Advances in Bioscience and Biotechnology
Vol.4 No.2(2013), Article ID:28428,8 pages DOI:10.4236/abb.2013.42033
Determination of amino-acidic positions important for Ocimum basilicum geraniol synthase activity

1Unit Mixte Recherche Santé Vigne & Qual Vins, Métab Second Vigne, Institut National de Recherche Agronomique, Université de Strasbourg, Colmar, France
2Institut de Biologie Moléculaire des Plantes, Université de Strasbourg, Strasbourg, France
Email: *fischer@colmar.inra.fr
Received 3 December 2012; revised 5 January 2013; accepted 13 January 2013
Keywords: Geraniol Synthase; Heterologous Expression; Monoterpenols; Point Mutations; Yeast
ABSTRACT
Terpenes are one of the largest and most diversified families of natural compounds. Although they have found numerous industrial applications, the molecular basis of their synthesis in plants has, until now, not been fully understood. Plant genomes have been shown to contain dozens of terpene synthase (TPS) genes, however knowledge of their amino-acidic protein sequence in not sufficient to predict which terpene(s) will be produced by a particular enzyme. In order to investigate the structural basis of a TPS specificity, we performed site directed mutations in the geraniol synthase from Ocimum basilicum. The results obtained suggest that a specific region on the catalytic site plays an important role in GPP transformation, either by stabilizing the GPP substrate on the catalytic site, or by enabling its transformation into a monoterpenol via an intermediate carbocation.
1. INTRODUCTION
The immense variety of chemical compositions encountered in aromatic plants has always attracted human interest, and such plants are believed to have been cultivated since the Neolithic for their fragrance, food improvement abilities, and traditional medicinal properties. Each of these plants can contain unusually high concentrations of chemicals and several hundreds of volatile compounds, with one or two of them being major compounds. Monoterpene aroma are amongst the most basic and prevalent fragrance representatives. They belong to the family of plant isoprenoids, which are amongst the most diversified compounds, with over 40,000 molecules having been described [1]. Their diversification started with ancestral plants, which were almost devoid of terpenoids, as illustrated by the fact that the Physcomitrella patens moss has only one terpene synthase gene, encoding a diterpene precursor from the synthesis of the gibberellic acid hormone [2]. Amplification of the terpenoid synthase gene family occurred via gene duplication and is still an ongoing process [3]. The economic importance of some isoprenoids is based on their perfume, food flavor improvement properties, and pharmaceutical or industrial properties. Recent plant genome analyses have highlighted hundreds of putative monoterpenoid synthase genes. The functional characterization of monoterpenoid synthases has therefore generated a large amount of data, and shown that a large proportion of the characterized enzymes catalyze the synthesis of several end products [4,5]. However, it has until now been impossible to predict which monoterpenoid(s) will be produced by a particular enzyme. The tremendous range of possible variations in the carbocationic reactions (cyclizations, hydride shifts, rearrangements, and termination steps) catalyzed by terpenoid synthases explains the wide range of possible products [6]. Such studies have usually been carried out using heterologous truncated cDNA expression in Escherichia coli (since the native plant enzyme targets the chloroplast and bacteria cannot process introns), followed by enzymatic assays with crude cell extracts.
Some plants, such as basils, have been more extensively studied for their aromatic properties and can be considered as model plants for monoterpenoid synthase studies. The basil genus comprises approximately 150 species, which are found mainly in the tropics, and which were probably first cultivated in India [7]. Many varieties of these aromatic plants are now grown worldwide, particularly in Southeast Asia, Mediterranean countries and California. Sweet basil (Ocimum basilicum) has various cultivars, varying in shape, color and fragrance. Differences in fragrance have been linked to variations in compounds such as geranial, neral, methylchavicol, eugenol [8,9]. The functional characterization of sweet basil monoterpenoid synthases expressed in E. coli has enabled the identification of a geraniol synthase (GES) [9] and a (R)-linalool synthase ((R)-LIS) [10]. Geraniol and linalool are the only products synthesized after heterologous expression by O. basilicum GES and (R)-LIS. Both genes are very similar, with a 78% identity on the nucleotide sequence and a 82% identity on the aminoacid sequence. Iijima et al. [9] made the first functional/structural analysis of these genes by investigating whether a hybrid protein could be affected in terms of either its activity or its specificity. A GES-(R)-LIS construct exhibited a dual geraniol/linalool synthase activity, whereas a (R)-LIS-GES construct was inactive.
