Advances in Nanoparticles
Vol.2 No.2(2013), Article ID:31884,11 pages DOI:10.4236/anp.2013.22024
Synthesis and Characterization of Various Protein α-Lactalbumin Nanotubes Structures by Chemical Hydrolysis Method
1Biomedical Material Department, Institute of Pharmacy, Martin Luther University, Halle-Wittenberg, Germany
2College of Chemical Engineering, University of Tehran, Tehran, Iran
3Process Simulation and Control Lab, Chemical Engineering Collage, Iran University of Science
and Technology (IUST), Tehran, Iran
4Department of Chemistry, College of Science, Alzahra University, Tehran. Iran
Email: *fakhroueian@ut.ac.ir
Copyright © 2013 Pegah Esmaeilzadeh et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received January 31, 2013; revised March 1, 2013; accepted March 8, 2013
Keywords: Nanobiotechnology; Milk Protein Nanotubes; Helical Self-Assembly; Water-Based Nanofluids; Chemical Hydrolysis; Nanomedicine
ABSTRACT
New water-based nanofluids including unparalleled milk protein α-lactalbumin hollow nano-bio-tubes using low cost, available and advanced partial chemical hydrolysis strategy in bottom-up nano-assembly have been employed in this work. The aqueous sol-gel chemistry in nanotechnology which we selected for this goal offers new fabrication as interesting smart protein nanotubes. The kinds of nanometer sized tubular structures such as waved, helically coiled, bent, bamboo-shaped, bead-like and branched single-walled protein nanotubes (SWPNTs) with a range of 3 - 8 nm in outer diameters were produced by this method. Complete characterization for natural produced nanotubes including SEM, TEM images, G bond and D bond in Raman spectroscopy, XRD patterns, DLS (Dynamic Light Scattering) and FTIR analysis were evaluated which they are most significant experiments in synthesized protein nanotubes soluble in clear water nanofluids and stabilization of transparent nanofluids was proved within more than one year after preparation. Various necessary ligand ion salts such as Mn2+, Zn2+ and Ca2+ or mixtures as bridge makers and producing biological selfassembly hollow SWPNTs were performed and we focused on new chemical technology under specific acidic hydrolysis method not conventional enzymatic proteolysis and applying surfactants, pH reagent, Tris-HCl buffer, polar solvent which could be produced by β-sheet stacked hydrolysed protein α-lactalbumin mechanism under appropriate conditions to achieving high efficiency new protein nanotubes skeleton. They can be promising materials applied in food science, diet nutrition, nanomedicine, nano-biotechnology and surgery.
1. Introduction
Besides widespread advancements in synthesis of nanotubes from non-biological materials (mainly carbon nanotubes) attempts to mimic the ingeniously natural bottom-up nanofabrication with the aim of achieving the design and synthesis of man-made natural nanotubes, open the door to a new technology of important applications in the scope of nanobiotechnology. Natural bimolecular assemblies exhibit the selectivity and specificity which can adopt a wide variety of structures that perform an array of functions, and finally they are based on various chemistries (nucleic acids, peptides and proteins, lipids, and carbohydrates) that promote self-assembly in water at neutral pH and ambient temperatures [1]. On the road to these emerging opportunities, proteins represent fertile territory for nanobiotechnology, because they have properties ideal for nano-engineering purposes [2].The building blocks self-assemble and in the presence of ion Ca2+ salt bridges connect the individual building blocks, resulting in a hollow structure that previously, the kinetics of nanotube formation has been studied as a function of Ca2+ concentration. The most important functional properties of whey proteins are solubility, viscosity, gel formation, emulsification, foaming, and like many other albumins, the capacity to form nanoparticles (Mehravar et al., 2009) [3] and (H. Hernandez-Sanchez et al., 2012) could obtain nanoparticles of bovine α-lactalbumin α-LA products in the 100 to 200 nm size range so that they could be used as carriers of bioactive compounds [4]. Proteins also are the fundamental building blocks of all biological matter that exhibit such finesse in architectural design [5]. In just one reviewin 2006, J. F. GravelandBikker, et al., succeeded to create and synthesis novel alpha-lactalbumin (a α/β globular milk protein) nanotubesvia enzymatic proteolysis in presence of Bacillus licheniformis protease (BLP) for the first time which selfassembled into nanometer-sized tubular structures at appropriate conditions. Linear assemblies such as rod, fiber and tube-like structures are particular interest, because they have unique properties with respect to applications [6] and these findings represent important characteristic for the development of a nanotechnological application. Proteins and peptides can be synthesized by chemical or biological means. In