Journal of Cancer Therapy
Vol. 4 No. 5 (2013) , Article ID: 33799 , 9 pages DOI:10.4236/jct.2013.45114
Constitutional Mismatch Repair-Deficiency Syndrome Is a Rare Cause of Cancer Even in a Highly Consanguineous Population*
![]()
1Human Cancer Genomic Research, Research Center, King Faisal Specialist Hospital and Research Cancer, Riyadh, Saudi Arabia; 2Department of Pediatric Hematology/Oncology, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia; 3Department of Pathology, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia; 4Developmental Genetics Unit, Department of Genetics, King Faisal Specialist Hospital and Research Center, Riyadh, Saudi Arabia.
Email: †kkuraya@kfshrc.edu.sa
Copyright © 2013 Rong Bu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 17th, 2013; revised June 18th, 2013; accepted June 25th, 2013
Keywords: Constitutional Mismatch Repair-Deficiency (CMMR-D); Lynch Syndrome (LS); MLH1; MSH2; MSH6 and PMS2; Biallelic Germline Mutations
ABSTRACT
Biallelic germline mutations in the mismatch repair genes, including MLH1, MSH2, MSH6 or PMS2, lead to a recessive constitutional mismatch repair-deficiency (CMMR-D) syndrome characterized by early onset malignancies in children and young adults. Because consanguinity unmasks autosomal recessive disorders, we hypothesized that the frequency of CMMR-D is inflated in the highly consanguineous population of Saudi Arabia. In this study, 371 pediatric and young adult patient samples from Saudi Arabia that cover the tumor spectrum of CMMR-D syndrome were analyzed for biallelic germline mutations in the MLH1, MSH2, MSH6 and PMS2 with the use of direct genomic sequencing. However, none of the 371 patients involved in the study was found to have biallelic pathological mutations of MLH1, MSH2, MSH6 or PMS2. This result indicates that CMMR-D is exceptionally rare among pediatric cancer patients and adult early onset cancer patients, even in the highly consanguineous Saudi population. Our findings suggest that larger cohorts will be needed, particularly in outbred populations, to determine the frequency of CMMR-D and that routine screening for this syndrome among cancer patients is not warranted.
1. Introduction
The mismatch repair (MMR) system contributes to genome integrity and DNA replication fidelity; and the MLH1, MSH2, MSH6 and PMS2 genes play a major role in this process [1]. MMR primarily remedies base-base mismatches and small insertion deletion loops (IDLs) that may occur during DNA replication [2]. In order to achieve this goal this mismatch repair machinery has to distinguish and to be directed to the newly synthesized strand from the template that carries the erroneous genetics information [3].
Heterozygous germline mutations in MLH1, MSH2, MSH6 and PMS2 cause Lynch syndrome (LS), an autosomal dominant cancer syndrome. It is the most common cause of hereditary colon cancer and is characterized by an increased risk for colorectal and endometrial cancers, with recent estimates of lifetime risks ranging from 22% to 66% and 14% to 39%, respectively [4-7]. Additional LS-associated cancers may occur most notably ovarian, gastric and renal pelvis cancers although lifetime risk is typically less than 10% for these tumors [8].
While LS patients harbor heterozygous mutant MMR alleles, there are 78 patients from 46 families diagnosed as CMMR-D, a recently recognized autosomal recessively inherited cancer syndrome caused by biallelic germline mutations in one of the MMR genes [9]. The tumour spectrum in CMMR-D syndrome includes childhood malignancies (mainly hematological malignancies and/or brain tumors) and very early onset LS-associated tumors [9-14]. 74% of cases show signs of neurofibromatosis type 1 (NF1), mainly café-au-lait spots (CLS) [14]. In addition, some individual cases have also been reported to be diagnosed with other cancers, including neuroblastoma, Wilms tumors, ovarian neuroectodermal tumor, myofibromatosis and sarcoma [15-18].
