Journal of Cancer Therapy
Vol. 3 No. 6A (2012) , Article ID: 25171 , 7 pages DOI:10.4236/jct.2012.326120
sTRAIL Serum Levels and TRAIL 1595 Genotypes: Associations with Progress and Prognosis of Colorectal Carcinoma
![]()
1Department of Molecular Medicine, Institute of Experimental Medicine, Istanbul University, Istanbul, Turkey; 2Surgery Clinic, Istanbul Education and Research Hospital, Istanbul, Turkey.
Email: *ilhanyaylim@gmail.com
Received October 1st, 2012; revised October 31st, 2012; accepted November 9th, 2012
Keywords: TRAIL; sTRAIL; Colorectal Cancer; Risk; Progression; Polymorphism
ABSTRACT
Programmed cell death called apoptosis, plays an important role in the development and maintenance of tissue homeostasis and abnormalities in apoptotic function have been known as important events in the pathogenesis of many cancer types, such as colorectal cancer. It has been shown that both the membrane-bound TRAIL and sTRAIL can induce apoptosis in several tumor types by activating death receptors. Our study was to investigate the existence of TRAIL 1595 C/T SNP in colorectal cancer patients and possible effects of this substitution on serum levels of sTRAIL. In the present study, TRAIL 1595 C/T polymorphism was genotyped in 76 patients with colorectal cancer and 98 healthy subjects using polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis. There were no significant differences in the distribution of TRAIL 1595 C/T genotypes and frequencies of the alleles between colorectal cancer patients and controls. The increased frequency of TRAIL 1595 homozygotes genotypes in patients who had advanced tumour stage was statistically significant (p = 0.0082). Serum sTRAIL levels in the colorectal patients with CT genotype were lower than those of patients with early tumor stage (p = 0.028). The decreased sTRAIL levels were observed in the patients with distant metastasis and CT genotype (p = 0.023).Our findings have suggested that TRAIL 1595 C/T genotypes and sTRAIL levels might be associated with the progression of colorectal cancer in Turkish population.
1. Introduction
Colorectal cancer is one of the common cancers affecting thousands of people every year Worldwide. 103, 170 new cases are estimated by the end of the 2012 in United States and 51,690 of those will be dead [1]. Although age, nutrition habits, obesity and sedentary life style are important risk factors of the colorectal cancer; family history of genetic alterations are important as well. In addition to inherited mutations, single nucleotide polymorphisms (SNPs) of various genes are defined to be related with colorectal cancer risk and disease progression as well as the treatment response [2].
Polymorphisms of certain genes which are required for cell cycle regulation and apoptosis are primarily important in cancer progression. TNF-related apoptosis ligand (TRAIL) is a member of tumor necrosis factor (TNF) family and capable of inhibit or induce apoptosis depending on the receptor type that is bounded. Ability of TRAIL in inducing apoptosis in cancer cells, without disturbing normal cells, is the reason why this ligand is taking attention for cancer therapy. This ligand can potentially bind to five different receptors. The soluble receptor is called as osteoprotegerin and the other four receptors are classified in two groups due to their effects on apoptosis. Agonist receptors are death receptor 4 and 5 (DR4, DR5) and antagonist receptors are decoy receptor 1 and 2 (DcR1, DcR2). Both of these receptor groups are expressed in normal cells while cancer cells only express agonist receptors DR4 and DR5 which makes it a promising cancer therapy agent [3,4]. Apart from being a candidate for cancer therapy, TRAIL is a key component of cellular signal mechanism. It has been shown that, T cells mediated apoptosis in tumor cells occur via TRAIL. Activation of several signal pathways such as NFkB, PI3K and MAPK, are triggered by TRAIL in specific cell lines [5-7].