this regard S. Banta et al. in 2007 researched about engineering of protein and peptide building blocks for nanotechnology usage [7] and the interactions that lead to the formation of these structures include chiral dipole-dipole interactions, π-π stacking, hydrogen bonds, nonspecific Van der Waals interactions, hydrophobic forces, electrostatic interactions, and repulsive steric forces [8]. Biological molecules such as peptides and proteins undergo self-assembly processes in vivo and in vitro, and these monomers are assembled into various nanometer-scale structures at room temperatures and atmospheric pressure [9]. The self-assembled protein and peptide nanostructures can be further organized to form various nanowires, nanotubes, and nanoparticles via their molecular-recognition functions. It seems that, the most special feature of α-lactalbumin protein nanotubes is due to its cavities and hollow cores. Because of their cavities, they could well serve as vehicles for drug or other encapsulated molecules such as vitamins and enzymesand also for doping of diverse ligand ions like Fe, Mn, Ca and Zn melts encapsulated in diet nutrition. During the headway of this research authors perceived that the unique stupendous linear self-assembly of α-lactalbumin building blocks in specific nanofluids using chemical hydrolysis and sol-gel matrix derived nanomaterials along with the matchless power of self-assembly can create thermodynamically stable nano-bio-tubes and rods products [10- 12]. This strategy also represents low cost example of bottom-up assembly and chemistry is entering the field of nanoscale science which is originally physics-oriented. This interesting well-orchestrated mechanism offers obvious advantages as the smart protein nanostructures as well as new authors’ researches in this activated field. In this work we donot need touse any special catalyst, and we did not applyany enzymatic proteolysis [13,14] system and these green obtained protein nanofluids were fabricated by chemical sol-gel method as nanofluids in water solutions using bovine α-lactalbumin protein as precursor which hydrolyses under various conditions which it is a platform on nanomedicine. They showed very clear and stable protein cylinder nanotubes after one year, confirmed by DLS measurement and TEM images.
2. Experimental Details
2.1. Materials
Bovine holo α-Lactalbumin (L6010) Sigma, Tris hydroxyl methyl aminomethane, acetyl acetone (acac), urea, citric acid, HCl 37%, Triethanolamine (TEA), calcium chloride (2H2O), manganese chloride (4H2O), zinc acetate, pure isopropyl alcohol. All chemicals were supplied from Merck, and domestic deionized water was obtained in this work.
2.2. Methods
Sample 1) The mixture of acacas nonionic surfactant, 10 ml isopropyl alcohol, 10 ml deionized water, 0.5 - 2 mg MnCl2·4H2O as connector and 0.5 - 2 mg protein α-lactalbumin was adjusted to acidic pH 2 - 3 using citric acid 1 M, HCl 2 M and urea 1 M solutions. The reaction solution was held in shaking water bath (Grant, OLS 200 from U.K.) at 42˚C with 60 - 80 rpm during 1 - 2 weeks and then was filtered using 0.1 μl filter paper diameter (Millipore company). Finally the filtrate solution was held at 42˚C during one week again and was kept at 4˚C in refrigerator about 8 h (the consideration of acidic pH, Mn2+ ion metal at 42˚C).
Sample 2) The mixtures of nonionic acac, isopropyl alcohol in deionized water were maintained at alkaline pH with aid of TEA (triethanolamine) as cationic surfactant and Tris-HCl buffer (pH 8 - 9, 75 mM) solutions. Then 100 μl from this solution with 100 μl of 0.5 - 2 mg protein α-lactalbumin dissolved in deionized water were mixed and eventually 0.001 - 0.005 mg of zinc acetate salt[Zn(CH3COO) 2]as metal ion salt was added. It was kept during 2 - 3 days in incubator at 37˚C and was stored at 4˚C for 3 days (alkaline pH, Zn2+ ion at 37˚C).
Sample 3) First of all, we should prepare the mixture of 3-5ml nonionic cacac with 20 ml isopropyl alcohol in deionized water. This precursor would be able to regulate in final neutral pH by urea 1M solution (pH controllerreagent) and Tris-HCl buffer (pH 8 - 9, 0.075 M) solution (Tris hydroxyl methyl amino methane a cationic surfactant) with mild stir up. Then ion salts solution of MnCl2·4H2O or CaCl2·H2O as the self-assembly essential stimulant motor was added to them. In the end of work, 0.5 - 2 mg protein α-lactalbumin solution was mixed. This product was kept between one-two weeks in shaking water bath at 42˚C and 60 - 80 rpm (mixed Mn + Ca ions, alkaline pH, at 42˚C).
Sample 4) 5 - 7 ml urea (1 M) with 4 ml isopropyl alcohol and 4 ml deionized water were mixed well together. After adding protein α-lactalbumin and MnCl2·4H2O the pH of solution adjust in final alkali pH by 0.075 M Tris hydroxyl methyl amino methane. The obtained reaction solution was incubated at 37˚C between one or two weeks and then transferred to 4˚C for almost one day (alkaline pH, Mn2+ ion, 37˚C, without acac).