Middle Eastern populations in general and Saudi Arabia in particular have high rates of consanguinity, with an average prevalence of first cousin marriages (the most common form of consanguinity) of 57.7% [19,20]. Such high consanguinity rates are known to result in unmasking of autosomal recessive disorders including those that are exceedingly rare. Thus, we reasoned that CMMR-D will be relatively more common in our Saudi population. Since no data exists on the frequency of CMMR-D among patients with the various forms of cancer that are known to be associated with this disorder, we set out to systematically examine a large series of patients with these malignancies for evidence of CMMR-D. Our data suggest that CMMR-D syndrome is exceptionally rare among pediatric and young adult cancer patients even in the highly consanguineous Saudi population.
2. Material and Methods
2.1. Patients Selection
371 Saudi pediatric and young adult archived cancer samples from the tumors spectrum of CMMR-D that have been reported in the literature were selected in this study. Cancer samples were selected based on the presence of any of the following: early onset cancer and patients with multiple tumor types and patients with affected siblings (Table 1). All archived samples included in this study were approved by Institutional Review Board (IRB) at King Faisal Specialist Hospital and Research Centre for the following Research Advisory Council (RAC) projects RAC# 2120017, RAC# 2040004 and RAC# 2090012.
2.2. Detection of Mutation and DNA Sequencing Analysis
To search for germline mutations, DNA (obtained from blood or normal tissue) was amplified using PCR primers. Primer pairs that were designed with “Primer 3” software are listed in Supplementary Table 1. All exons of the MLH1, MSH2, MSH6 and PMS2 genes, including splice junctions, were amplified by polymerase chain reaction (PCR). PCR was performed in 25 μl reactions volume containing 0.2 mM dNTPs, 0.2 units of Hot Star DNA polymerase (Qiagen), 0.2 μl forward and reverses primer respectively, 2.3 mM MgCl2 and 20 ng of DNA. The
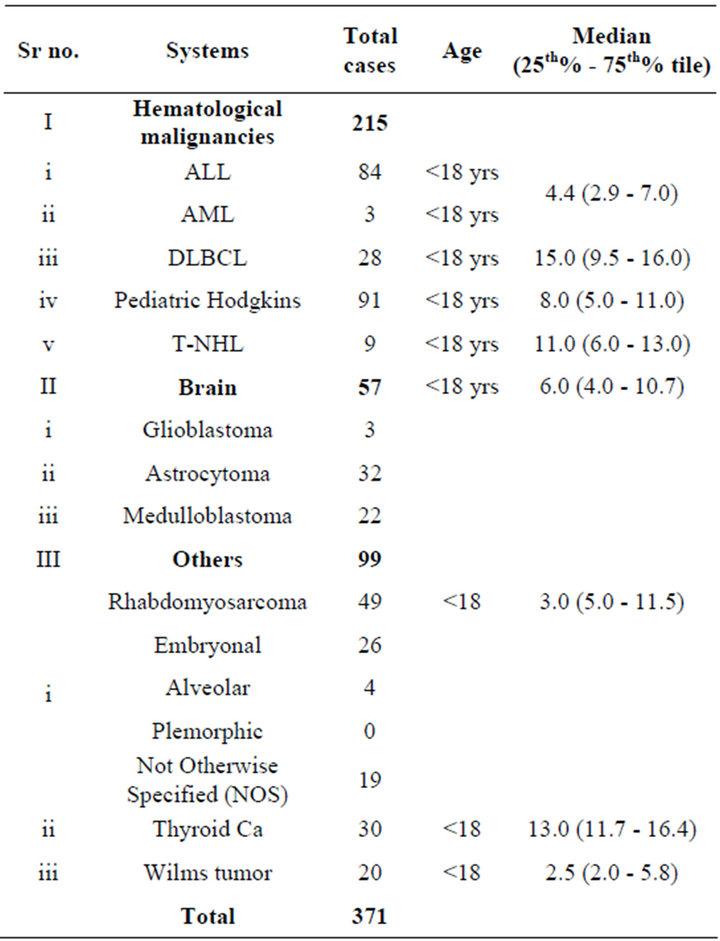
Table 1. Patient selection criteria and tumor type.