TRAIL gene was mapped in 3q26 with 5 exons and 20 kb length [8]. Isolation and characterization of the 5’- upstream region of the gene was carried out and 4 SNPs was reported in this region, using human colorectal cancer cell lines [9] and four SNPs were reported in 3’UTR of TRAIL gene [10,11]. One SNP in the 3’UTR [12] and four SNPs in the polymorphic region [13] reported in multiple sclerosis. Moreover, SNPs of TRAIL have been reported in fatty liver disease [14], breast cancer [15], sporadic breast tumor [5] and prostate cancer [16].
Increased soluble TRAIL (sTRAIL) concentrations are shown to be related with specific clinical conditions. In multiple sclerosis (MS) patients sTRAIL levels were suggested as potential response marker for interferonbeta treatment [17]. Higher sTRAIL concentrations were detected in hepatisis B virus (HBV) infection [18]. It is not surprising that gene polymorphisms may affect gene expression thus, serum levels of gene product. Such a relation was reported in a Chinese patient group with fatty liver disease. A correlation between sTRAIL concentrations, 1595 C/T polymorphism and Fatty liver disease susceptibility was shown [14].
A report on TRAIL polymorphisms and sTRAIL levels are lacking in colorectal cancer patients. In this study, we aimed to investigate the existence of TRAIL 1595 C/T SNP in colorectal cancer patients and possible effects of this substitution on serum levels of sTRAIL.
2. Material and Methods
2.1. Participants
76 colon cancer patients with mean age 60.3 (age range between 25 - 87) and 98 healthy controls with mean age 54.8 (age range between 24 - 85) were included in the study. All participants provided written consent prior to study. The patients were selected from Istanbul Research and Education Hospital, Surgery Clinic. Questionnaires, medical records, and pathological reports were received to confirm the diagnosis and cancer status. The patients received a standard questionnaire regarding the diagnosis time, family history, treatments and other medical issues. The blood samples were collected before any chemotherepeutic or radiation therapy treatment had been started. Pathological staging information on all colorectal cancer diagnoses were confirmed by manual review of the pathology reports and clinical charts. Nodal status was categorized as no regional lymph nodes affected (N0) or at least one nodal metastasis. The control subjects were selected among the volunteers of the proper age group who do not use any regular medications by the time of study. The control group was selected from patients attending the general surgery and who were treated for non-neoplastic diseases such as inguinal hernia, trauma or pilonidal sinus.
2.2. DNA Isolation
Blood samples were collected in tubes containing EDTA. Genomic DNA was isolated using standard salting our procedure [19].
2.3. SNP Detection
TRAIL 1595 C/T polymorphism detection was carried out by restriction fragment length polymorphism (RFLP) method. Polymerase chain reaction (PCR) method was used for amplification of the TRAIL gene using forward primer 5’-TGA GCA CTA CAG CAA ACA TGA-3’ and reverse primer 5’-GCA CCA CTA AAA GAT CGC AGT-3’. A PCR reaction mix, which contains 150 ng DNA tamplate, 15 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl and 200 µM each of deoxynucleotides and Taq DNA polymerase (MBI Fermantas, Vilnius, Lithuania) was used. The PCR reactions were thermally cycled with an initial danuration step at 95˚C for 5 min, followed by 30 cycles of 94˚C for 45 s, 56˚C for 45 s and 72˚C for 45 s, and then a final extension step at 72˚C for 5 min. The 391-base pair PCR product was digested by RsaI (MBI Fermantas) at 37˚C for 2.5 hours. After enzyme digestion the homozygous individuals for C allele (CC genotype) were identified by the presence of 59 and 332 bp. The homozygous individuals for the T allele (TT genotype) were identified by the presence of 59, 146, 186 bp fragments. Digestion products were visualized under ultraviolet light after agarose gel electrophoresis.
2.4. TRAIL Assay
Fresh-blood samples were immediately centrifuged at 3000 rpm for 5 min to separate serum and samples were kept frozen at –20˚C until the study. Serum TRAIL levels were determined with commercially available sandwich ELISA kit (Platinium ELISA, Bender MedSystems GmbH, Vienna).
