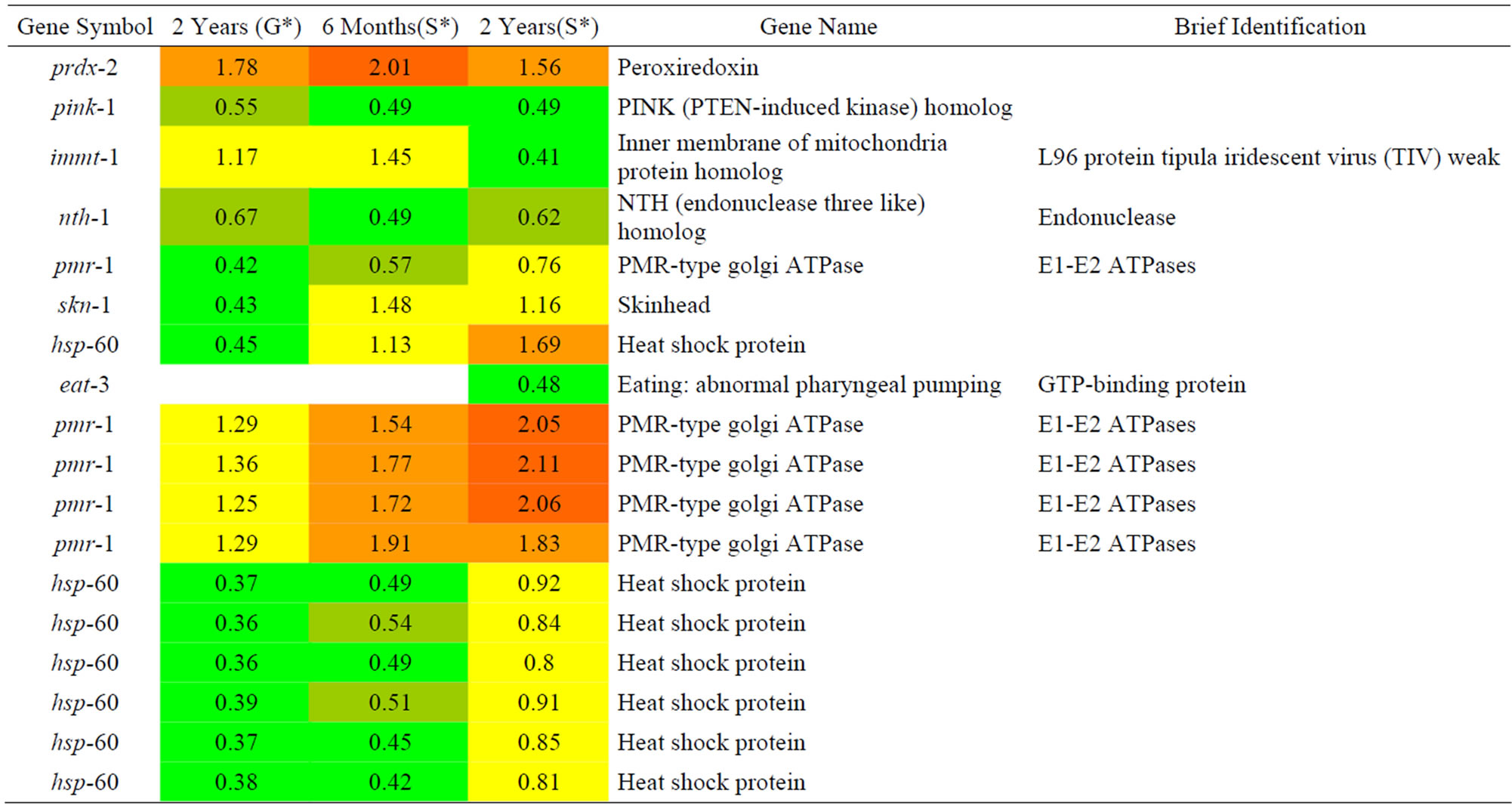
Table 7. Results of a microarray analysis that shows how genes that are related to oxidative stress were up/down-regulated after one dose of simulated gamma ray irradiation at each dose rate.
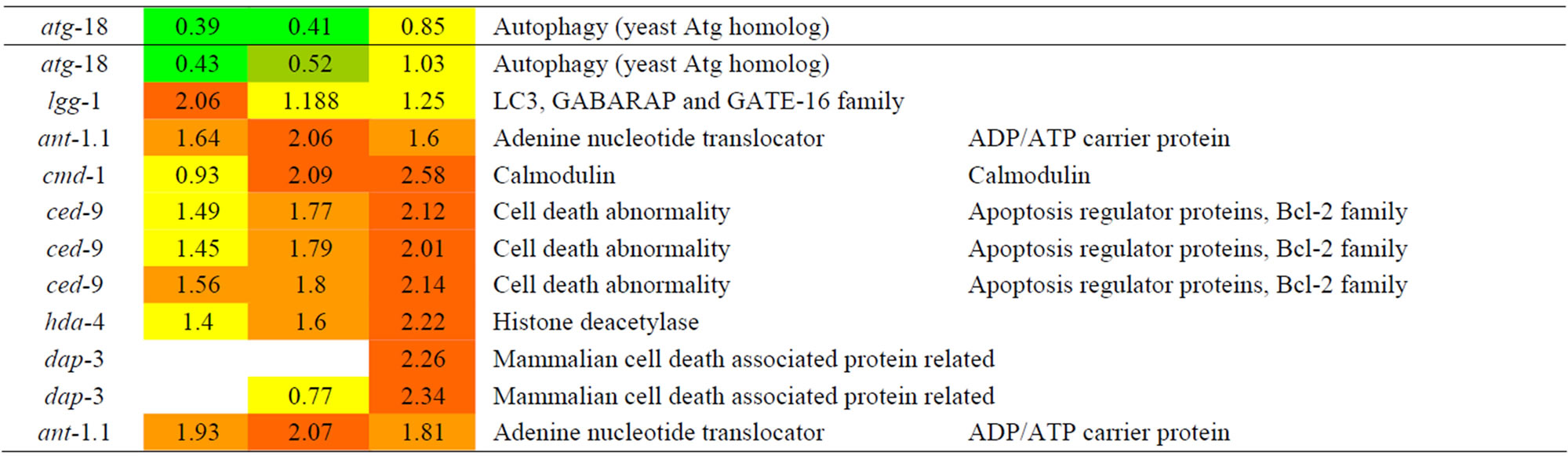
Table 8. Results of a microarray analysis that shows how genes that are related to cell death were up/down-regulated after one dose of simulated gamma ray irradiation at each dose rate.