PCR conditions was as follows: 95˚C for 10 min followed by 35 cycles of 95˚C for 45 s, 60˚C for 45 s and 72˚C for 1 min, and ending with an elongation step at 72˚C for 10 min. The PCR products were purified with AMPure (Agencourt, Danvers, MA) Kit and then subjected to automated cycle sequencing with the ABI Prism Big Dye Terminator Cycle v3.1 sequencing kit (Applied Biosystems, Foster City, CA). Sequence analysis was performed using the Mutation Surveyor v3.24 (Soft-genetics LLC, State College, PA) software. Reference sequence of all genes was downloaded from GenBank.
3. Results
Previous studies suggest that the clinical features tend to differ in patients carrying biallelic germline mutation in different MMR genes. MLH1 and MSH2 mutation appear to present more frequently than MSH6 and PMS2 mutation in patients with hematologic malignancies. In contrast, the MSH6 and PMS2 biallelic germline mutation is more commonly detected in patient with brain tumor. Furthermore, the MLH1 and MSH2 mutation carriers have tendency to develop tumor earlier than those with MSH6 and PMS2 mutations. Despite the clear predominance of different type of MMR genes in different tumor types, to fully screen for CMMR-D in our study we sequenced all the 4 MMR genes that are known to be involved in the CMMR-D tumor spectrums. However, in spite of this comprehensive approach none of the 371 patients analyzed were found to have biallelic pathological mutations of MLH1, MSH2, MSH6 or PMS2.
4. Discussion
Careful search for CMMR-D Syndrome among pediatric cancer patients is warranted by the accumulation of reports describing more patients suffering from this syndrome [11]. The recognition of the actual incidence of this syndrome allows to: 1) understand the role of impaired MMR in the pathogenesis of sporadic cancers such as hematological malignances, brain tumors, sarcomas, which deserves further investigation; 2) develop specific therapeutic considerations for the patient with CMMR-D syndrome due to the MMR-defect caused resistance to certain chemotherapeutic agents [21-24]; 3) establish surveillance/guidelines for CMMR-D syndrome families e.g. parents are obligate carriers; and finally 4) prevent this new syndrome and misdiagnosis of this new syndrome and NF1. Given the significant clinical implications for diagnosis, prognosis, therapy and family counseling and the scarcity of data regarding CMMR-D among general population [9], the prevalence of CMMRD among young cancer patients requires further characterization.
The results of the current study demonstrate that the prevalence of CMMR-D among young cancer patients in Saudi population is exceedingly low. There are no available data to compare our result with, since our knowledge of the CMMR-D syndrome originates primarily from case reports. To our knowledge, this is the largest study ever done to delineate the prevalence of CMMR-D among pediatric and young adult cancer patients. An additional strength of our study is the inclusion of samples from the tumor spectrum of CMMR-D that have been reported in the literature. These include four groups of tumors: 1) hematological malignancies [10]; 2) brain tumors [25,26]; and others [17]. We also chose to sequence all four genes known to cause CMMR-D instead of resorting to easier screening methods like microsatellite instability (MSI) and or immunohistochemistry (IHC). Typically, confirmation of the diagnosis of CMMRD involves the analysis of MSI and/or IHC followed by mutational analysis [10,27] but since MSI analysis can be unreliable in CMMR-D-related brain tumors and IHC can show normal results in CMMR-D cases with underlying missense mutation [9,11,26,28], we decide to go directly to confirmatory mutation analysis of all MMR genes known to be involved in CMMR-D including MLH1, MSH2, MSH6, PMS2 despite the markedly higher cost associated with this approach.