2.5. Statistical Analysis
Statistical analyses were performed using the SPSS version 11.0 for Windows (SPSS Inc. Chicago, IL, USA). Differences in the frequencies of the TRAIL 1595 C/T polymorphism between CRC patients and the control group and between clinical data within the CRC subgroup were analyzed using the Chi-square test. The Hardy-Weinberg equilibrium was tested for all polymorphisms. Differences in sTRAIL sera levels between patients and control subjects were examined using the Mann-Whitney U test.
3. Results
Table 1 shows the demographic characteristics of our

Table 1. Characteristics of patients with colorectal cancer.
colorectal cancer patients. Genotypes and allele frequencies for TRAIL 1595 C/T polymorphism in colorectal cancer patients and controls are listed in Table 2. Genotype distributions for TRAIL 1595 C/T polymorphism in both groups were in agreement with Hardy-Weinberg equilibrium (for patients χ2 = 0.064, p = 0.79; for controls χ2 = 0.036, p = 0.848). TRAIL 1595 C/T genotype and allele frequencies between colorectal cancer patients and controls were statistically significant (p = 0.536, p = 0.280, respectively). Distributions of TRAIL 1595 C/T genotypes according to clinical parameters and tumor characteristics of colorectal cancer patients are summarized in Table 3. Patients with the advanced tumor stage (T3/T4) colorectal cancer showed a higher prevalence of CC genotypes and homozygotes (CC + TT) (72%, 76%) when compared to the patients with T1/T2 stage (41.2%, 41.2%). (p = 0.039; OR: 1.749; 95% CI: 0.966 - 3.167; p = 0.008; OR: 1.846; 95% CI: 1.024 - 3.327), respectively.

Table 2. Genotypes and allele frequencies for TRAIL 1595 C/T.

Table 3. Distribution of TRAIL 1595 C/T genotypes with clinicopathological features in colorectal cancer patients.
Among colorectal cancer patients, there were no significant association between the TRAIL 1595 genotypes and other clinical and tumor parameters including age at diagnosis, family history, smoking, lymph node metastasis, distant metastasis, periganglionar invasion, angiolymphatic invasion and also tumor differentiation (Data are not shown). Among colorectal cancer patients, there were no significant association for sTRAIL levels between the patients and healthy controls. In our colorectal patients who have distant metastasis, angiolymphatic invasion, advanced tumor stage, TRAIL 1595 CT heterozygote genotype was significantly associated with decreased sTRAIL levels. There were no significant difference for sTRAIL levels between patients with advanced tumor stage (817.32 ± 187.3 pg/ml) and early tumor stage (1002.7 ± 209.4 pg/ml) (p = 0.119). However, serum sTRAIL levels in colorectal patients with CT genotype (276 ± 116.5 pg/ml) were lower than those of patients with early tumor stage (T1 + T2) (756.4 ± 174.3) (p = 0.028). There was also an association for the presence of distant metastasis in sTRAIL levels. According to this data, sTRAIL levels in the patients with distant metastasis and with CT genotype were lower (18.1 ± 4.4 pg/ml) than those with the absence distant metastasis (594 ± 123.5) (p = 0.023). Serum sTRAIL levels in colorectal patients with CT genotype and who have angiolymphatic invasion (202.9 ± 112.5 pg/ml) were lower than those of the absence of this kind of invasion (704.7 ± 186.2) (p = 0.036) (Table 4).
4. Discussion
TRAIL is known to selectively induce apoptosis in cancer cell lines and responsible in the apoptotic mechanism of the cell. Soluble form of this death ligand is also known to be biologically active and involved in the pathophysiology of different disease states such as cancer, viral infections, autoimmune diseases and inflammation [17,18,20-24].
Polymorphisms of TRAIL gene was related with a variety of diseases so far. It was shown that CC genotype of –716 C/T promotor polymorphism rendered risk for sporadic breast cancer in compression to CT and TT genotype [5]. Another study on the –716 polymorphism reported no significant correlation between genotypes and disease risk in the prostate cancer with a Chinese patient group [16].