Table 9. Results of a microarray analysis that shows how up/down-regulated genes that are related to DNA repair were influenced by G-forces. A G-force simulation test was repeated 3 times under the same condition (p value < 0.01 using a hypergeometric test).

Table 10. Results of a microarray analysis that shows how up/down-regulated genes that are related to oxidative stress were influenced by G-forces. A G-force simulation test was repeated 3 times under the same condition (p < 0.01 using a hypergeometric test).
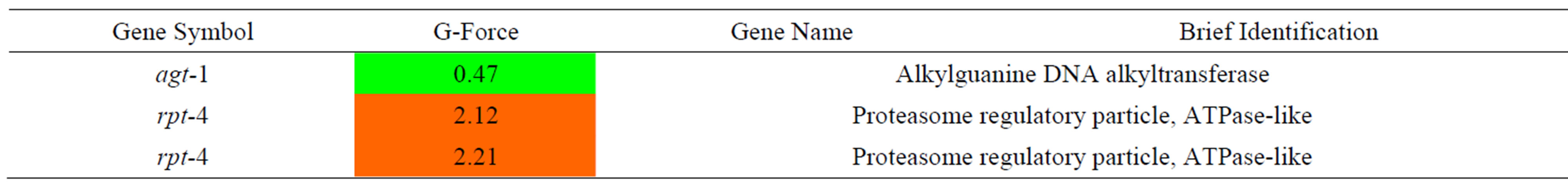
Table 11. Results of a microarray analysis that shows how up/down-regulated genes that are related to cell death were influenced by G-forces. A G-force simulation test was repeated 3 times under the same condition (p < 0.01 using a hypergeometric test).
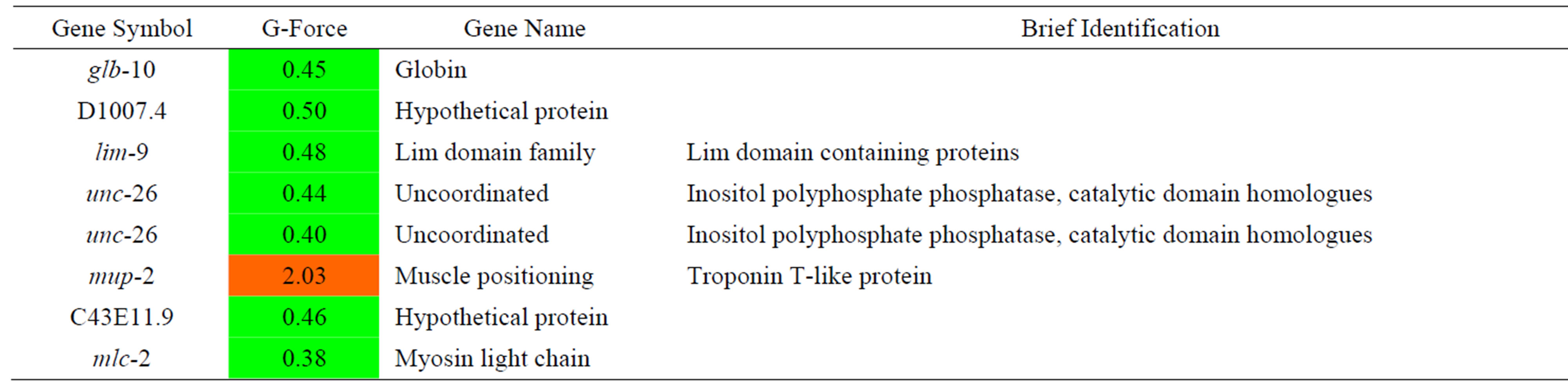
Table 12. Results of a microarray analysis that shows how up/down-regulated genes that are related to muscle were influenced by G-forces. A G-force simulation test was repeated 3 times under the same condition (p < 0.01 using a hypergeometric test).
branching and nucleotide excision repair. The msh-5 gene encodes a germline-specific homolog of the yeast and mammalian DNA mismatch repair protein MSH5 (MutS-family) that is required for crossing over and chiasma formation during C. elegans meiosis. The rpt-4 gene encodes a predicted ATPase subunit of the 19S regulatory complex of the proteasome. It has been reported that proteasome is related to DNA repair function and protein degradation. In addition, we found 3 genes lig-1, mei-1, and cdc-48.2 that were down-regulated by proton exposure. The mei-1 gene is important for making the meiosis I spindle, and cdc-48.2 is believed to be a member of the p97/AAA family, and is particularly essential during C. elegans meiosis. The facts that DNA repair genes (rad-23, msh-5) had their expression increased and the meiosis genes (mei-1, cdc-48.2) had their expression decreased further supports the view that a DNA repair mechanism was reduced due to proton exposure.
3.2.2. Oxidative Stress
We analyzed changes in the genes due to the effects to protons radiation on C. elegans that were related to the oxidative stress as shown in Table 4 and only found down-regulated genes. The skn-1 gene showed decreased expression according to increased proton radiation. According to other researchers [18], several proteasome subunit genes are regulated by SKN-1 protein, and it is known that SKN-1 responds to oxidative stress by upregulating genes that detoxify and defend against free radicals and other reactive molecules. Therefore, increased expression of the stress-resistant gene was increased by SKN-1 and expression of the stress-sensitivity gene against stress decreased. The hsp-60 gene encodes a mitochondrial-specific chaperone that imparts resistance on mitochondria against oxidative stress through increased expression. In addition, decreased skn-1 and hsp-60 expression seemed to cause sensitivity to oxidative stress.