Although a size of 371 samples that represent the spectrum of CMMR-D-related cancers and of the right age appears large, we concede that our sample size calculation was complicated by the lack of any published case series that systemically investigate the frequency of CMMR-D. So while it is clear that a larger cohort is required to identify CMMR-D, the minimal size required is difficult to predict. Nevertheless, our data show that the frequency is <0.0027 (<1/371) and since this estimate is based on a consanguineous population in which autosomal recessive disorders are usually higher in frequency, we expect that the frequency will be even lower in outbred populations. Another implication from our study is that CMMR-D is not an important causal factor in the unexplained trend of high frequency of CRC in our population and the younger age distribution [29-31]. This leaves open the possibility of other recessive loci contributing to this pattern and/or perhaps other environmental factors, an area of research that we are currently pursuing. Until then, our data suggest that routine screening for CMMR-D among Saudi pediatric cancer patients is not recommended.
5. Acknowledgements
We thank, Ms. Mehar Sultana for technical help for ALL project; and Ms. Maha Al-Rasheed and Khadija A. S. AlObaisi for their technical assistance, Dr Sarita Prabhakaran, and Zeeshan Qadri for data abstraction and analysis.
REFERENCES
- B. Liu, N. C. Nicolaides, S. Markowitz, J. K. Willson, R. E. Parsons, J. Jen, N. Papadopolous, P. Peltomaki, A. de la Chapelle, S. R. Hamilton, et al., “Mismatch Repair Gene Defects in Sporadic Colorectal Cancers with Microsatellite Instability,” Nature Genetics, Vol. 9, No. 1, 1995, pp. 48-55. doi:10.1038/ng0195-48
- G. Marra and J. Jiricny, “DNA Mismatch Repair and Colon Cancer,” Advances in Experimental Medicine and Biology, Vol. 570, 2005, pp. 85-123. doi:10.1007/1-4020-3764-3_4
- C. P. Kratz, C. M. Niemeyer, E. Juttner, M. Kartal, A. Weninger, A. Schmitt-Graeff, U. Kontny, M. Lauten, S. Utzolino, J. Radecke, C. Fonatsch and K. Wimmer, “Childhood T-Cell Non-Hodgkin’s Lymphoma, Colorectal Carcinoma and Brain Tumor in Association With CafeAu-Lait Spots Caused by a Novel Homozygous PMS2 Mutation,” Leukemia, Vol. 22, No. 5, 2008, pp. 1078- 1080. doi:10.1038/sj.leu.2405008
- F. Alarcon, C. Lasset, J. Carayol, V. Bonadona, H. Perdry, F. Desseigne, Q. Wang and C. Bonaiti-Pellie, “Estimating Cancer Risk in HNPCC by the GRL Method,” European Journal of Human Genetics, Vol. 15, No. 8, 2007, pp. 831-836. doi:10.1038/sj.ejhg.5201843
- M. A. Jenkins, J. G. Dowty, J. L. Hopper and M. C. Southey, “Molecular Screening of All Colorectal Tumors Diagnosed before Age 50 Years Followed by Genetic Testing Efficiently Identifies Lynch Syndrome Cases,” International Journal of Cancer, Vol. 124, No. 5, 2009, pp. x-i. doi:10.1002/ijc.24173
- F. Quehenberger, H. F. Vasen and H. C. van Houwelingen, “Risk of Colorectal and Endometrial Cancer for Carriers of Mutations of the Hmlh1 and Hmsh2 Gene: Correction for Ascertainment,” Journal of Medical Genetics, Vol. 42, No. 6, 2005, pp. 491-496. doi:10.1136/jmg.2004.024299
- E. Stoffel, B. Mukherjee, V. M. Raymond, N. Tayob, F. Kastrinos, J. Sparr, F. Wang, P. Bandipalliam, S. Syngal and S. B. Gruber, “Calculation of Risk of Colorectal and Endometrial Cancer among Patients with Lynch Syndrome,” Gastroenterology, Vol. 137, No. 5, 2009, pp. 1621-1627. doi:10.1136/jmg.2004.024299