TRAIL 1595 C/T polymorphism was related with fatty liver disease (FLD) and a less severe form of steatosis non-alcoholic fatty liver disease (NAFLD) patients in Chinese population. Individuals with TT genotype were shown to have a lower risk for FLD and sTRAIL concentrations in NAFLD were significantly higher than those of controls. Thus, TRAIL 1595 polymorphism was suggested as a good marker for NAFLD patients [14]. This polymorphism was also associated with MS susceptibility in Japanese patients and allele frequencies were represented as 0.592 and 0.408 for C and T allele respectively, in the control group [12].
Frequencies of C allele in TRAIL 1595 were determined as 0.592 in Japanese population and 0.712 in Caucasians [10]. Distribution of different genotypes of the TRAIL 1595 among the control group correlated with the Gray’s findings on Caucasian population. Together with increased number of control subjects, our current results also verify our previous findings on group of Turkish healthy subjects [15]. Our previous study was the first report on TRAIL 1595 polymorphism and its relation to breast cancer. In current study we represent the first data on relation between TRAIL 1595 polymorphism, sTRAIL levels and colorectal cancer in a Turkish patient group. We found no significant difference in TRAIL 1595 genotypes with control and patient group. However a significant difference among the patient groups for those having advanced stage tumor was detected. Distribution of the CC genotype among the patients having T3-T4 advanced tumor stage was 1.75 fold higher than the other genotypes. Moreover, homozygosity among the advanced tumor stage patients was significantly higher than the CT genotype with 1.85 fold. We found no significant difference between genotypes for other prognostic parameters such as tumor node metastasis, distance metastasis, angiolymphatic or perineural invasion.
It has been shown that both membrane bounded and sTRAIL can induce apoptosis in a variety of tumor types and sTRAIL is used as a positive marker for apoptosis [25-28]. It was reported that sTRAIL could induce apoptosis of the tumor cells at pmol concentrations without harming the normal tissue cells [29]. sTRAIL levels have been associated with several pathological conditions. Low levels of sTRAIL were detected in MS patients [17]. Patients with cirrhosis and liver cancer were shown to have higher sTRAIL levels than healthy controls or hepatisis B virus carriers. This upregulation was positively correlated with the liver damage [18]. Another study showed that sTRAIL concentrations were significantly higher in NAFLD patients [14]. A recent study reported that plasma s TRAIL levels were significantly higher in the ulcerative colitis patients [30]. Decreased sTRAIL levels were reported in non-drug using type 2 diabetic patients [31]. High sTRAIL concentrations were
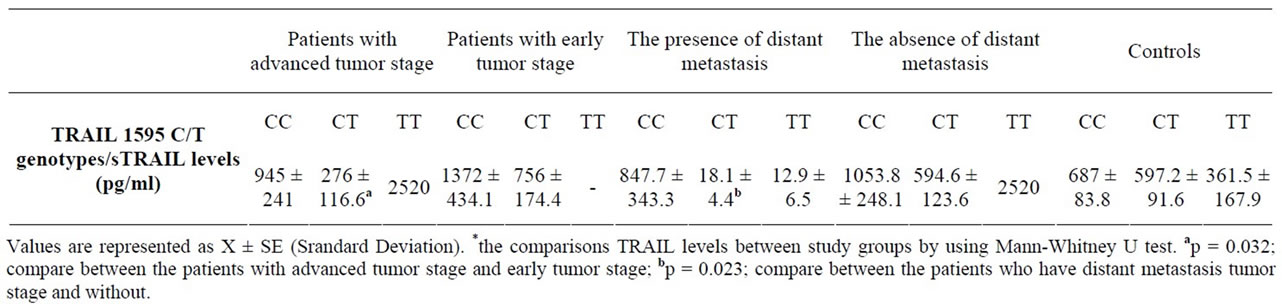
Table 4. The sTRAIL levels, TRAIL 1595 C/T genotypes of our study groups and some tumour charecterictics of colorectal cancer patients.
related with a better prognosis in hearth failture patients [32]. A higher baseline serum sTRAIL levels was associated with a better survival of peritoneal dialysis patients [33].