3.2.3. Cell Death
We analyzed changes in genes in C. elegans that are related to death functions due to the effects to protons radiation as shown in Table 5 and found 4 up-regulated genes. Among the 4 genes, 2 were predicted to have a clear role in anti-apoptosis. They included icd-1 (C. elegans inhibitor of cell death-1) and ced-9 (C. elegans cell-death inhibitor Bcl-2). The hrdl-1 gene encodes an E3 ubiquitin ligase (orthologues of gp78) that plays essential roles in an unfolded protein response (UPR), endoplasmic reticulum-associated degradation (ERAD) pathways, and resistance to ER stress [19]. The fourth up-regulated gene, gsa-1, encodes an alpha subunit of heterotrimeric G proteins. gsa-1 is an essential gene: a loss-of-function due to a mutation in gsa-1 can cause death during the first stage of larval development if gsa-1 is over-expressed. The expression of a constitutively active form of G alpha(s) from an inducible promoter results in the hyper-contraction of bodywall muscle cells as well as vacuolization and degeneration of neurons within hours of induction (Korswagen et al., 1997). On the contrary, cdl-1, which is the homolog of human hairpin (stem-loop) binding proteins (HBP/ SLBP), is downregulated. In addition, it is an essential gene for normal rapid apoptosis. These data suggest that protons reduce cell death apoptosis based on increased expression of icd-1 and ced-9 as anti-apoptosis proteins and decrease the expression of cdl-1. Moreover, increased hrdl-1 will results in increased ER-stress.
3.3. Changes in C. elegans Genes Due to the Effects of Gamma Rays
We analyzed changes in the genes of C. elegans due to the effects of gamma rays as shown in Table 6. lig-1 as DNA ligase I, nth-1 as endonuclease, C04E6.7, and F56F11.4 as hypothetical protein expression were decreased due to gamma ray irradiation. Compared with proton irradiation, lig-1 was decreased due to gamma rays. There were no considerable changes in any other mechanisms except for lig-1.
3.3.1. Oxidative Stress
We analyzed changes in the genes of C. elegans caused by the effects of gamma rays on C. elegans that are related to oxidative stress as shown in Table 7. Among them, pink-1 is related to PINK-1 (PTEN-induced kinase-1) and its expression tended to increase along with the gamma dose rate. The expression of hsp-60, which encodes a mitochondrial-specific chaperone, decreased along with irradiated proton. Based on the decreasing expression of pink-1 and hsp-60, we predicted that C. elegans is more sensitive to oxidative stress.
3.3.2. Cell Death
We analyzed the changes in the genes of C. elegans caused by the effects of gamma rays as shown in Table 8. K08F11.5, which is predicted to be a Ras related/RacGTP binding protein, showed increased expression depending on gamma ray dose. vps-41 encodes the C. elegans homolog of the Saccharomyces cerevisiae vacuolar sorting protein Vps41p, which is essential for apoptosis. The expression of vps-41 was increased with increasing gamma rays. ndx-4, which encodes a homo-dimeric diadenosine tetraphosphatehydrolase, showed increased expression. ndx-4 has a key role in regulating the intracellular Ap(4)A levels and hence potentially the cellular response to metabolic stress, differentiation, or apoptosis via the Ap(3)A/Ap(4)A ratio[21]. NDX-4 contributes to genomic stability in vivo in C. elegans. Phenotypic analyses of an ndx-4 mutant reveal that loss of NDX-4 leads to upregulation of key stress responsive genes [22]. In addition, crt-1, which encodes an ortholog of calreticulin and is induced by stress, showed increased expression. A deficiency in crt-1 in C. elegans does not affect it, but it is essential for specific necrosis [23]. We assumed that cell death mechanism is upregulated. vps-41, which is a protein that is related to apoptosis; crt-1, which is a protein that is related to necrosis.
3.4. Changes in C. elegans Caused by the Effects of Protons
We found 956 up-regulated genes that showed more than a 2-fold change. The Venn diagram in Figure 4 shows the number of regulated genes that exhibited increased expression through exposure to protons or gamma rays. One hundred eighty-eight genes (20%) were commonly expressed between protonand gamma ray-exposed groups. The number of 2-fold up-regulated genes was large and therefore, we selected the top 10 GO terms categories to determine the GO terms that were over-represented, as shown in Figure 5. In GO terms, embryonic
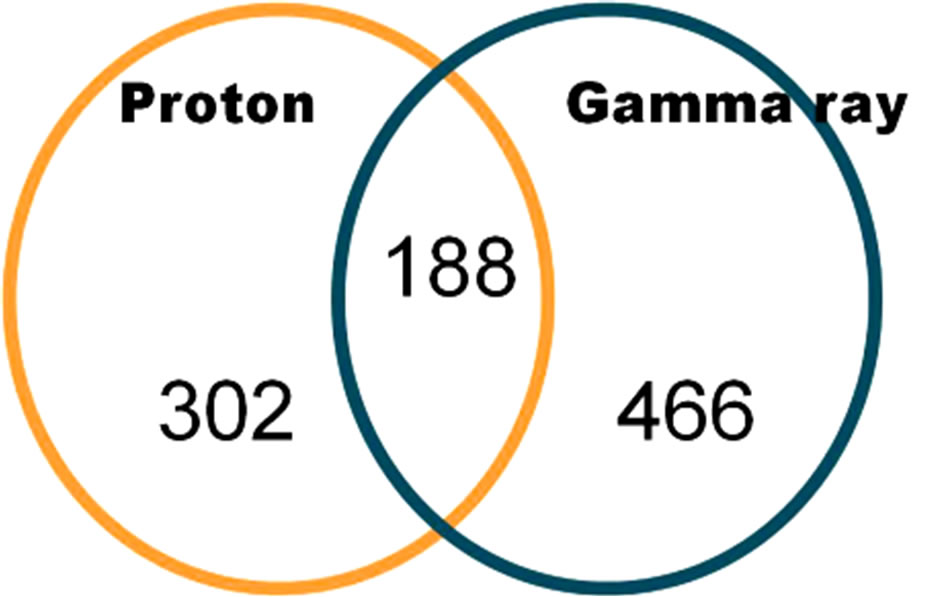
Figure 4. 2-fold up-regulated genes in common between protonand gamma-irradiated of C. elegans (p < 0.01 using a hypergeometric test).