An alternative approach to the investigation of terpene synthase specificities is to use 3D modeling, combined with site directed mutagenesis. This method has been used only once before, on two cyclic monoterpene synthases, the 1,8 cineole and sabinene synthases from Salvia fruticosa [11]. In the present study, our aim was to characterize the geraniol synthase catalytic site via a similar site-directed mutation approach, based on the specificities of geraniol or linalool synthases. We first used an array of functionally characterized GES and LIS from several plants, to determine the amino-acid signatures on the catalytic site that would correlate with the GES or LIS activity (Figure 1). Both GES and LIS have been functionally characterized in O. basilicum and Chinese basil (Perilla frutescens) [12]. We therefore used an alignment of O. basilicum, P. frutescens GES and (R)- LIS cDNA, combined with a 3D model construction, to define putative GES/LIS-specific amino-acids for sitedirected mutagenesis. In a previous study we engineered Saccharomyces cerevisiae to convert it into a suitable heterologous expression system for truncated O. basilicum GES cDNA [13]. We used this system to test whether any O. basilicum GES modifications would result in a modification of the GES enzyme specificity.
2. MATERIALS AND METHODS
2.1. Directed Mutagenesis of GES
Plasmid pGB7/PMA1 was constructed by integrating a 700 pb PMA1 promoter in the NotI-SacI digested plasmid pGB7 [14]. The pSM5 plasmid was constructed through integration of O. basilicum geraniol synthase (GES), by means of recombination in the pGB7/PMA1 plasmid, in fusion with the green fluorescent protein (GFP) from pGB7. The pSM5 plasmid was then used for site-directed mutations of GES. The plasmids carrying the mutations were created using the QuikChange SiteDirected Mutagenesis Kit from Stratagene, in accordance with the manufacturer’s instructions. The complementary Forward and Reverse primers were designed as described in Table 1. All of the plasmids were verified by sequencing.
2.2. Yeast Transformation
The haploid G strain (Mata, his3D, leu2∆0, ura3-, trp1∆63, YJL167W::kanMX4 [pFL44erg20K197G]) [13] was transformed with a pSM5 plasmid series expressing wild type and mutated GES. A 1.5 mL aliquot of G cells, grown to the stationary phase YNB medium supplemented with the appropriate auxotrophic amino acids, was centrifuged (1 min, 10,000 g) and 10 mL of mutated plasmid pSM5 (125 ng) was added to the yeast pellet. Transformed cells were selected for uracil prototrophy and G418 resistance on a minimal medium.
2.3. Monoterpenoid Extraction and Quantification
The cells from a stationary phase culture were harvested by centrifugation (5000 g for 5 min). Octanol (4 mg) and Ethylheptanoate (4 mg) were added as an internal standard to 20 mL culture medium supernatant. Monoterpenoids were extracted using a stir bar (Twister; Gerstel, Mülheim a/d Ruhr; Germany) sorptive extraction liquid desorption gas chromatograhy mass spectrometry (SBSELD-GC-MS) [15] with the following modifications: acetonitrile as solvent and a 1 µL injection volume.