Apart from being a prognostic parameter sTRAIL levels can be used to determine the efficiency of the therapy. Recently, Kargi et al. reported that increased sTRAIL levels after treatment were correlated with survival in colorectal cancer patients [34,35].
Taken together, these studies indicate that altered sTRAIL levels are observed either before or after the therapy in specific diseases. Our current study represents the differences between sTRAIL levels in patients having different stage tumors. Although differences in sTRAIL levels are variable according to disease; a higher sTRAIL level for survival, better prognosis or treatment efficiency is mostly common. In our study, we detected lower sTRAIL levels in the patients having advanced stage tumor and bad prognosis. The lowest sTRAIL levels were observed in the patients having CT genotype, distant metastasis, advanced tumor stage and angiolymphatic invasion together with statistical significance. Taken together, it is reasonable to suggest that CT genotype with low sTRAILl levels are related with disregulation of apoptosis in advanced stage colorectal cancer.
The mechanism underlying the effect of the 1595 C/T polymorphism on the sTRAIL levels is not clear yet however, we found a significant difference between CT genotype, bad prognosis and sTRAIL levels. Even a more detailed study is needed to clarify this relation; our study represents a different perspective on the effect of TRAIL 1595 C/T variant and sTRAIL levels on colon cancer progression.
5. Acknowledgements
This study was supported by a grant from Istanbul University, Research Foundation (Project BYP-8823), Turkey.
REFERENCES
- R. Siegel, D. Naishadham and A. Jemal, “Cancer Statistics, 2012,” CA: A Cancer Journal for Clinicians, Vol. 62, No. 1, 2012, pp. 10-29. doi:10.3322/caac.20138
- S. Al-Sohaily, A. Biankin, R. Leong, M. Kohonen-Corish and J. Warusavitarne, “Molecular Pathways in Colorectal Cancer,” Journal of Gastroenterology and Hepatology, Vol. 27, No. 9, 2012, pp. 1423-1431.
- E. W. Duiker, C. H. Mom, S. de Jong, P. H. Willemse, J. A. Gietema, A. G. van der Zee and E. G. de Vries, “The Clinical Trail of TRAIL,” European Journal of Cancer, Vol. 42, No. 14, 2006, pp. 2233-2240. doi:10.1016/j.ejca.2006.03.018
- Y. X. Zou, X. D. Zhang, Y. Mao, G. C. Lu, M. Huang and B. J. Yuan, “Acute Toxicity of a Single Dose DATR, Recombinant Soluble Human TRAIL Mutant, in Rodents and Crab-Eating Macaques,” Human & Experimental Toxicology, Vol. 29, No. 8, 2010, pp. 645-652. doi:10.1177/0960327109357214
- R. Pal, S. Gochhait, S. Chattopadhyay, P. Gupta, N. Prakash, G. Agarwal, A. Chaturvedi, N. Husain, S. A. Husain and R. N. Bamezai, “Functional Implication of TRAIL-716 C/T Promoter Polymorphism on Its in Vitro and in Vivo Expression and the Susceptibility to Sporadic Breast Tumor,” Breast Cancer Research and Treatment, Vol. 126, No. 2, 2011, pp. 333-343. doi:10.1007/s10549-010-0900-5