Sample 5) Chemical hydrolysis solution was designed and prepared for use of 10 ml isopropyl alcohol, 0.075 M Tris-HCl buffer (pH 7.5), acac (acetyl acetone) and 4 - 6 ml one molar urea solution, the pH was adjusted (6.0 to 7.5). 200 μl of this stock solution was added to 200 μl from 0.5 - 2 mg protein α-lactalbumin solution. Then MnCl2·4H2O as a ligand forming ion metal was added, and the hydrolysis reaction was performed. The final solution was incubated for 3 - 5 days in incubator at 37˚C and stored at 4˚C in one day (neutral pH, Mn2+ ion, 37˚C).
2.3. Sample Preparation for Transmission Electron Microscopy (TEM)
Final solusions were taken into Ultrasound for 5 - 10 min, then 15 µL of the samples were placed on carbon coated grids with help of micropipet and be allowed to airdrying. Examination was done using a Philips TEM CM 120.
3. Results and Discussion
3.1. SEM, TEM, XRD, Raman, DLS and FTIR Characterizations for Protein Nanotubes
Natural milk protein nanotubes characterized by SEM images and nanotubes morphology was observed and TEM pictures showed actually diameter of these nanotubes. XRD pattern indicated special structures according standard card and Raman spectroscopy investigated D and G bonds for exclusive nanotubes. Repeatability and stability of synthetic nanotubes structures in transparent water based solution was confirmed byqualitative dynamic light scattering (DLS) analysis and distribution size curves after long times from making. Doping of divalent ions-bridging between specific amino acids and carboxyl groups and finally the special vibrational bands for milk protein nanotubes are so considered by FTIR spectroscopy.
3.1. Characterization of Single Walled Protein Nanotubes Sample 1
In combined Figure 1 SEM (Philips, XL30) and TEM (Philips, CM 120) images and also FTIR spectroscopy (Thermo Nicolet Nexus 870, USA) for sample 1 exhibitedvery complete natural single walled protein nanotubes (5 - 7.5 nm). The critical calculation predicted that various forms of helically coiled structures are possible and those structures are energetically and thermo dynamically stable. Helical and spring structures of carbon nanotubes may have been observed as early as the 1950s as filamentary carbon structures, or fibers [15].
TEM images show very stable protein nanotubes soluble after 3 months. FTIR spectrum shows very sharp transmission band centered at 1724.49 cm−1 should be attributed to the C=O ketone or carboxylic acid stretch and 1624.88 cm−1 frequency for C=O amide or contaminating water in carbon skeleton in PNTs structure respectively. The signal at 2921.52 cm−1 is attributed to –CH3 stretching vibration band. The peak at 1411.22 cm−1 can be assigned to stretching vibration of C=C in aromatic rings, the medium peaks at 1280.30 cm−1 and short peaks in range of 615.79 - 544.84 cm−1 are belong to special PNTs skeleton. A very weak signal at 825.92 cm−1 can be assigned to stretching vibration of C-O-C groups andthe weak peak at 1151.41 cm−1 can be interpreted to the characteristic of primary alcohol [16]. In addition, the very strong and sharp transmission band centered at 431.73 cm−1 may be for ion salt Mn2+ as bridging complex ligand.
3.2. Characterization of SWPNTs Sample 2 by SEM and TEM
SWPNTs (4 - 6 nm) for sample 2 was illustrated in Figure 2 through SEM images of regularly bent during growth which are able to display the breaking of very thin film layers in boundary of nanofluids and exposured nano products from down over up. TEM images of branched products confirmed nearly 4 - 6 nm diameters for single-walled protein nanotubes.
In this important evidence, we can suggest the sharp G-bond and broad D-bond in limited ranges in Figure 3, because they can be different of pure carbon nanotubes (CNTs) and pure protein too.
3.3. Characterization of Beaded SWPNTs Sample 3 in Mixtures of Mn + Ca and Urea
We could prove that this product was kept stable after 3 months, by repetition of its Raman spectroscopy. (a) SEM, (b) TEM, (c) XRD(Seifert, Germany, 3003 PTS) and (d) FTIR were shown in Figure 4.
SEM and TEM images confirm the existence of very nice PNTs products soluble in nanofluid with diameter of (6.5 - 7.5) nm for SWPNTs. The XRD pattern of PNTs showed max intensity d100 plane in 21.5˚ (2θ) and another one in 70.73˚ (2θ) which is similar to CNTs. These results have made the Raman spectrumsuch a good tool to characterize PNTs that could prove these proofs. G-bond indicated at limited ranges between 1590.18 cm−1 (strong), –1427.01 cm−1 (short), and D-bond was seen as a
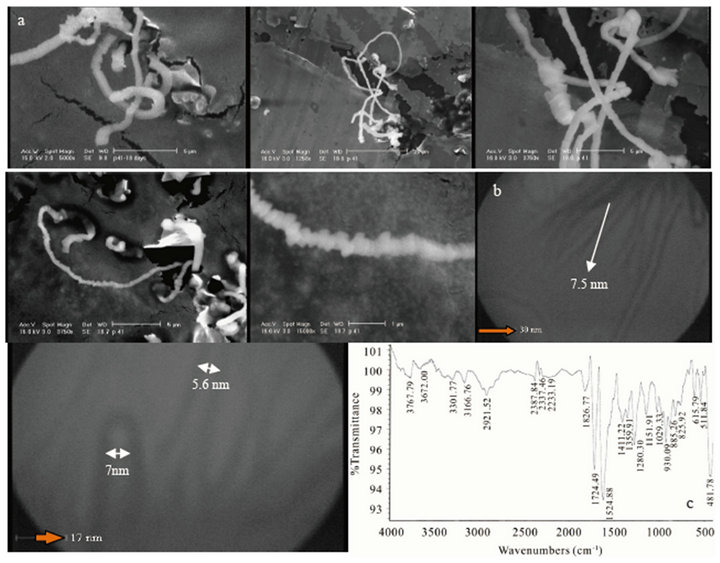
Figure 1. SEM (a) images of new helically coiled, (b) images of branched protein nanotubes, and FTIR spectroscopy (c) of SWNTs (5 - 7.5 nm) of PNTs.