- M. Aarnio, J. P. Mecklin, L. A. Aaltonen, M. NystromLahti and H. J. Jarvinen, “Life-Time Risk of Different Cancers in Hereditary Non-Polyposis Colorectal Cancer (HNPCC) Syndrome,” International Journal of Cancer, Vol. 64, No. 6, 1995, pp. 430-433. doi:10.1002/ijc.2910640613
- K. Wimmer and J. Etzler, “Constitutional Mismatch Repair-Deficiency Syndrome: Have We So Far Seen Only the Tip of an Iceberg?” Human Genetics, Vol. 124, No. 2, 2008, pp. 105-122. doi:10.1007/s00439-008-0542-4
- T. Ripperger, C. Beger, N. Rahner, K. W. Sykora, C. L. Bockmeyer, U. Lehmann, H. H. Kreipe and B. Schlegelberger, “Constitutional Mismatch Repair Deficiency and Childhood Leukemia/Lymphoma—Report on a Novel Biallelic MSH6 Mutation,” Haematologica, Vol. 95, No. 5, 2010, pp. 841-844. doi:10.3324/haematol.2009.015503
- K. Wimmer and C. P. Kratz, “Constitutional Mismatch Repair-Deficiency Syndrome,” Haematologica, Vol. 95, No. 5, 2010, pp. 699-701. doi:10.3324/haematol.2009.021626
- K. Jasperson, W. Samowitz and R. Burt, “Constitutional Mismatch Repair-Deficiency Syndrome Presenting as Colonic Adenomatous Polyposis: Clues from the Skin,” Clinical Genetics, Vol. 80, No. 4, 2010, pp. 394-397.
- K. E. Felton, D. M. Gilchrist and S. E. Andrew, “Constitutive Deficiency in DNA Mismatch Repair,” Clinical Genetics, Vol. 71, No. 6, 2007, pp. 483-498. doi:10.1111/j.1399-0004.2007.00803.x
- Q. Wang, C. Lasset, F. Desseigne, D. Frappaz, C. Bergeron, C. Navarro, E. Ruano and A. Puisieux, “Neurofibromatosis and Early Onset of Cancers in hMLH1-Deficient Children,” Cancer Research, Vol. 59, No. 2, 1999, pp. 294-297.
- S. Kruger, M. Kinzel, C. Walldorf, S. Gottschling, A. Bier, S. Tinschert, A. von Stackelberg, W. Henn, H. Gorgens, S. Boue, K. Kolble, R. Buttner and H. K. Schackert, “Homozygous PMS2 Germline Mutations in Two Families with Early-Onset Haematological Malignancy, Brain Tumours, HNPCC-Associated Tumours, and Signs of Neurofibromatosis Type 1,” European Journal of Human Genetics, Vol. 16, No. 1, 2008, pp. 62-72. doi:10.1038/sj.ejhg.5201923
- M. De Vos, B. E. Hayward, R. Charlton, G. R. Taylor, A. W. Glaser, S. Picton, T. R. Cole, E. R. Maher, C. M. McKeown, J. R. Mann, J. R. Yates, D. Baralle, J. Rankin, D. T. Bonthron and E. Sheridan, “PMS2 Mutations in Childhood Cancer,” Journal of the National Cancer Institute, Vol. 98, No. 5, 2006, pp. 358-361. doi:10.1093/jnci/djj073
- C. P. Kratz, S. Holter, J. Etzler, M. Lauten, A. Pollett, C. M. Niemeyer, S. Gallinger and K. Wimmer, “Rhabdomyosarcoma in Patients with Constitutional MismatchRepair-Deficiency Syndrome,” Journal of Medical Genetics, Vol. 46, No. 6, 2009, pp. 418-420. doi:10.1136/jmg.2008.064212
- J. W. Poley, A. Wagner, M. M. Hoogmans , F. H. Menko, C. Tops, J. M. Kros, R. E. Reddingius, H. Meijers-Heijboer, E. J. Kuipers and W. N. Dinjens, “Biallelic Germline Mutations of Mismatch-Repair Genes: A Possible Cause for Multiple Pediatric Malignancies,” Cancer, Vol. 109, No. 11, 2007, pp. 2349-2356. doi:10.1002/cncr.22697
- M. A. El-Hazmi, A. R. Al-Swailem, A. S. Warsy, A. M. Al-Swailem, R. Sulaimani and A. A. Al-Meshari, “Consanguinity among the Saudi Arabian Population,” Journal of Medical Genetics, Vol. 32, No. 8, 1995, pp. 623-626. doi:10.1002/cncr.22697
- M. I. El-Mouzan, A. A. Al-Salloum, A. S. Al-Herbish, M. M. Qurachi and A. A. Al-Omar, “Regional Variations in the Prevalence of Consanguinity in Saudi Arabia,” Saudi Medical Journal, Vol. 28, No. 12, 2007, pp. 1881-1884.