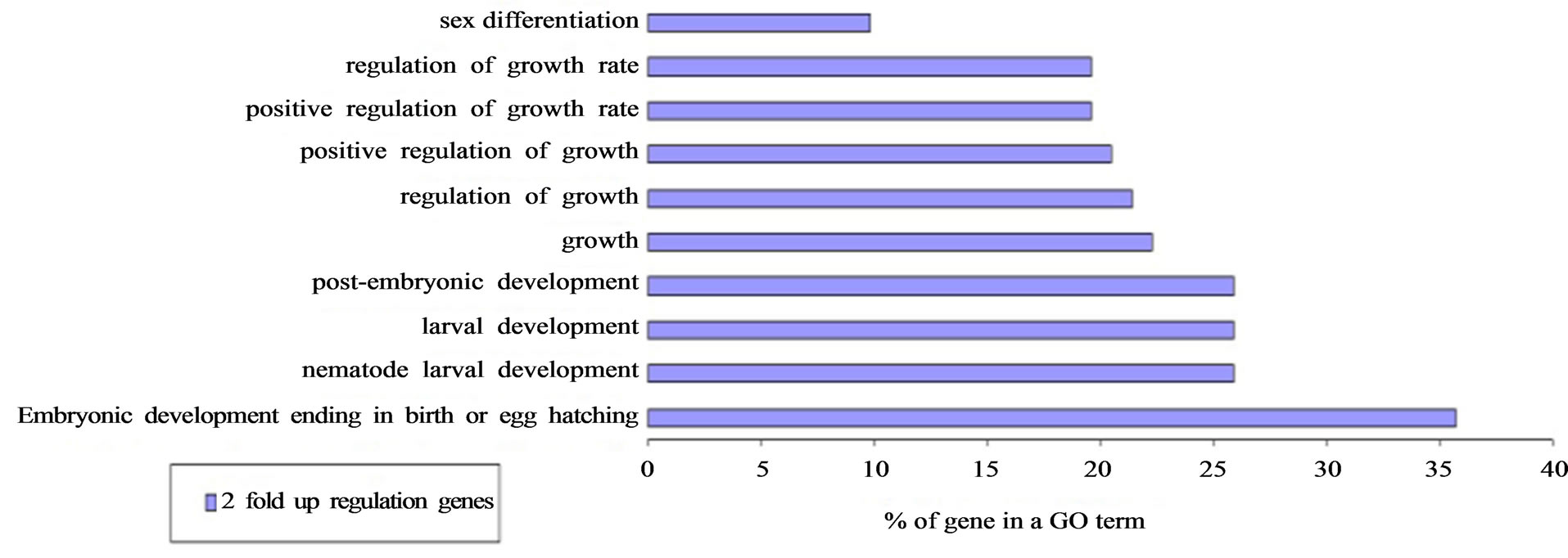
Figure 5. Two-fold up-regulation of GO term genes in common between protonand gamma ray-exposed groups.
development ending occurred at a high frequency (35.7%), and larval development and regulation of the growth GO term were significantly increased in commonly expressed genes between protonand gamma ray-exposed groups.
Out of the 34,037 reliable genes, whose expressions were found to have been decreased by irradiation, we found 1122 down-regulated genes that exhibited more than a 2-fold change. As shown in Figure 6, 122 commonly expressed genes were affected by protons and gamma rays. In the GO term, embryonic development ending in birth or egg hatching occurred at a high frequency (22.7%) and the other GO term had relatively few matches with the genes that exhibited decreased expression after both proton and gamma ray irradiation, as shown in Figure 7.
In a previous study, 170 of 17,871 genes were upregulated and 390 of 17,871 genes were down-regulated after 3 h of exposure to 3 Gy to protons or gamma rays (Nelson et al., 2002). When comparing the profile, protons and gammas existed in different clusters. Therefore, it is expected that they have different effects on the properties of C. elegans. Nelson performed an experiment using 3 Gy but that amount of radiation was much larger than what could be expected for space radiation. However, our study reached the same conclusion that protons and gamma rays have different effects on C. elegans.
3.5. Effects of G-Forces on C. elegans
We analyzed the upor down-regulated C. elegans genes that experienced the same force of gravity as that experienced during a Soyuz rocket launch. Muscle atrophy during space flight is a serious problem, and therefore, we added one more GO term, muscle function, compared to the GO terms in the former analysis of DNA repair, oxidative stress, and cell death. We then checked if there

Figure 6. Genes that showed 2-fold down-regulation due to protons and gamma rays in irradiated C. elegans (p value < 0.01 using a hypergeometric test).
were common genes that showed the same changes in expression due to G-forces and space radiation.
3.5.1. DNA Repair
We analyzed the changes in gene expression in C. elegans that were related to DNA repair during G-force simulation as shown in Table 9. A DNA repair enzyme, agt-1, showed decreased expression during G-force simulation. The expression of rpt-4 was increased when we applied not only proton and gamma ray irradiation but also simulated G-forces. However, its expression was significantly increased during G-force simulation compared with proton and gamma ray irradiation. The rpt-4 gene encodes a proteasome 19s subunit and is known to degrade DNA repair enzymes in yeast [20,24]. As agt-1, which is a DNA repair enzyme, was only decreased and rpt-4, which is a proteasome of DNA repair protein, was increased, we predicted that there would be a marked decline in DNA repair under the G-force simulation.
3.5.2. Oxidative Stress
We analyzed the changes in gene expression in C. elegans that were related to oxidative stress caused by G-force simulation as shown in Table 10 and found only down-regulated genes. A C. elegans mitofilin homolog, immt-1, exhibited decreased expression. This gene is related to reproduction, physiology, and mitochondrial
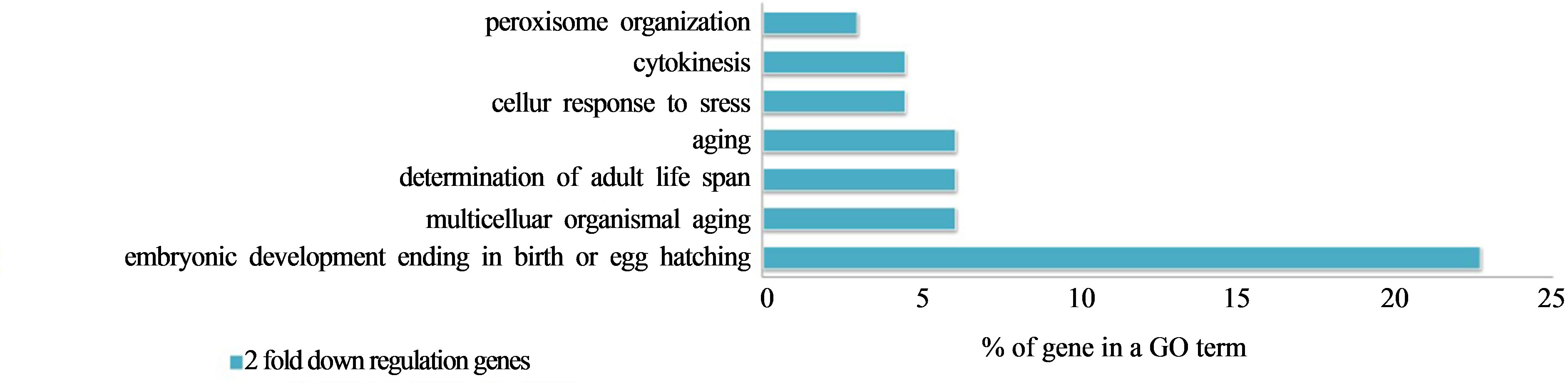
Figure 7. Two-fold down-regulated genes in GO terms in common between protonand gamma-irradiated C. elegans.
cristae formation. Mutations in immt-1 and immt-2 cause reduced motility, increased levels of reactive oxygen species, decreased mitochondrial mass, and imparted resistance to oxidative stress [25]. Therefore, we predicted that the oxidative stress mechanism did not change much except for immt-1.