Monoterpenol extracts were analyzed by GC-MS using an Agilent 6890N gas chromatograph equipped with a Gerstel MPS2 sampler, coupled to an Agilent 5975B inert MSD (Agilent Technologies). The gas chromatograph was fitted with a DB-Wax capillary column (60 m × 0.32 mm i.d. × 0.50 µm film thickness, J&W Scientific) and helium was used as the carrier gas (1 mL×min−1 constant flow). The GC oven temperature was programmed to increase from 45˚C to 82˚C at a rate of 20˚C×min−1, and then to increase to 235˚C at a rate of 2.7˚C×min−1 (hold for 15 min). The injector was set to 250˚C and used in pulsed splitless mode (25 psi for 0.50 min). The temperatures of the interface, MS ion source and quadrupole were 270˚C, 230˚C and 150˚C, respectively. The mass spectrometer was operated in the electron impact ionization mode (EI, 70 eV), and the masses were scanned over a m/z range of 29 - 300 amu. Agilent MSD ChemStation software (G1701DA, Rev D.03.00) was used for instrument control and data processing. The mass spectra were compared with the Wiley’s library reference spectral bank. Total amounts of geraniol, linalool, citronellol, nerol and a-terpineol were determined using linear calibration curves, leading to a value of 0.98 for R2, over the range of concentrations from 0 to 200 mg/mL.
2.4. GES Model Building
Homology modeling was implemented using Modeller [16] with Mentha spicata (4S)-limonene synthase as template [17]. The O. basilicum GES sequence was aligned with Mentha spicata (4S)-limonene using ClustalX
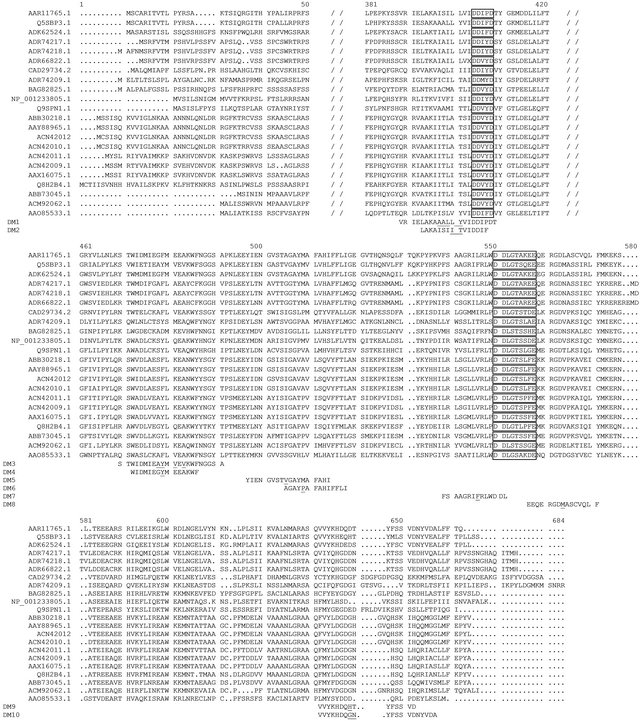
Figure 1. Partial multi-alignment from functionally characterized GES and LIS, made with Multalin (http://multalin.toulouse.inra.fr/multalin/). GenBank accession code for GES: O. basilicum (AAR11765.1), Vitis vinifera (ADR74217.1, ADR66822.1, ADR74218.1, ADR74217.1), Cinnamomum tenuipile (CAD29734.2), P. Frutescens (ABB30218.1), P. Frutescens var. hirtella GES (GenBank: ACN42012), Perilla setoyensis (ACN42010.1), Perilla citriodora (AAY88965.1), Phyla dulcis (ADK62524.1). GenBank accession code for LIS: P. Frutescens var. hirtella (ACN42011.1), Lavandula angustifolia (ABB73045.1), Perilla setoyensis (ACN42009.1), Perilla citriodora (AAX16075.1), Osmanthus fragrans var. thunbergii (ACM92062.1), Solanum lycopersicum (NP_001233805.1), Ocimum basilicum (Q5SBP3.1), Artemisia annua (Q9SPN1.1), Arabidopsis thaliana (AAO85533.1), Backhousia citriodora (BAG82825.1), Mentha aquatic (Q8H2B4.1), Vitis vinifera (ADR74209.1). FARM (first Asp-rich motif) and SARM (second Asp-rich motif) domains are surrounded. Directed mutations made on O. basilicum GES (AAR11765.1) are specified (DM1-10) and the amino acids modified in each directed mutation are underlined.
Table 1. Specificities of forward oligonucleotides used to modify the O. Basilicum GES. Mutated codons are underlined.

[18]. Visual analysis of the model and the illustrations were made using PyMOL [19].