- R. H. Scott, T. Homfray, N. L. Huxter, S. G. Mitton, R. Nash, M. N. Potter, D. Lancaster and N. Rahman, “Familial T-Cell Non-Hodgkin Lymphoma Caused by Biallelic MSH2 Mutations,” Journal of Medical Genetics, Vol. 44, No. 7, 2007, p. e83. doi:10.1136/jmg.2007.048942
- J. Jiricny, “The Multifaceted Mismatch-Repair System,” Nature Reviews Molecular Cell Biology, Vol. 7, No. 5, 2006, pp. 335-346. doi:10.1038/nrm1907
- A. Peters, H. Born, R. Ettinger, P. Levonian and K. B. Jedele, “Compound Heterozygosity for MSH6 Mutations in a Pediatric Lymphoma Patient,” Journal of Pediatric Hematology/Oncology, Vol. 31, No. 2, 2009, pp. 113- 115. doi:10.1038/nrm1907
- R. H. Scott, S. Mansour, K. Pritchard-Jones, D. Kumar, F. MacSweeney and N. Rahman, “Medulloblastoma, Acute Myelocytic Leukemia and Colonic Carcinomas in a Child with Biallelic MSH6 Mutations,” Nature Clinical Practice Oncology, Vol. 4, No. 2, 2007, pp. 130-134. doi:10.1038/nrm1907
- H. Toledano, Y. Goldberg, I. Kedar-Barnes, H. Baris, R. M. Porat, C. Shochat, D. Bercovich, E. Pikarsky, I. Lerer, I. Yaniv, D. Abeliovich and T. Peretz, “Homozygosity of MSH2 c.1906G→C Germline Mutation Is Associated with Childhood Colon Cancer, Astrocytoma and Signs of Neurofibromatosis Type I,” Familial Cancer, Vol. 8, No. 3, 2009, pp. 187-194. doi:10.1007/s10689-008-9227-3
- L. Giunti, V. Cetica, U. Ricci, S. Giglio, I. Sardi, M. Paglierani, E. Andreucci, M. Sanzo, M. Forni, A. M. Buccoliero, L. Genitori and M. Genuardi, “Type A Microsatellite Instability in Pediatric Gliomas as an Indicator of Turcot Syndrome,” European Journal of Human Genetics, Vol. 17, No. 7, 2009, pp. 919-927. doi:10.1038/ejhg.2008.271
- J. Mueller, I. Gazzoli, P. Bandipalliam, J. E. Garber, S. Syngal and R. D. Kolodner, “Comprehensive Molecular Analysis of Mismatch Repair Gene Defects in Suspected Lynch Syndrome (Hereditary Nonpolyposis Colorectal Cancer) Cases,” Cancer Research, Vol. 69, No. 17, 2009, pp. 7053-7061. doi:10.1038/ejhg.2008.271
- G. Bougeard, F. Charbonnier, A. Moerman, C. Martin, M. M. Ruchoux, N. Drouot and T. Frebourg, “Early Onset Brain Tumor and Lymphoma in MSH2-Deficient Children,” The American Journal of Human Genetics, Vol. 72, No. 1, 2003, pp. 213-216. doi:10.1086/345297
- W. H. Isbister, M. Murad and Z. Habib, “Rectal Cancer in the Kingdom of Saudi Arabia: The King Faisal Specialist Hospital experience,” Australian & New Zealand Journal of Surgery, Vol. 70, No. 4, 2000, pp. 269-274. doi:10.1086/345297
- “Eight Year Cancer Incidence among Nations of the GCC States. 1998-2005 Cancer Incidence Report of Gulf Cooperation Council States,” Excutive Board of the Health Ministers’ Council, Riyadh, King Fahd National Library Cataloging-in-Publication Data, 2009.