- C. Schmaltz, O. Alpdogan, B. J. Kappel, S. J. Muriglan, J. A. Rotolo, J. Ongchin, L. M. Willis, A. S. Greenberg, J. M. Eng, J. M. Crawford, G. F. Murphy, H. Yagita, H. Walczak, J. J. Peschon and M. R. van den Brink, “T Cells Require TRAIL for Optimal Graft-versus-Tumor Activity,” Nature Medicine, Vol. 8, No. 12, 2002, pp. 1433- 1437. doi:10.1038/nm1202-797
- R. W. Johnstone, A. J. Frew and M. J. Smyth, “The TRAIL Apoptotic Pathway in Cancer Onset, Progression and Therapy,” Nature Reviews Cancer, Vol. 8, No. 10, 2008, pp. 782-798. doi:10.1038/nrc2465
- S. R. Wiley, K. Schooley, P. J. Smolak, W. S. Din, C. P. Huang, J. K. Nicholl, G. R. Sutherland, T. D. Smith, C. Rauch, C. A. Smith, et al., “Identification and Characterization of a New Member of the TNF Family That Induces Apoptosis,” Immunity, Vol. 3, No. 6, 1995, pp. 673-682. doi:10.1016/1074-7613(95)90057-8
- Q. Wang, Y. Ji, X. Wang and B. M. Evers, “Isolation and Molecular Characterization of the 5’-Upstream Region of the Human TRAIL Gene,” Biochemical and Biophysical Research Communications, Vol. 276, No. 2, 2000, pp. 466-471. doi:10.1006/bbrc.2000.3512
- H. L. Gray, E. L. Sorensen, J. S. Hunt and C. Ober, “Three Polymorphisms in the 3’ UTR of the TRAIL (TNF-Related Apoptosis-Inducing Ligand) Gene,” Genes and Immunity, Vol. 2, No. 8, 2001, pp. 469-470. doi:10.1038/sj.gene.6363806
- K. Unoki, T. Kasuga, E. Matsushima and K. Ohta, “Attentional Processing of Emotional Information in Obsessive-Compulsive Disorder,” Psychiatry and Clinical Neurosciences, Vol. 53, No. 6, 1999, pp. 635-642. doi:10.1046/j.1440-1819.1999.00618.x
- S. Kikuchi, R. Miyagishi, T. Fukazawa, I. Yabe, Y. Miyazaki and H. Sasaki, “TNF-Related Apoptosis Inducing Ligand (TRAIL) Gene Polymorphism in Japanese Patients With Multiple Sclerosis,” Journal of Neuroimmunology, Vol. 167, No. 1-2, 2005, pp. 170-174. doi:10.1016/j.jneuroim.2005.06.021
- A. Weber, K. P. Wandinger, W. Mueller, O. Aktas, O. Wengert, E. Grundström, S. Ehrlich, C. Windemuth, T. Kuhlmann, T. Wienker, W. Brück and F. Zipp, “Identification and Functional Characterization of a Highly Polymorphic Region in the Human TRAIL Promoter in Multiple Sclerosis,” Journal of Neuroimmunology, Vol. 149, No. 1-2, 2004, pp. 195-201. doi:10.1016/j.jneuroim.2003.12.014
- X. Yan, L. Xu, J. Qi, X. Liang, C. Ma, C. Guo, L. Zhang, W. Sun, J. Zhang, X. Wei and L. Gao, “sTRAIL Levels and TRAIL Gene Polymorphisms in Chinese Patients with Fatty Liver Disease,” Immunogenetics, Vol. 61, No. 8, 2009, pp. 551-556. doi:10.1007/s00251-009-0389-4
- Y. Yildiz, I. Yaylim-Eraltan, S. Arikan, H. A. Ergen, S. Küçücük and T. Isbir, “Is There Any Correlation between TNF-Related Apoptosis-Inducing Ligand (TRAIL) Genetic Variants and Breast Cancer?” Archive of Medical Science, Vol. 6, No. 6, 2010, pp. 932-936. doi:10.5114/aoms.2010.19304
- Y. Y. Mi, J. M. Li, N. Shao, Z. C. Min, B. Xu, L. X. Hua and N. H. Feng, “TRAIL Gene Polymorphism and Genetic Susceptibility to Prostate Cancer in the Chinese Han Population of Nanjing,” Zhonghua Nan Ke Xue, Vol. 17, No. 3, 2011, pp. 242-246.