3. RESULTS AND DISCUSSIONS
3.1. Monoterpenoids Quantification
Although the product specificities of monoterpenol synthases have been extensively studied using heterologous expression, the manner in which their amino acidic sequence relates to the specific enzymatic function still remains largely unknown, with the notable exception of the sabinene and 1,8 cineole synthases from S. fruticosa [11]. Although classical sequence comparisons led to the identification of seven subfamilies [20], further analysis of their enzymatic activity was not possible, due to the history of this protein’s family. The original plant terpenoid synthase was a di-terpene synthase, and it is thought that monoterpenoid synthases originated via the loss of the domain of approximately 200 amino-acids [21]. This loss did not take place in a single ancestral event, but occurred several times during plant evolution, leading to an ongoing process of mono and sesqui-TPS generation [22]. In the present study, to enable catalytic site analysis, we positioned each residue differing from O. basilicum and P. frutescens GES and LIS within the predicted catalytic site, and used an in vivo characterization approach to investigate the functional consequences of amino-acidic substitutions at the putative catalytic site of O. basilicum GES. We discovered that 3 mutations resulted in an abolished function of GES and one mutation resulted in a partial loss of function, whereas 5 mutations did not affect the original function.
Expression of GES in the erg20K197G yeast strain resulted in the biosynthesis of a number of monoterpenols (geraniol, linalool, citronellol, nerol) in an YNB medium (Table 2), whereas almost no production of a-terpineol was detected. Untransformed control erg20K197G cells exhibited a much lower production of monoterpenols. The production level of geraniol was 16 times higher arising from expression of GES. An increase in linalool, citronellol and nerol was also detected, but their overall amounts represented only 4% - 12% of the geraniol detected. Expression of 4 mutated GES K197G DM1, K197G DM2, K197G DM5 and K197G DM7 resulted in a monoterpenol profile very similar to that observed with the untransformed control. The expression of 5 other GES mutants (K197G DM3 – K197G DM4 – K197G DM6 – K197G DM8 – K197G DM9) resulted in a monoterpenol profile, very similar to that observed with erg20K197G yeast cells transformed with wild type GES. Only one strain (K197G DM10) exhibited an intermediate monoterpenol profile, between that of erg20K197G transformed with wild type GES and that of the untransformed control.
3.2. Correlation between Model Prediction and Experimental Results
It is of interest to compare our results with previous structure function studies of O. basilicum GES. Using a GES-(R)-LIS protein chimeric approach, the function was evaluated by Iijima et al. [10]. Replacement of the first 360 aa on GES by their (R)-LIS homologues resulted in a loss of function. Our catalytic site modeling indicates that only one area in the first 360 aa on GES could potentially be involved in the catalytic site, suggesting a significant role for this region in the proposed GES function (Figures 2(A) and (B)). The site-directed modification of this area corresponds to DM1, and the resulting K197G DM1 strain indeed exhibits a dramatic loss of GES activity (Table 2). This result prompted us to test another mutation, DM2, replacing the 2 amino-acids
Table 2. Monoterpenol content (mg×L−1) of FPPS mutated yeast strains, untransformed or transformed by control or mutated GES plasmid. Terpenoids were extracted from a minimal medium at their stationary growth phase. The results are the mean of 3 experiments ± standard deviation.
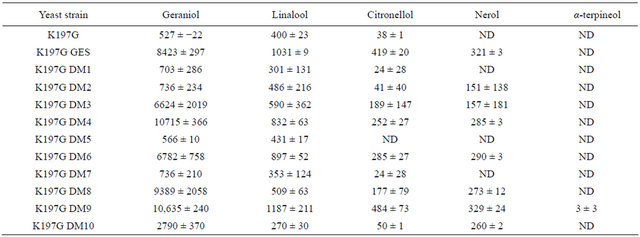
ND: Not Detected.