3.5.3. Cell Death
We analyzed changes in gene expression in C. elegans due to G-force simulation that are related to cell death. The pqn-60 gene, which is a glutamine-asparagine (Q/N)- rich (prion-like) protein that is expressed during germ line apoptosis, and dad-1, which is an ortholog of human DAD1 in apoptosis, exhibited decreasing expression.
3.5.4. Muscle Function
We analyzed changes in gene expression caused by Gforce simulation that are related to muscle function. The mup-2 gene, which is a troponin T-like protein, showed increased expression, while many other genes showed decreased expression. The genes related to muscle function (glb-10, globin subunit, D100.4, hypothetical protein, and lim-9, muscle fixation) were decreased in expression. In addition, unc-26, which acts as a synaptojanin in neuron vesicles, and mlc-2, which is a component of the myosin light chain, exhibited decreased expression. Generally, we found that the genes that are related to muscle function showed decreasing expression and therefore we could infer that G-forces influence the motility of C. elegans.
3.6. Effects of G-Forces and Space Radiation on C. elegans
We compared common 2-fold up/down-regulated genes under both space radiation and G-force simulation, and space radiation including both results for proton and gamma ray irradiation. We found that 755 genes showed more than a 2-fold change in expression out of 35,007 reliable genes that exhibited increased expression due to space radiation and G-forces. As shown in Figure 8, 122 genes were commonly expressed genes that were affected by space radiation and G-force simulation in C.
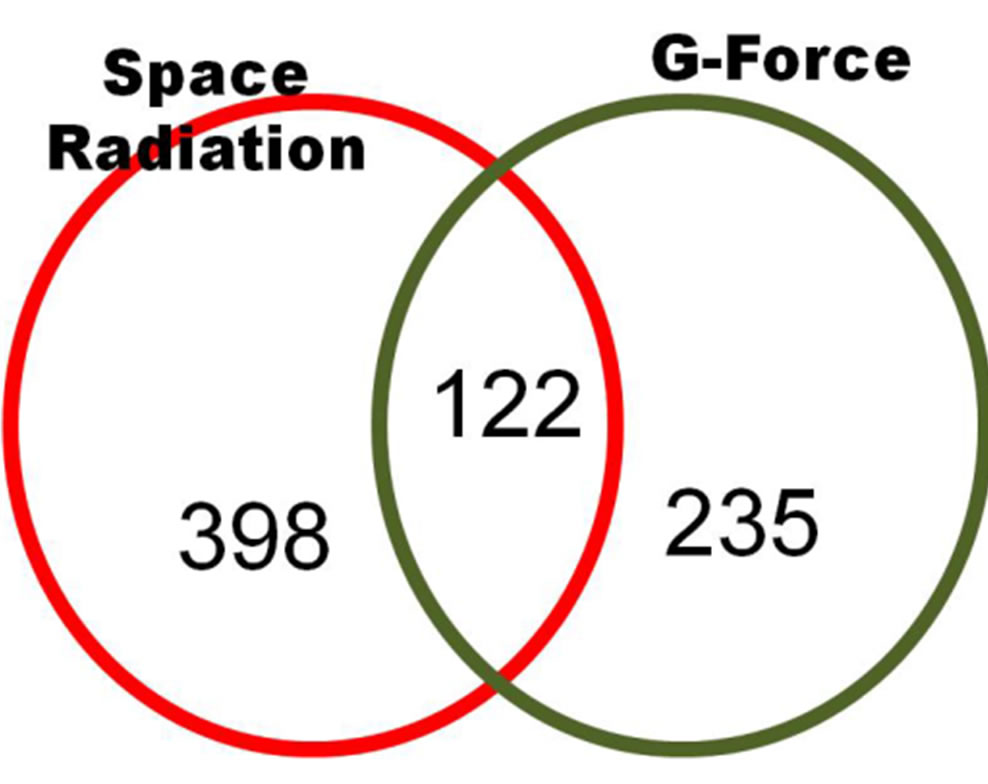
Figure 8. 2-fold up-regulated genes in common between space radiation and G-force treated C. elegans (p < 0.01 using a hypergeometric test).
elegans. In the GO term, embryonic development ending in birth or egg hatching occurred at a high frequency (25.7%) and the nematode larval development, larval development, and post-embryonic development GO terms were increased in commonly expressed genes between proton and gamma ray irradiation as listed in Figure 9.
We found that 862 genes showed changes in more than 2-fold expression out of 35,007 reliable genes due to space radiation and G-forces that had expression genes by the space irradiation. As shown in Figure 10, 104 genes in common showed decreased gene expression due to both space radiation and G-force simulation. In particular, in this field, the change in the GO term was relatively low at less than 6% as shown in Figure 11.