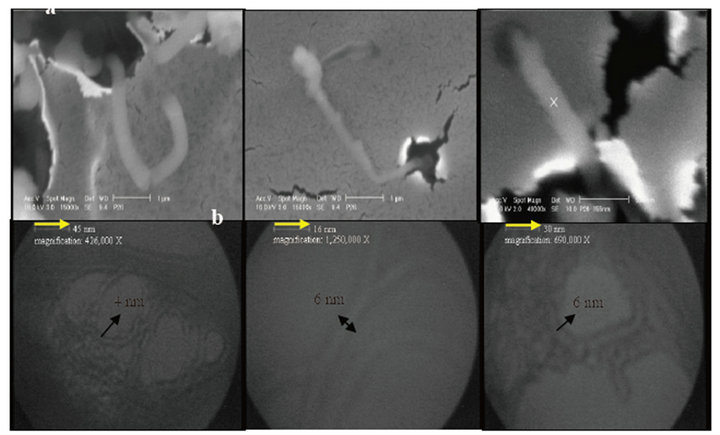
Figure 2. (a) SEM; (b) TEM (4 - 6 nm diameters) images of the helically coiled for sample 2.
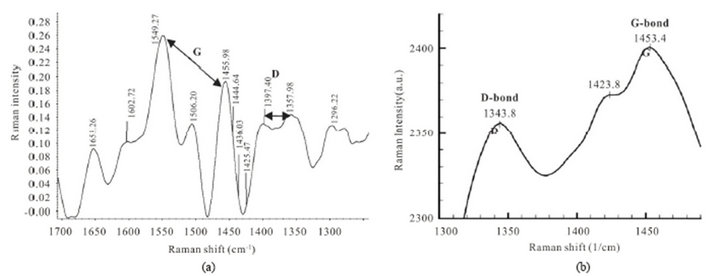
Figure 3. (a) Raman spectrum for PNTs of sample 2. G-bond spectrum is observed as doublet at ranges of (1549.27 - 1455.98 cm−1) and broad D-bond is showed at (1397.40 - 1357.98 cm−1); (b) G-bond (1453.4 cm−1) and D bond (1343.8 cm−1) in Raman spectroscopy for pure protein.

Figure 4. SEM and TEM pictures, XRD pattern and FTIR spectrum of bead-like single-walled protein nanotubes of sample 3 in appearance of two ion metal connectors.
weak bond at 1368.97 cm−1 (Figure 5(e)). All of these peaks have shifts related to CNTs. We could observe same peaks after 3 months in Raman spectroscopy (Figure 5(f)) which it showed very stability during this period. This morphology for sample 3 would produce under same conditions with Ca2+ cation ligand too. The characteristic vibrational modes of PNTs are: strong triplet peaks at 3464.67, 3361.50 and 3213.04 cm−1 for –OH and –NH stretching vibration modes, 1679.23 cm−1 for C=O stretching amide, 1625.44 cm−1 is attributed to contaminating water, the vibrational sharp signal centered at 1451.67 cm−1 is connected with the –OH bending deformation in carboxylic acids and phenolic groups and also it is belongs to –C=C– symmetric stretching vibration in CNTs. Two short and low broad peaks centered at 587.01 and 528.89 cm−1 can be accordance to inorganic bonds (Mn2+-O) or (Ca2+-O) ions metal in product. The extra peaks at around 878.04 - 786.82 cm-1 can be assigned to stretching vibration of C-O-C groups or specific of CNTsPNTs [17]. It was found that all of these PNTs vibrational modes have depicted fewer amounts of replacement frequencies and shifts in infrared spectra in comparison of CNTs.
