Open Journal of Veterinary Medicine
Vol. 2 No. 4 (2012) , Article ID: 25424 , 4 pages DOI:10.4236/ojvm.2012.24037
The Use of RIDA®COUNT for Monitoring the American Foulbrood Pathogen
1Faculty of Science, Charles University in Prague, Prague, Czech Republic
2Bee Research Institute at Dol, Libcice nad Vltavou, Czech Republic
3Biology Centre of the Academy of Sciences of the Czech Republic, v. v. i., Institute of Soil Biology, Ceske Budejovice, Czech Republic
4Faculty of Agrobiology, Food and Natural Resources, Czech University of Life Science, Prague, Czech Republic
Email: *stepan.ryba@gmail.com
Received September 26, 2012; revised November 2, 2012; accepted November 12, 2012
Keywords: AFB; Apis mellifera L.; Bacterial Disease; Diagnosis; Diseases of Honeybees; MYPGPn Medium; Paenibacillus larvae
ABSTRACT
American Foulbrood (AFB) is currently one of the most dangerous diseases in honeybees due to its high virulence and worldwide spread. Quick evaluation of the diagnosis of this disease is crucial. Successful eradication in the area indicates a need to test all bee colonies, but this is expensive and time consuming. A new method of detecting Paenibacillus larvae using the RIDA®COUNT test (R-Biopharm AG, Germany) was verified in the present study. The test is based on the principle of the cultivation test with MYPGPn medium, coloration of the bacteria with TTC (2,3,5-triphenyltetrazoliumchloride) chromophore, and heat treatment of the sample. Using this new method, color-highlighted colonies of P. larvae can be established on the seventh day after inoculating the spores. An identical number of colonies grown with the classic cultivation test on Petri dishes containing MYPGPn medium or RIDA®COUNT-P. larvae (RC-PL) sheets were verified.
1. Introduction
The honeybee (Apis mellifera L.) is inevitably attacked by pathogens that can weaken the bee colony, leading to the loss of entire bee colonies. American Foulbrood (AFB) is a highly infectious disease of bee colonies that has spread worldwide, becoming a serious threat for bee keeping. The disease has also caused a significant decrease in the number of bee colonies [1].
AFB is caused by the gram-positive rod-shaped bacteria Paenibacillus larvae, which is found only in larval and pupal bee stages. During the infection, many oval spores are created and spread by bees to the hive, beekeeping tools, contaminated combs, bee wax, pollen, and honey. After the infection by a spore in the digestive track, larvae and pupae turn brown and the final glue-like colloid liquid that comes from their decaying bodies is full of new spores. Spores are highly resistant to heat and chemicals and can survive for many years [2-5].
Bee colonies infected by P. larvae spores are not defeated immediately by the disease. After several months or years, the infection changes into the clinical stadium. The bee colony is weakened, unable to clean infected cells, and the entire colony dies. During this long period spores can be spread to surrounding bee colonies [1,6].
The main objective of this study was to verify the possibility of detecting the AFB bacteria and the sensitivity of the method based on the cultivation test performed on RIDA®COUNT sheets (R-Biopharm AG, Darmstadt, Germany) specific for cultivation of P. larvae spores. The method of P. larvae detection using RIDA®COUNT sheets was compared to classic cultivation tests on Petri dishes using MYPGPn agar [7]. The bacteria P. larvae was confirmed on Petri dishes and also using PCR [8]. The coloring principle of the method is based on the cultivation of microorganisms on standard nutrients in combination with a specific chromogenic detection system. TTC (2,3,5-triphenyltetrazoliumchloride) chromogen was used to visualize the growing bacteria on RIDA®COUNT sheets. Colonies are generated during the growth phase of the microorganisms, and the presence of a specific enzyme substrate changes the color to make colony visible [9]. The color change of colonies on RIDA®COUNT sheets allows to count colonies of bacteria grown.
2. Materials and Methods
2.1. Production of a Spore Suspension in Water
Samples of dried scales from P. larvae spores were vigorously mixed with water and dissolved into known concentration of spores, 105/ml, based on the number of spores estimated by the cultivation test on MYPGPn agar.
2.2. Preparation of MYPGPn Broth Medium
MYPGPn broth medium consisted of 3.5 g beef extract, 6 g acid casein hydrolyzate, 0.5 g soluble starch, 15 g yeast autolyzate, 3 g K2HPO4, 1 g Na-pyruvate, 1000 ml distilled H2O, and 2 g glucose plus nalidix acid (30 mg/l) to depress the growth of microbes except P. larvae. All of the substances (except glucose) were dissolved in water and sterilized for 20 minutes at 121˚C. The glucose was sterilized by filtration and added to partly cooled medium. NaOH was used to adjust the pH of the medium to 7.2 - 7.5.