- K. P. Wandinger, J. D. Lünemann, O. Wengert, J. Bellmann-Strobl, O. Aktas, A. Weber, E. Grundström, S. Ehrlich, K. D. Wernecke, H. D. Volk and F. Zipp, “TNFRelated Apoptosis inducing Ligand (TRAIL) as a Potential Response Marker for Interferon-Beta Treatment in Multiple Sclerosis,” Lancet, Vol. 361, No. 9374, 2003, pp. 2036-2043. doi:10.1016/S0140-6736(03)13641-0
- L. H. Han, W. S. Sun, C. H. Ma, L. N. Zhang, S. X. Liu, Q. Zhang, L. F. Gao and Y. H. Chen, “Detection of Soluble TRAIL in HBV Infected Patients and Its Clinical Implications,” World Journal of Gastroenterology, Vol. 8, No. 6, 2002, pp. 1077-1080.
- S. A. Miller, D. D. Dykes and H. F. Polesky, “A Simple Salting out Procedure for Extracting DNA from Human Nucleated Cells,” Nucleic Acids Research, Vol. 16, No. 3, 1988, p. 1215. doi:10.1093/nar/16.3.1215
- P. M. Holland, “Targeting Apo2L/TRAIL Receptors by Soluble Apo2L/TRAIL,” Cancer Letters, 2011. doi:10.1016/j.canlet.2010.11.001
- N. B. Liabakk, A. Sundan, S. Torp, P. Aukrust, S. S. Frøland and T. Espevik, “Development, Characterization and Use of Monoclonal Antibodies against sTRAIL: Measurement of sTRAIL by ELISA,” Journal of Immunological Methods, Vol. 259, No. 1-2, 2002, pp. 119-128. doi:10.1016/S0022-1759(01)00501-4
- M. N. Lub-de Hooge, E. G. de Vries, S. de Jong and M. Bijl, “Soluble TRAIL Concentrations Are Raised in Patients with Systemic Lupus Erythematosus,” Annals of the Rheumatic Diseases, Vol. 64, No. 6, 2005, pp. 854-858. doi:10.1136/ard.2004.029058
- A. Bisgin, E. Terzioglu, C. Aydin, B. Yoldas, V. Yazisiz, N. Balci, H. Bagci, R. M. Gorczynski, C. A. Akdis and S. Sanlioglu, “TRAIL Death Receptor-4, Decoy Receptor-1 and Decoy Receptor-2 Expression on CD8+ T Cells Correlate with the Disease Severity in Patients with Rheumatoid Arthritis,” BMC Musculoskeletal Disorders, Vol. No. 11, 2010, p. 192. doi:10.1186/1471-2474-11-192
- C. Aydin, A. D. Sanlioglu, A. Bisgin, B. Yoldas, L. Dertsiz, B. Karacay, T. S. Griffith and S. Sanlioglu, “NF-κB Targeting by Way of IKK Inhibition Sensitizes Lung Cancer Cells to Adenovirus Delivery of TRAIL,” BMC Cancer, Vol. 10, 2010, p. 584. doi:10.1186/1471-2407-10-584
- J. Shi, D. Zheng, Y. Liu, M. H. Sham, P. Tam, F. Farzaneh and R. Xu, “Overexpression of Soluble TRAIL Induces Apoptosis in Human Lung Adenocarcinoma and Inhibits Growth of Tumor Xenografts in Nude Mice,” Cancer Research, Vol. 65, No. 5, 2005, pp. 1687-1692. doi:10.1158/0008-5472.CAN-04-2749
- A. D. Sanlioglu, E. Dirice, O. Elpek, A. F. Korcum, M. Ozdogan, I. Suleymanlar, M. K. Balci, T. S. Griffith and S. Sanlioglu, “High TRAIL Death Receptor 4 and Decoy Receptor 2 Expression Correlates with Significant Cell Death in Pancreatic Ductal Adenocarcinoma Patients,” Pancreas, Vol. 38, No. 2, 2009, pp. 154-160. doi:10.1097/MPA.0b013e31818db9e3