3.4. Dynamic Light Scattering (DLS) Technique Analysis for Stable Nanofluid of Sample 3
The prepared protein nanofluids of product 3 can be maintained higher than 10 - 11 months (one year), which it was confirmed as only qualitative manner by DLSmeasurement. Furthermore, Figure 6 shows three curves which they were measured by Nano-ZS Malvern system, (a) displays the size distribution by number in which 100% of these nanotubes have diameter of 5.48 nm, (b) the curve of size distribution by intensity that can help us determine the diameter size of 6nm for 78% intensity with 0.8 nm width, and finally (c) shows size distribution by volume in which total of nanotubes have shown the diameter sizes of 5.75 nm with 0.97 nm width into 100% volume. On the other hand, the size distribution of these nanotubes has remained under 10 nm after one year in 4˚C, and these documents are very remarkable in performance of them.
3.5. Representation of Some Pictures of Protein Nanotubes of Product 3
Figure 7 shows schematic pictures of some PNTs nanofluid of product 3. (a) the evidence of beginning of synthesis and (b) physical transparency phenomenon after more than 11 months (one year) maintaining in 4˚C.
3.6. Characterization of SWPNTs Sample 4 Containing Various Concentrations of Urea Reagent
The other final characterizations of natural protein nanotubes (NPNTs) in this research are about the production of single walled NPNTs for sample 4 which was shown in Figure 8. Bead-like nanotubes were fabricated by chemical hydrolysis including urea concentrations and alkaline pH value. The diameter of nanotubes was determined by TEM characterization in Figure 8(b).
SEM was measured at 37˚C as narrow cylinder nanotubes of protein. TEM was shown this claim in diameters. XRD pattern determined all diffraction peaks which could be indexed in the protein nanotubes (PNTs) and also it compared with XRD of pure protein. The max sharp intensity for PNTs and broad pure protein curve were obtained in 22.17 (2θ) and 25 or 29.70 (2θ) respectively [18]. The strong and sharp peaks indicated that product has had good crystallinity. G-Bond (1515.03 cm−1) and D-bond (1352.58 cm−1) were also confirmed by Raman spectroscopy for these rolling amino acids nanostructures at 37˚C. It seems that these new characterization are especial for PNTs. FTIR spectroscopy (d) is as well as sample 3 contains very sharp absorption at 564.22 cm−1 for in organic metal ion in PNTs structure.
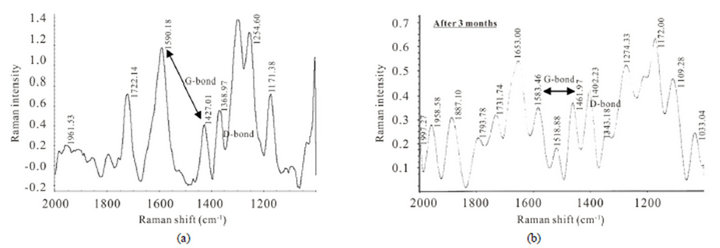
Figure 5. (a) and (b) Raman spectrum for PNTs of sample 3 (before and after 3 months). G-bond and D-bond spectra were observed at new frequencies, which they are different from of CNTs.
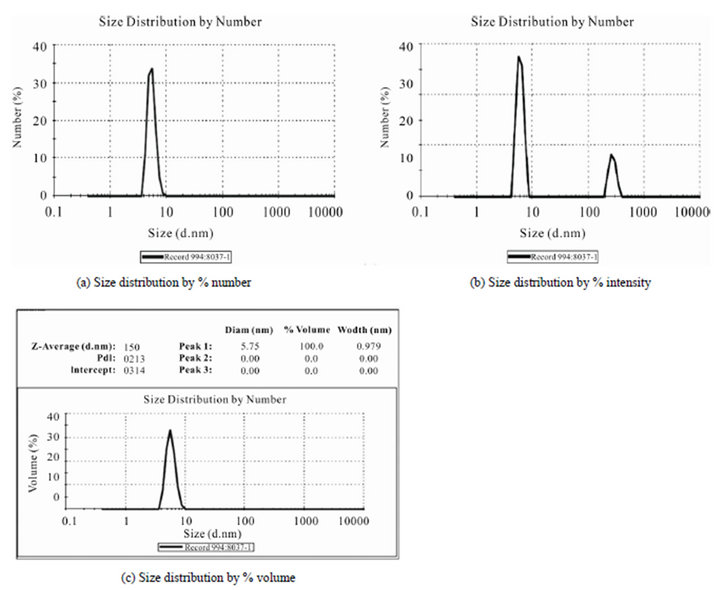
Figure 6. The various DLS curves of qualitative size distribution of sample 3 by (a) % number; (b) % intensity; (c) % volume versus diameter size (nm).

Figure 7. The exhibition of very clear and stability of α-lactalbum in nano-bio-tubes in two conditions, (a) in beginning of product synthesis and (b) after aging at 4˚C during one year.