4. Conclusions
We used microarrays to identify changes in gene expression in C. elegans in a simulated space environment: space radiation during long-term flight and G-forces in a spacecraft during launch. We examined C. elegans through ground tests such as irradiating it with protons and gamma rays to simulate long-term space flight and simulating G-forces by using a mini-centrifuge. Our results revealed that protons and gamma rays induce similar genes that are related to DNA damage responses, oxidative stress, and cell death. These 2 types of radiation regulated (induced and reduced) gene expression that was related to embryonic development ending in birth or egg hatching. We confirmed that protons induced
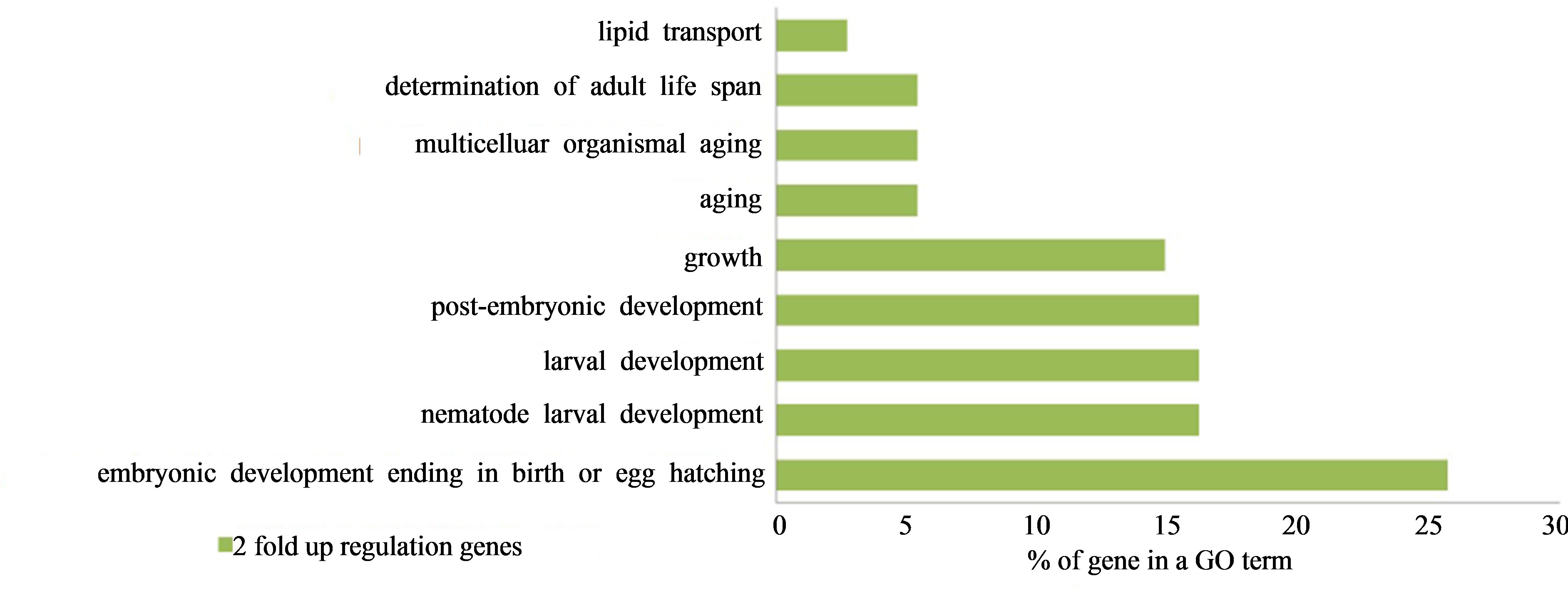
Figure 9. Two-fold up-regulated GO term genes in common between space radiation and G-force treated C. elegans.
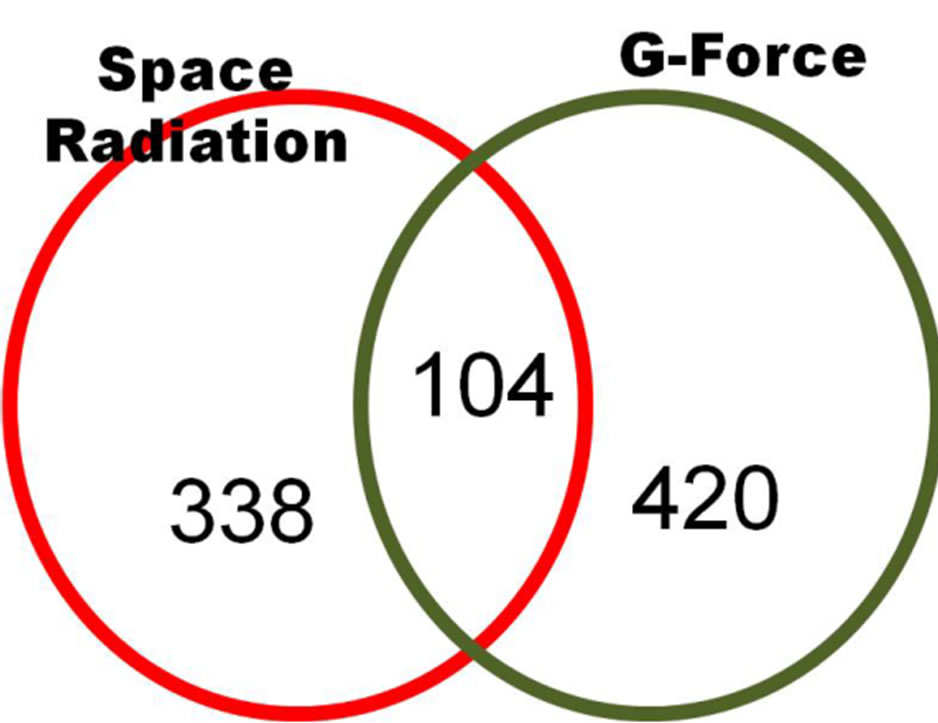
Figure 10. Two-fold down-regulated genes of C. elegans in common between space radiation and G-force simulation (p < 0.01 using a hypergeometric test).
expression of genes that are related to the DNA damage response and anti-apoptosis, whereas gamma rays are induced apoptosis. Therefore, we predicted that C. elegans experiences some damage due to irradiation during longterm space flight. When we exposed C. elegans to the G-forces that occur during spacecraft launch situations, genes related to muscle function displayed decreasing expression. In addition, there were common changes in expression in genes between the G-force and space radiation simulations.
According to Nelson [26], 599 of 17,871 genes analyzed with a microarray showed differential expression 3 h after exposure to 3 Gy of radiation: protons, iron, and gamma rays. One-hundred-seventy genes were up-regulated by protons and gamma rays, and 390 genes were down-regulated. However, the amount 3 Gy was too high in terms of space radiation aboard the ISS and so we carried out our simulations using less than 3 Gy. We also confirmed the same results as Nelson found: protons and gamma rays have different effects on C. elegans gene pattern through GO analysis. Short term (10 d) exposure to actual space radiation and microgravity on ICE-FIRST revealed that apoptosis proceeds normally [9] and that no significant increase in the rate of mutation occurs [10]. Decreased levels of myogenic transcription factor were observed [27]. Long-term studies (for 6 months) on the ISS showed that 1228 of 2265 genes were differentially expressed: 906 genes were up-regulated and 322 genes were down-regulated [13]. DAF-16 downstream genes (dod-19, dod-6, and dod-3), a proapoptic gene (ced-3), a radiation sensitive gene (rad-51), and glutathione S-transferase genes (gst-13, gst-17, gst-26, gst-27, and gst-36) were up-regulated. Down-regulation of a muscle growth gene (mua-3) and up-regulation of a myosin heavy-chain reaction gene (myo-3) were observed. These results showed that long term exposure of C. elegans to space radiation and microgravity involves changes in ageing, oxidative stress, and muscle growth.