2.3. Cultivation of P. larvae Spores on RIDA®COUNT Sheets
TTC (30 mg/l) and 10 µl of spore suspension (heat treated for 10 min at 80˚C) of known concentration was added to the prepared MYPGPn broth medium to correspond to the final concentrations of 101, 102, or 103 of spores per ml. The prepared MYPGPn broth medium and TTC and P. larvae inoculum was applied to a specified RIDA®COUNT sheet (R-Biopharm AG, Germany, company marking 041026-2, without the addition of nutrient medium). The RIDA®COUNT sheets were treated for 10 min at 80˚C to depress the growth of undesirable microorganisms.
2.4. Isolation of DNA, PCR Amplification, and Electrophoresis
Bacterial DNA was isolated using a QIAamp DNA Mini Kit (Qiagen). DNA was eluted using 50 μl of buffer and stored at –20˚C. Desalted primers were used to amplify a 451 bp fragment of a P. larvae gene (AF1f: GCT CTG TTG CCA AGG AAG AA; AF1r: AGG CGG AAT GCT TAC TGT GT) [8]. Samples of the amplicons were separated by electrophoresis on a 1.0% agarose gel. The approximate product size was determined using a 100-bp ladder.
3. Results
A sensitive and easy method for detecting P. larvae spores on RIDA®COUNT sheets using MYPGPn broth medium was tested in the laboratory. The P. larvae spores grew on the RIDA®COUNT sheets and with TTC chromophore, and grown P. larvae colonies exhibited a coloring reaction in the form of red dots (Figure 1). The P. larvae colonies grown on RIDA®COUNT sheets were well read on the seventh day as the coloring reaction was sufficient and other color-highlighted colonies did not grow. A quantitative comparison between the classic cultivation test on Petri dishes and the new test on RIDA®COUNT sheets specific for P. larvae detection showed equal quantities of inoculated spores on dishes and sheets. Detection of P. larvae colonies by the RIDA®COUNT test and Petri dishes using PCR showed the presence of colonies in both cases (data not shown). Negative and positive controls were performed for all types of tests.
4. Discussion
Due to the high rate of infection and rapid spread of AFB, a fast and reliable method for P. larvae detection and early recognition of infected bee colonies before the onset of a clinical stage is important. This ability can prevent not only bigger losses, but also the spread of the infection to broader areas. Due to disease screening, the overall costs for global eradication are reduced. Methods of detecting P. larvae based on PCR and nested PCR,
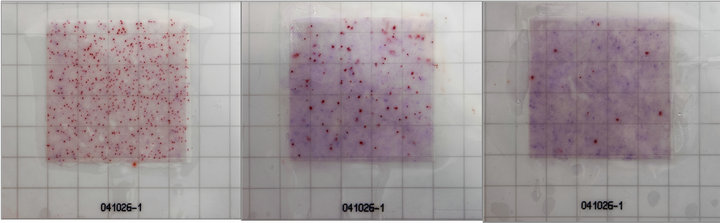 (a) (b) (c)
(a) (b) (c)
Figure 1. RIDA®COUNT—Paenibacillus larvae test. (a) 1000; (b) 100; and (c) 10 inoculated Paenibacillus larvae spores with the red coloring reaction (7th day) using TTC chromophore on liquid MYPGPn broth medium.
which use bee brood, honey, or bee debris, are known [3,10-12]. Nevertheless, these methods require a laboratory background and qualified laboratory staff. The standard cultivation tests on Petri dishes using MYPGPn agar medium are an alternative for testing the AFB pathogen. These methods require knowledge of laboratory routine and a technical background for test performance. The principle and application of knowledge of RIDA®COUNT tests (R-Biopharm AG, Germany, http://www.r-biopharm.com) was first described by H. Morita et al. [9].
Since being developed as alternatives to conventional agar plate methods, RIDA®COUNT tests have been used to detect the microorganisms, particularly in the clinical diagnostics and food industry. The use of RIDA®COUNT tests in cave research was described by Mulec et al. [13]. RIDA®COUNT tests can be used to screen both bacterial and fungal pathogens. The number of visible colonies indicates the number of CFUs grown on a specific medium during a specified incubation period at approximately 37˚C. MYPGPn medium is designed for the specific detection of P. larvae, which is able to resist in medium containing nalidixic acid (can be substituted with pipemidic acid). The elimination of undesired microorganisms is achieved by heat shock after inoculation of the RIDA®COUNT sheet. To verify the use of RIDA®COUNT sheets in practice, subsequent robust experiments with healthy and diseased bee colonies need to be performed. Screening samples from the hive using specific RIDA®COUNT tests for P. larvae detection may become a useful tool for fast and efficient detection of the pathogen in the environment. The total number of grown colored colonies could be an indicator of microbial status and could be used to assess the level of P. larvae infection in the hive. The paper RIDA®COUNT sheets are storage and they do not take up much space. The cards can be stored in the freezer for later use, to revitalize the colonies.