3.7. Characterization of SWPNTs Sample 5 at pH 6 -7.5 after 3-5 days Annealing Time
Figure 9 shows new typical SEM images for protein nanotubes which they could grow as homogeneously perpendicular to the substrate like sweep-shape morphology (a). It is very interesting evidence, because the yield of synthetic products was reported very high and we can see them by TEM pictures (b). Figure 10 indicates FTIR spectrum (c) which is similar to the sample 4. It consists
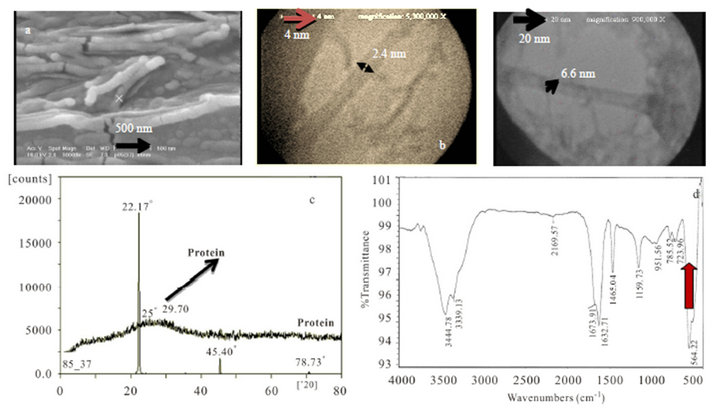
Figure 8. (a) SEM; (b) TEM images; (c) XRD pattern of nanotubes comparison with pure protein structure; (d) FTIR spectroscopy for synthetic SWPNTs sample 4.
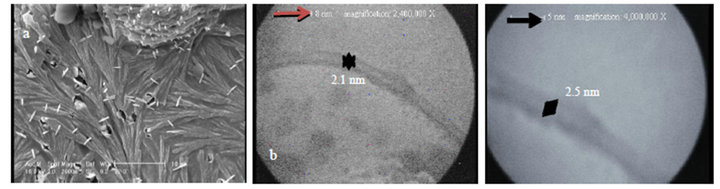
Figure 9. Typical new pictures of SWNPTs (3 nm) for PNTs, (a) SEM images; (b) TEM pictures of sample 5.
of two-branched and strong peaks belonged to –OH and –NH stretching bands at 3456.12 - 3360.87 cm−1, another sharp absorption amide band (C=O) was seen at 1671.67 - 1628.49 cm−1 which they attribute to protein structure, and finally the special short peaks at 587.65 - 522.48 and 469.40 cm−1 that they can be assigned to inorganic ligand metal Mn2+ and perfect PNTs nanostructures. The XRD pattern (d) shows max intensity phase d100 in 22.5˚ (2θ) with relatively sharp and strong peak, the other peaks at 37, 46, and 71˚ contain very low intensities, which imply and prove nanofine of nanotubes products. Raman spectrum (e) can be consider as doublet feature G-bond at 1565.01 - 1418.97 cm−1 ranges which was produced from the high degree of symmetry and order of carbon materials. They were generally used to identify well-ordered CNTs [19]. Generally the intensity of the E2g modes of graphitic materials are sharp and strong when the sample is highly crystalline, while disordered graphites and carbons show a feature around 1353.13 cm−1 as singlet short D-bond. On the other hand, the single peak at 1353.13 cm−1 might come from symmetry-lowering effect or defect effect in new products.
4. The Proposed Formation Mechanism of Protein Nanotubes
Based on our difficult experiments in Lab., we found the optimum useful concentration and incubation temperatures and aging time for achievement of only various morphologies of protein nanotubes. Our suggested method should be able to hydrolysis of protein α-lactalbumin actually towards cleavage sites (13Asp-X and
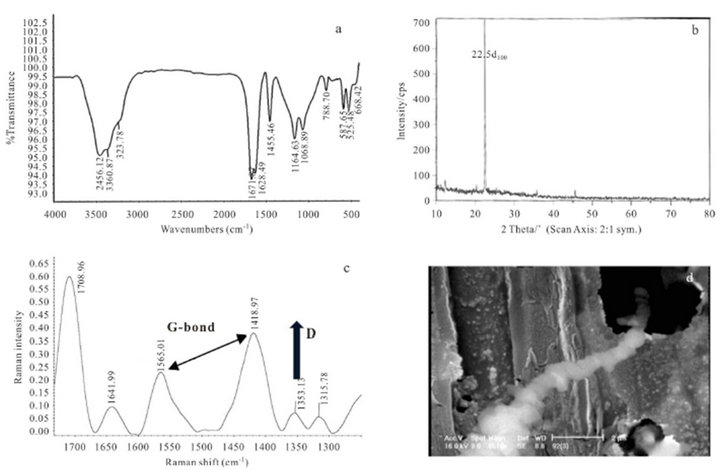
Figure 10. (a) FTIR spectroscopy; (b) XRD pattern; (c) Raman spectroscopy for sample 5; (d) shows SEM analysis of bamboo nanotubes form after 13 days keeping at 4˚C.