- H. S. Al-Eid, “Cancer Incidence Report Saudi Arabia 1999-2000,” King Faisal Specialist Hospital and Research Centre, Riyadh, 2004.
Supplementary
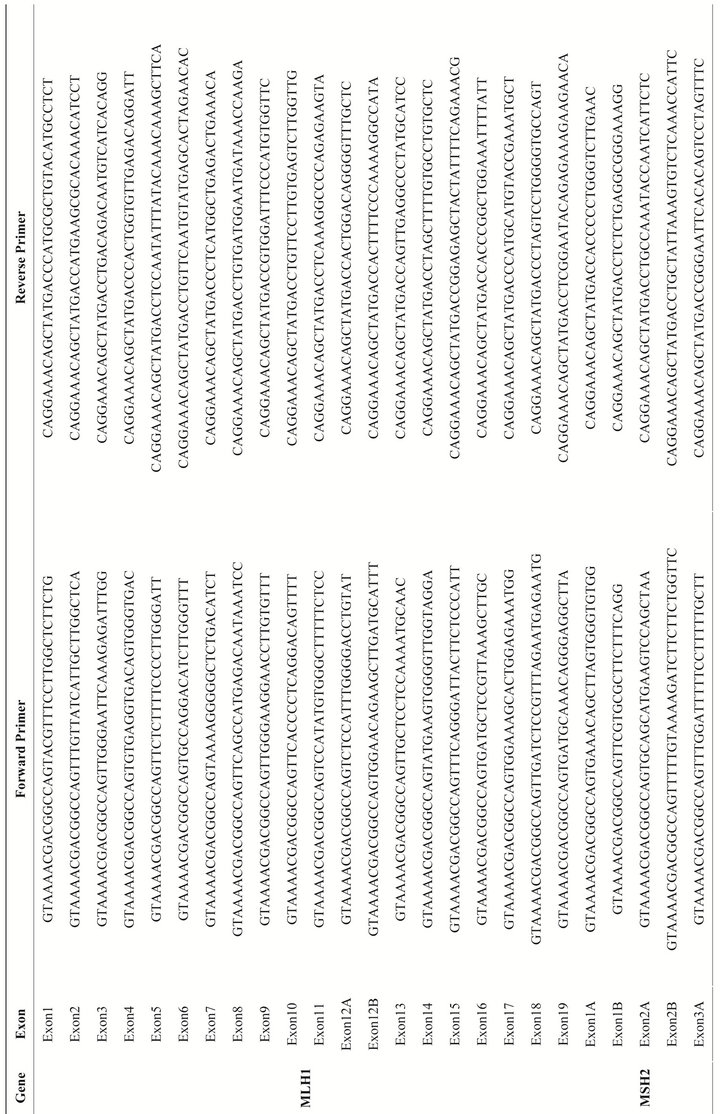



Table 1. Primer sequence used in MLH1, MSH2, MSH6 and PMS2 mutation analysis.Each primer includes the M13 tail in order to facilitate subsequent sequencing using universal M13 primers.
NOTES
*The acute Lymphoblastic Leukemia (ALL) part (RAC2120 017) of this study was supported by Sanad Children’s Cancer Support Society, Riyadh, Saudi Arabia.
#These authors contributed equally.
†Corresponding author.