We used microarrays to identify gene expression changes in C. elegans under a simulated space environment that entailed space radiation during long-term flight and G-forces that occur in a spacecraft during launch. We examined C. elegans through ground tests such as irradiating it with protons and gamma rays to simulate long-term space flight and G-force simulations using a mini-centrifuge. In addition, changes in genes were observed. This study observed changes in genes involved in DNA repair, oxidative stress, cell death, and muscle function. When irradiated, differential gene expression in C. elegans was associated with DNA repair, oxidative stress, and cell death. However, genes that affect muscle function did not change. Our results revealed that protons and gamma rays induce similar changes in genes that are related to the DNA damage response, oxidative stress, and cell death. These types of radiation regulate (induce and reduce) in common gene expression that is related to embryonic development ending in birth or egg hatching. We confirmed that protons induce the expression of genes that are related to the DNA damage response and anti-apoptosis, while gamma rays induce apoptosis. Therefore, we predicted that C. elegans would experience some damage from irradiation during long-term space flight. When we exposed C. elegans to G-forces
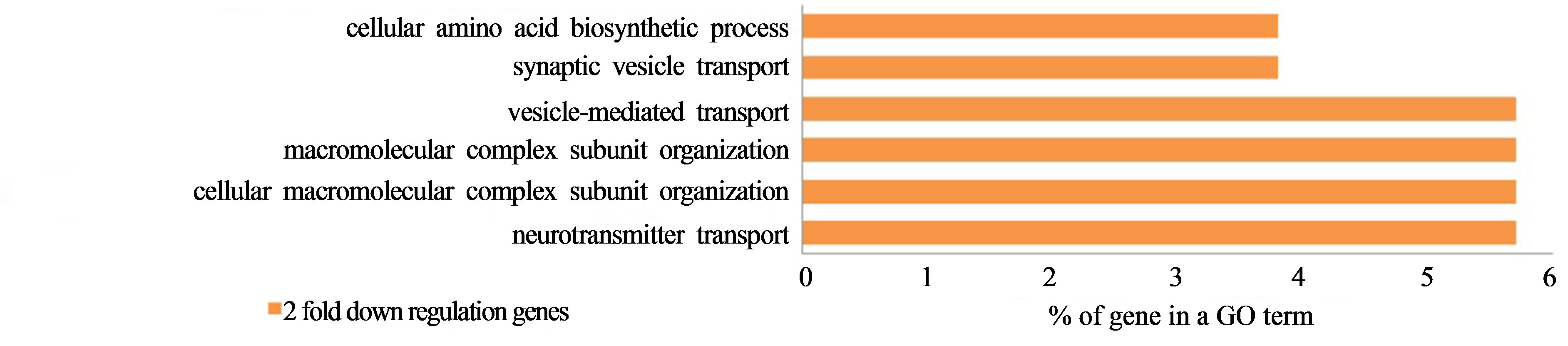
Figure 11. Two-fold down-regulated GO term genes of C. elegans in common between space radiation and G-force simulation (p < 0.01 using a hypergeometric test).
that are present during a spacecraft launch situation, genes that are related to muscle function displayed decreasing expression. In addition, there were common changes in the expression of genes between G-forces and space radiation.
5. Acknowledgements
This work was supported by the Korea Cancer Center Hospital, the Korea Atomic Energy Research Institute, and the Advanced Radiation Technology Institute to simulate space radiation. We give our specials thanks to Ju-Young Lee of KAIST, who supported this research by providing us with C. elegans culturing techniques and related protocols.
REFERENCES
- G. Reitz, R. Beaujean, E. Benton, S. Burmeister, Ts. Dachev, S. Deme, M. Luszik-Bhadra and P. Olko, “Space Radiation Measurements On-Board ISS—The DOSMAP Experiment,” Radiation Protection Dosimetry, Vol. 116, No. 1-4, 2005, pp. 374-379. doi:10.1093/rpd/nci262
- NASA Space Monitoring Data Center, http://smdc.sinp.msu.ru/index.py?nav=model-ap8ae8
- Vanderbilt University School of Engineering, https://creme.isde.vanderbilt.edu/CREME-MC
- T. Sakashita, T. Takanami, S. Yanase, N. Hamada, M. Suzuki, T. Kimura, Y. Kobayashi, N. Ishii and A. Higashitani, “Radiation Biology of Caenorhabditis elegans: Germ Cell Response, Aging and Behavior,” Journal of Radiation Research, Vol. 51, No. 2, 2010, pp. 107-121. doi:10.1269/jrr.09100
- Y. Zhao, R. Johnsen, D. Baillie and A. Rose, “Worms in Space? A Model Biological Dosimeterm,” Gravitational and Space Biology, Vol. 18, No. 2, 2005, pp. 11-16.