7-Glu-X) of peptide bonds, then amino acids and carboxylates ions were degraded by chemical hydrolysis into nanoemulsion environment in atmospheric pressure. Various cations, such as divalent ions Mn2+, Zn2+, Ca2+ and Fe2+ or mixed of them can induce tubular self-assembly of partially hydrolysed α-lactalbumin and the molar cation ratio determines which one is faster than the other. Our aims are to do nucleation and growth mechanism towards helical regular structures. Apparently, they all possess a structural element that is required for helical self-assembly. Ions size ligands and coordination number are likely to play an additional role, the ion is required to act as a connector between two building blocks via a salt bridge. Then Ca2+ ion has a high affinity for the wellknown (primary) Ca2+ binding site and stabilizes the native conformation upon binding and a secondary binding site was located 0.8 nm away from the primary binding site [20]. The folding and subsequent assembly of polypeptide chains reaction between the molecules is believed to be the sum of β-sheet stacking involving Hbonds (β-sheet) and Ca2+ ion bridges between specific carboxylates and amino acids, fixing the molecules in the right orientation in the assembly [21]. Reliance on magic power of self-assembly, actually we just provide specific conditions via chemical hydrolysis and surfactants in nanoemulsions for produced protein units that they canthermo dynamically reach to the lowest free energy. Self-assembly phenomena binds head-to-tail of building blocks together, then they turn laterally selfassemble to make two-dimensional arrays that twist to form hollow cylindrical filaments, with a diameter of about 3 - 8 nm single walled nanotubes and finally stacking of ringshaped cyclic peptide subunits leads to protein nanotube formation [6]. Of course, it may have been made nanoparticles, nanorods, and many others shape, but we try to sophisticate and elaborate our proposal towards formation nanotubes [22].
5. Conclusion
Single-walled natural milk protein nanotubes including new morphologies were fabricated by a chemically direct rolling process that achieved in this work along with high efficiency, low cost and control ability over chemically sol-gel method, not by enzymatic proteolysis and without using individual special and traditional catalysts in synthesis and any need to high temperatures for the first time (in comparison with ordinary CNTs). These synthetic natural nanotubes with the property of multiple-responsiveness to temperature, pH, formulation of nanofluid, ligands binding, surfactants and aging time are envisioned to have immense applications, especially for preparing new medicine nanofluids or nutritional drugs containing clear water-based solutions consist of various Ca2+, Zn2+ and Mn2+ ions in food science and pharmaceutics. So there are important points using Mn2+ and (Mn2+ + Ca2+) at different pH that are able to produce helically coiled and bead-like nanotubes shapes in samples 1 and 3 (pH is major) and the effect of annealing times at two pH indicated very influence factors in formation of bottom-up structures in samples 4 and 5 ( annealing time is important). The Zn2+ metal ion at alkaline pH could provide low yield branched production in sample 2.
6. Acknowledgements
The authors wish to express our sincere and deep gratitude to Mr. Rezai (SEM analysis), Dr. Sepehriseresht (TEM analysis), Mr. Naemi (XRD analysis), Mr. Ahmadi Taba (Raman and FTIR spectroscopy) and Mr. Bahari from RIPI (DLS analysis) for all vivacious times which we experienced by them.
REFERENCES
- M. G. Ryadnov and D. N Woolfson, “Fiber Recruiting Peptides: Noncovalent Decoration of an Engineered Protein Scaffold,” Journal of the American Chemical Society, Vol. 126, No. 24, 2004, pp. 7454-7455. doi:10.1021/ja048144r
- Y. Astie, H. Bayley and S. Howorka, “Protein Components for Nanodevices,” Current Opinion in Chemical Biology, Vol. 9, No. 6, 2005, pp. 576-584. doi:10.1016/j.cbpa.2005.10.012
- R. Mehravar, M. Jahanshahi and N. Saghatoleslami, “Production of Biological Nanoparticles from α-Lactalbumin for Drug Delivery and Food Science Application,” African Journal of Biotechnology, Vol. 8, No. 24, 2009, pp. 6822-6827.
- I. J. Arroyo-Maya, J. O. Rodliles-Lopez, M. CornejoMazon, G. F. Gutierrez-Lopez, A. Hernandez-Arana, C. Toledo-Nunez, G. V. Barbosa-Canovas, J. O. FloresFlores and H. Hernandez-Sanchez, “Effect of Different Treatments on the Ability of α-Lactalbumin to Form Nanoparticles,” Journal of Dairy Science, Vol. 95, No. 11, 2012, pp. 6204- 6214. doi:10.3168/jds.2011-5103
- S. Keten and M. J. Buehler, “Large Deformation and Fracture Mechanics of a Beta-Helical Protein Nanotube: Atomistic and Continuum Modeling,” Computer Methods in Applied Mechanics and Engineering, Vol. 197, No. 41, 2008, pp. 3203-3214. doi:10.1016/j.cma.2007.11.028
- J. F. Graveland-Bikker and C. G. de Kruifa, “Unique Milk Protein Based Nanotubes: Food and Nanotechnologymeet,” Trends in Food Science & Technology, Vol. 17, No. 5, 2006, pp. 196-203. doi:10.1016/j.tifs.2005.12.009
- S. Banta, Z. Megeed, M. Casali, K. Rege and M. L. Yarmush, “Engineering Protein and Peptide Building Blocks for Nanotechnology,” Journal of Nanoscience and Nanotechnology, Vol. 7, No. 2, 2007, pp. 387-401. doi:10.1166/jnn.2007.153
- J. D. Hartgerin and E. Beniash, S. Stupp, “Peptide-amphiphile nanofibers: A versatile scaffold for the preparation of self-assembling materials,” Proceedings of the National Academy Sciences, Vol. 99, 2002, pp. 133-138.