- I. T. Koksal, A. D. Sanlioglu, B. Karacay, T. S. Griffith and S. Sanlioglu, “Tumor Necrosis Factor-Related Apoptosis Inducing Ligand-R4 Decoy Receptor Expression Is Correlated with High Gleason Scores, Prostate-Specific Antigen Recurrence, and Decreased Survival in Patients with Prostate Carcinoma,” Urologic Oncology: Seminars and Original Investigations, Vol. 26, No. 2, 2008, pp. 158-165. doi:10.1016/j.urolonc.2007.01.022
- P. Aukrust, N. B. Liabakk, F. Müller, E. Lien, T. Espevik and S. S. Frøland, “Serum Levels of Tumor Necrosis Factor-Alpha (TNF Alpha) and Soluble TNF Receptors in Human Immunodeficiency Virus Type 1 Infection— Correlations to Clinical, Immunologic, and Virologic Parameters,” The Journal of Infectious Diseases, Vol. 169, No. 2, 1994, pp. 420-424. doi:10.1093/infdis/169.2.420
- L. H. Han, W. S. Sun, C. H. Ma, L. N. Zhang, S. X. Liu, Q. Zhang, L. F. Gao and Y. H. Chen, “Detection of Soluble TRAIL in HBV Infected Patients and Its Clinical Implications,” World Journal of Gastroenterology, Vol. 8, No. 6, 2002, pp. 1077-1080.
- Y. Jiang, X. Q. Lin, S. G. Cao, C. L. Xu, J. Z. Wang, Z. M. Huang, X. R. Chen, L. Song and Z. X. Xue, “Correlations of genetic Polymorphisms of Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand Gene and Its Plasma Phenotype with Ulcerative Colitis,” Zhonghua Yi Xue Za Zhi, Vol. 92, No. 18, 2012, pp. 1244-1248.
- A. Bisgin, A. D. Yalcin and R. M. Gorczynski, “Circulating Soluble Tumor Necrosis Factor Related Apoptosis Inducing-Ligand (TRAIL) Is Decreased in Type-2 Newly Diagnosed, Non-Drug Using Diabetic Patients,” Diabetes Research and Clinical Practice, Vol. 96, No. 3, 2012, p. e84-e86. doi:10.1016/j.diabres.2012.02.028
- A. Niessner, P. J. Hohensinner, K. Rychli, S. Neuhold, G. Zorn, B. Richter, M. Hülsmann, R. Berger, D. Mörtl, K. Huber, J. Wojta and R. Pacher, “Prognostic Value of Apoptosis Markers in Advanced Heart Failure Patients,” European Heart Journal, Vol. 30, No. 7, 2009, pp. 789- 796. doi:10.1093/eurheartj/ehp004
- P. Y. Poon, C. C. Szeto, B. C. Kwan, K. M. Chow, C. B. Leung and P. K. Li, “Relationship between Serum Levels of Tumour Necrosis Factor-Related Apoptosis-Inducing Ligand and the Survival of Chinese Peritoneal Dialysis Patients,” Nephrology (Carlton), Vol. 17, No. 5, 2012, pp. 466-471. doi:10.1111/j.1440-1797.2012.01605.x
- A. Kargi, A. D. Yalcin, N. Erin, B. Savas, H. H. Polat and R. M. Gorczynski, “IL8 and Serum Soluble TRAIL Levels following Anti-VEGF Monoclonal Antibody Treatment in Patients with Metastatic Colon Cancer,” Clinical Laboratory, Vol. 58, No. 5-6, 2012, pp. 501-505.
- A. Bisgin, A. Kargi, A. D. Yalcin, C. Aydin, D. Ekinci, B. Savas and S. Sanlioglu, “Increased Serum sTRAIL Levels Were Correlated with Survival in Bevacizumab-Treated Metastatic Colon Cancer,” BMC Cancer, Vol. 12, 2012, p. 58. doi:10.1186/1471-2407-12-58
NOTES
*Corresponding author.