5. Acknowledgements
We are very grateful to R-Biopharm AG (Darmstadt, Germany) for kindly providing the test cards. The work was supported by the project Knowledge and Technology Transfer in Selected Regions based on European educational model “Technology Transfer Manager” (reg. No. CZ.1.07/2.4.00/12.0082), and by grant NAZV QH, reg. No. 72144 of the National Agency for Agriculture Research, Czech Republic. The authors would like to acknowledge Janez Mulec (Karst Research Institute, Scientific Research Centre of the Slovenian Academy of Sciences and Arts, Postojna, Slovenia) for initial information on the use of RIDA®COUNT tests. Manuscript was edited by San Francisco Edit.
REFERENCES
- H. Hansen and C. J. Brodsgaard, “American Foulbrood: A Review of Its Biology, Diagnosis and Control,” Bee World, Vol. 80, No. 1, 1999, pp. 195-201.
- D. C. de Graaf, A. M. Alippi, M. Brown, J. D. Evans, M. Feldlaufer, A. Gregorc, M. Hornitzky, S. F. Pernal, D. M. Schuch, D. Titera, V. Tomkies and W. Ritter, “Diagnosis of American Foulbrood in Honey Bees: A Synthesis and Proposed Analytical Protocols,” Letters in Applied Microbiology, Vol. 43, No. 6, 2006, pp. 583-590. doi:10.1111/j.1472-765X.2006.02057.x
- D. C. de Graaf, P. De Vos, M. Heyndrickx, S. Van Trappen, N. Peiren and F. J. Jacobs, “Identification of Paenibacillus larvae to the Subspecies Level: An Obstacle for AFB Diagnosis,” Journal of Invertebrate Pathology, Vol. 91, No. 2, 2006, pp. 115-123. doi:10.1016/j.jip.2005.10.010
- E. Genersch, E. Forsgren, J. Pentikainen, A. Ashiralieva, S. Rauch, J. Kilwinski and I. Fries, “Reclassification of Paenibacillus larvae subsp. pulvifaciens and Paenibacillus larvae subsp. larvae as Paenibacillus larvae without Subspecies Differentiation,” International Journal of Systematic and Evolutionary Microbiology, Vol. 56, No. 3, 2006, pp. 501-511. doi:10.1099/ijs.0.63928-0
- U. Riessberger-Galle, W. von der Ohe and K. Crailsheim, “Adult Honeybee’s Resistance against Paenibacillus larvae larvae, the Causative Agent of the American Foulbrood,” Journal of Invertebrate Pathology, Vol. 77, No. 4, 2001, pp. 231-236. doi:10.1006/jipa.2001.5032
- N. Bakhiet and D. P. Stahly, “Ultrastructure of Sporulating Bacillus larvae in a Broth Medium,” Applied and Environmental Microbiology, Vol. 50, No. 3, 1985, pp. 690- 692.
- D. Titera and M. Haklova, “Detection Method of Paenibacillus larvae Larvae from Beehive Winter Debris,” Apimondia, Vol. 38, 2003, pp. 131-133.
- T. Bakonyi, I. Derakhshifar, E. Grabensteiner and N. Nowotny, “Development and Evaluation of PCR Assays for the Detection of Paenibacillus larvae in Honey Samples: Comparison with Isolation and Biochemical Characterization,” Applied and Environmental Microbiology, Vol. 69, No. 3, 2003, pp. 1504-1510. doi:10.1128/AEM.69.3.1504-1510.2003
- H. Morita, M. Ushiyama, S. Aoyama and M. Iwasaki, “Sensitivity and Specificity of the Sanita-Kun Aerobic Count: Internal Validation and Independent Laboratory Study,” Journal of AOAC International, Vol. 86, No. 2, 2003, pp. 355-366.
- F. M. Lauro, M. Favaretto, L. Covolo, M. Rassu and G. Bertoloni, “Rapid Detection of Paenibacillus larvae from Honey and Hive Samples with a Novel Nested PCR pRotocol,” International Journal of Food Microbiology, Vol. 81, No. 3, 2003, pp. 195-201. doi:10.1016/S0168-1605(02)00257-X
- V. A. Govan, M. H. Allsopp and S. Davison, “A PCR Detection Method for Rapid Identification of Paenibacillus larvae,” Applied and Environmental Microbiology, Vol. 65, No. 5, 1999, pp. 2243-2245.
- S. Ryba, D. Titera, M. Haklova and P. Stopka, “A PCR Method of Detecting American Foulbrood (Paenibacillus larvae) in Winter Beehive Wax Debris,” Veterinary Microbiology, Vol. 139, No. 1-2, 2009, pp. 193-196. doi:10.1016/j.vetmic.2009.05.009
- J. Mulec, V. Kristufek and A. Chronakova, “Comparative Microbial Sampling from Eutrophic Caves in Slovenia and Slovakia Using RIDA®COUNT Test Kits. Use of RIDA®COUNT in Caves,” International Journal of Speleology, Vol. 41, No. 1, 2012, pp. 1-8. doi:10.5038/1827-806X.41.1.1
Abbreviations
RC-PL = RIDA®COUNT-Paenibacillus larvae;
TTC = 2,3,5-triphenyltetrazoliumchloride.
NOTES
*Corresponding author.