- X. Gao and H. Matsui, “Peptide-Based Nanotubes and Their Applications in Bionanotechnology,” Advanced Materials, Vol. 17, No. 17, 2005, pp. 2037-2050. doi:10.1002/adma.200401849
- P. Esmaeilzadeh, Z. Fakhroueian and A. A. Miran Beigi, “Synthesis of Biopolymeric α-Lactalbumin Protein Nanoparticles and Nanosphericals as Green Nanofluids Using in Drug Delivery and Food Technology,” Journal of Nano Research, Vol. 16, 2011, pp. 89-96. doi:10.4028/www.scientific.net/JNanoR.16.89
- P. Esmaeilzadeh, Z. Fakhroueian, M. Jahanshahi, M. Chamani, H. R. Zamanizadeh and B. Rasekh, “ A Synthetic Garden of State of the Art Natural Protein Nanoarchitectures Dispersed in Nanofluids,” Journal of Biomedical Nanotechnology, Vol. 7, No. 3, 2011, pp. 1-8. doi:10.1166/jbn.2011.1304
- P. Esmaeilzadeh, Z. Fakhroueian and Po. Esmaeilzadeh, “Producing the Controlled Hybrid Lattice of Natural Protein Nanotubes by Chemical Hydrolysis for Nanomedicine,” Journal of Nano Research, Vol. 21, 2013, pp. 7-13. doi:10.4028/www.scientific.net/JNanoR.21.7
- J. F. Graveland-Bikker, R, Ipsen, J. Otte and C. G. De Kruif, “Influence of Calcium on the Self-Assembly of Partially Hydrolyzed Alpha-Lactalbumin,” Langmuir, Vol. 20, No. 16, 2004, pp. 6841-6846. doi:10.1021/la049579v
- L. Chen, G. E. Remondetto and M. Subirade, “Food Protein-Based Materials as Nutraceutical Delivery Systems,” Trend in Food Science and Technology, Vol. 17, No. 5, 2006, pp. 272-283. doi:10.1016/j.tifs.2005.12.011
- M. Zhang and J. Li, “Carbon Nanotube in Different Shapes,” Materials Today, Vol. 12, No. 6, 2009, pp. 12- 18. doi:10.1016/S1369-7021(09)70176-2
- L. Chen and H. Xie, “Surfactant-Free Nanofluids Containing Doubleand Single-Walled Carbon Nanotubes Functionalized by a Wet-Mechanochemicalreaction,” Thermochemical Acta, Vol. 497, pp. 67-71.
- G. Jiang, L. Wang, C. Chen, X. Dong, T. Chen and H. Yu, “Study on Attachment of Highly Branched Molecules onto Multiwalled Carbon Nanotubes,” Material Letters, Vol. 59, No. 16, 2005, pp. 2085-2089. doi:10.1016/j.matlet.2005.01.085
- X. Huang, C. Pan and X. Huang, “Preparation and Characterization of γ-MnO2/CNTs Nanocomposite,” Materials Letters, Vol. 61, No. 4-5, 2007, pp. 934-936. doi:10.1016/j.matlet.2006.06.040
- D. Zhang, L. Shi, J. Fang, K. Dai and X. Li, “Preparation and Desalination Performance of Multiwall Carbon Nanotubes,” Materials Chemistry and Physics, Vol. 97, No. 2-3, 2006, pp. 415-419. doi:10.1016/j.matchemphys.2005.08.036
- M. Kronman, “Metal Ion Binding and the Molecular Conformational Properties of α-Lactalbumin,” Critical Reviews in Biochemistry and Molecular Biology, Vol. 24, No. 6, 1989, pp. 565-667. doi:10.3109/10409238909080054
- R. Ipsen and J. Otte, “Self-Assembly of Partially Hydrolysed α-Lactalbumin,” Biotechnology Advances, Vol. 25, No. 6, 2007, pp. 602-605. doi:10.1016/j.biotechadv.2007.07.006
- P. Esmaeilzadeh, Z. Fakhroueian, H. R. Zamanizadeh and B. Rasekh, “Single-Walled Natural Protein Nanotubes Soluble in Polar Solvent,” International Journal of Green Nanotechnology, Vol. 3, No. 1, 2011, pp. 37-46. doi:10.1080/19430892.2011.574529
NOTES
*Corresponding author.