- G. A. Nelson, W. W. Schubert, G. A. Kazarians, G. F. Richards, E. V. Benton, E. R. Benton and R. Henke, “Development and Chromosome Mechanics in Nematodes: Results from IML-1,” Advances in Space Research, Vol. 14, No. 8, 1994, pp. 209-214. doi:10.1016/0273-1177(94)90405-7
- G. A. Nelson, W. W. Schubert, G. A. Kazarians, G. F. Richards, E. V. Benton, E. R. Benton and R. Henke, “Radiation Effects in Nematodes: Results from IML-1 Experiments,” Advances in Space Research, Vol. 14, No. 10, 1994, pp. 87-91. doi:10.1016/0273-1177(94)90455-3
- N. J. Szewczyk, R. Mancinelli, W. Mclamb, D. Reed, B. S. Blumberg and C. A. Conley, “Caenorhabditis Elegans Survives Atmospheric Breakup of STS-107, Space Shuttle Columbia. Astrobiology,” Astrobiology, Vol. 5, No. 6, 2005, pp. 690-705. doi:10.1089/ast.2005.5.690
- A. Higashitani, A. Higashibata, Y. Sasagawa, T. Sugimoto, Y. Miyazawa, N. J. Szewcyk, M. Viso, G. Gasset, B. Eche, K. Fukui, T. Shimazu, N. Fujimoto, K. Kuriyama and N. Ishioka, “Checkpoint and Physiological Apoptosis in Germ Cells Proceeds Normally in SpaceFlown Caenorhabditis elegans,” Apoptosis, Vol. 10, No. 5, 2005, pp. 949-954. doi:10.1007/s10495-005-1323-3
- Y. Zhao, K. Lai, I. Cheung, J. Youds, M. Tarailo, S. Tarailo and A. Rose, “A Mutational Analysis of Caenorhabditis elegans in Space. Mutation Research—Fundamental and Molecular Mechanisms of Mutagenesis,” Mutation Research, Vol. 601, No. 1-2, 2006, pp. 19-29. doi:10.1016/j.mrfmmm.2006.05.001
- N. J. Szewczyk, I. A. Udranszky, E. Kozak, J. Sunga, S. K. Kim, L. A. Jacobson and C. A. Conley, “Delayed Development and Lifespan Extension as Features of Metabolic Lifestyle Alteration in C. elegans under Dietary Restriction,” The Journal of Experimental Biology, Vol. 209, 2006, pp. 4129-4139. doi:10.1242/jeb.02492
- N. J. Szewczyk, J. Tillman, C. A. Conley, L. Grangere, L. Segalate, A. Higashitani, S. Honda, Y. Honda, H. Kagawa, R. Adachi, A. Higashibata, N. Fujimoto, K. Kuriyama, N. Ishioka, K. Fukui, D. Baillie, A. Rose, G. Gasset, B. Eche, D. Chaput and M. Viso, “Description of International Caenorhabditis elegans Experiment First Flight (ICE-First),” Advances in Space Research, Vol. 42, No. 6, 2008, pp. 1072-1079. doi:10.1016/j.asr.2008.03.017
- E. A. Oczypok, T. Etheridge, J. Freeman, L. Stodieck, R. Johnsen, D. Baillie and N. J. Szewczyk, “Remote AutoMated Multi-Generational Growth and Observation of an Animal in Low Earth Orbit,” Journal of the Royal Society Interface, Vol. 9, No. 68, 2012, pp. 596-599. doi:10.1098/rsif.2011.0716
- http://www.geneontology.org/index.shtml
- http://www.biocarta.com
- http://www.genmapp.org
- http://david.abcc.ncifcrf.gov
- D. L. Miller, M. W. Budde and M. B. Roth, “HIF-1 and SKN-1 Coordinate the Transcriptional Response to Hydrogen Sulfide in Caenorhabditis elegans,” Aging Cell, Vol. 8, No. 5, 2009, pp. 524-541. doi:10.1371/journal.pone.0025476
- Y. Sasagawa, K. Yamanaka and T. Ogura, “Caenorhabditis elegans UBX Cofactors for CDC-48/p97 Control Spermatogenesis,” Genes Cells, Vol. 12, No. 9, 2007, pp. 1063- 1073.
- N. J. Krogan, M. H. Y. Lam, J. Fillingham, M. C. Keogh, M. Gebbia, J. Li, N. Datta, G. Cagney, S. Buratowski, A. Emili and J. F. Greenblatt, “Proteasome Involvement in the Repair of DNA Double-Strand Breaks,” Molecular Cell, Vol. 16, No. 6, 2004, pp. 1027-1034. doi:10.1016/j.molcel.2004.11.033
- S. Bailey, S. E. Sedelnikova, G. M. Blackburn, H. M. Abdelghany, P. J. Baker, A. G. McLennan and J. B. Rafferty, “The Crystal Structure of Diadenosine Tetraphosphate Hydrolase from Caenorhabditis elegans in Free and Binary Complex Forms,” Structure, Vol. 10, No. 4, 2002, pp. 589-600. doi:10.1016/S0969-2126(02)00746-3
- H. Yu, H. Zhao, L. E. Wang, Y. Han, W. V. Chen, C. I. Amos, T. Rafnar, P. Sulem, K. Stefansson, M. T. Landi, N. Caporaso, D. Albanes, M. Thun, J. D. McKay, P. Brennan, Y. Wang, R. S. Houlston, M. R. Spitz and Q. Wei, “DNA Repair Genotype and Lung Cancer Risk in the Beta-Carotene and Retinol Efficacy Trial,” DNA Repair (Amst), Vol. 7, 2011, pp. 176-187.
- B. J. Park, D. G. Lee, J. R. Yu, S. Jung, K. Choi, J. Lee, J. Lee, Y. S. Kim, J. I. Lee, J. Y. Kwon, J. Lee, A. Singson, W. K. Song, S. H. Eom, C. S. Do, H. Kim, J. Bandyopadhyay and J. Ahnn, “Calreticulin, a Calcium-Binding Molecular Chaperone, Is Required for Stress Response and Fertility in Caenorhabditis elegans,” Molecular and Cellular Biology, Vol. 12, No. 9, 2001, pp. 2835-2845. doi:10.1091/mbc.12.9.2835
- S. J. Russell, S. H. Reed, W. Huang, E. C. Friedberg and S. A. Johnston, “Proteasome Involvement in the Repair of DNA Double-Strand Breaks,” Molecular Cell, Vol. 3, No. 6, 1999, pp. 687-695. doi:10.1016/S1097-2765(01)80001-0
- J. Y. Mun, T. H. Lee, J. H. Kim, B. H. Yoo, Y. Y. Bahk, H. S. Koo and S. S. Han, “Caenorhabditis elegans Mitofilin Homologs Control the Morphology of Mitochondrial Cristae and Influence Reproduction and Physiology,” Journal of Cellular Physiology, Vol. 224, No. 3, 2010, pp. 748-756. doi:10.1002/jcp.22177
- G. A. Nelson, T. A. Jones, A. Chesnut and A. L. Smith, “Radiation-Induced Gene Expression in the Nematode Caenorhabditis elegans,” Journal of Radiation Research, Vol. 43, 2002, pp. 199-203.
- A. Higashibata, N. J. Szewczyk, C. A. Conley, M. Imamizo-Sato, A. Higashitani and N. Ishioka, “Decreased Expression of Myogenic Transcription Factors and Myosin Heavy Chains in Caenorhabditis elegans Muscles Developed during Spaceflight,” The Journal of Experimental Biology, Vol. 209, No. 16, 2006, pp. 3209-3218. doi:10.1242/jeb.02365
NOTES
*Corresponding author.