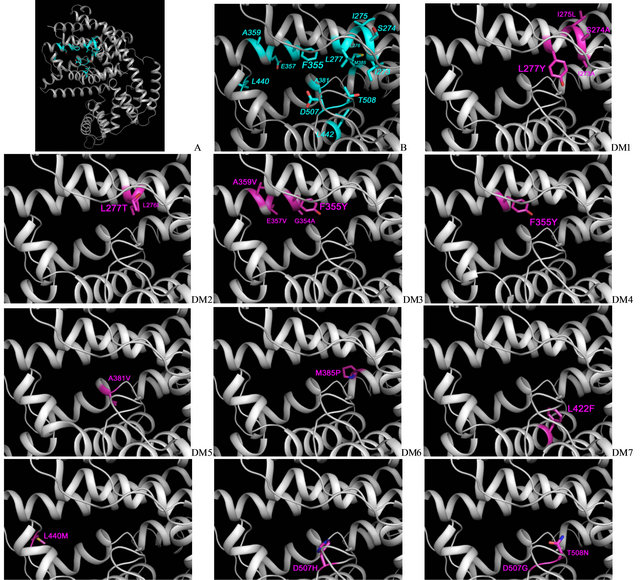
Figure 2. GES catalytic site, showing the amino acids in the catalytic site pocket. Native amino-acids are colorcoded in grey and cyan (A, B), and the amino-acids modified by directed mutagenesis are color-coded in purple for each construct (DM1-DM10).
corresponding to the difference between P. frutescens GES and (R)-LIS in O. basilicum GES. GES activity in the K197G DM2 strain is also suppressed. It is also noteworthy that P. frutescens belongs to the same family (Lamiaceae) as sweet basil. From this comparison it appears that there is a good correlation between DM1, DM2 and the GES-(R)-LIS inactive chimeric construct. One can infer that the loss of GES function obtained by Iijima et al. [10] was due to a modification of this area, defined by DM1 and DM2. The same area also appeared to be of interest in the sabinene and 1,8 cineole synthases analysis of S. fruticosa made by Kampranis et al. [11], since it corresponds to their Region 1. Kampranis et al. [11] suggested that these residues, located on helix a 14 and aligned along the pore of the catalytic centre, could be involved in the deprotonation of the water molecule, facilitating the attack on the intermediate a-terpinyl carbocation. The multiple changes they introduced in Region 1 resulted in a progressive shift from a sabinene or 1,8 cineole synthase activity to a wider range of detected products. On the basis of our results and those reported by Iijima et al. [10], we infer that any modification of size (Y) or charge (T) of the amino-acids in this region is likely to substantially alter the GES enzymatic function on the intermediate geranyl carbocation, whereas a simple change from a GES→LIS signature in this area may not be sufficient to affect the original GES enzymatic specificity.
Following replacement of the aa after position 360 on the GES, by their (R)-LIS homologues, Iijima et al. [10] found a dual geraniol/linalool synthase function. As monoterpenol biosynthesis proceeds via an intermediate carbocation [9], one could therefore infer that they affected the intermediate geranyl carbocation, thereby shifting the charges towards an intermediate potential geranyl or linalyl carbocation. One could therefore consider that mutations in the second half of the GES protein may either inactivate the enzyme or shift the specificity of GES towards mixed geraniol and linalool production. A comparison of O. basilicum GES, (R)-LIS, P. frutescens GES and (R)-LIS (Figure 1), combined with O. basilicum GES modeling (Figure 2), suggested 6 O. basilicum GES modeling (Figure 2), suggested 6 areas of potential interest.
DM3 and DM4 are in the middle of the catalytic site, and in the area where the 3 Mg2+ are thought to be located [17]. DM3 was intended to modify this area, making it more similar to O. basilicum (R)-LIS, and DM4 was used to induce a F→Y change since both O. basilicum and P. frutescens GES have an F at this position, whereas their (R)-LIS have a Y. The DM3 and DM4 modifications did not affect GES activity or specificity, suggesting that this area has only a minor in vivo role in directing the enzymatic function.
DM5 and DM7 are located in the region, which is predicted to interact with the first and middle parts of GPP, respectively [17]. Although both the V and F residues, introduced respectively into DM5 and DM7, are larger than the original A and L amino-acids, they remain hydrophobic. DM7 is targeted to the motif in order to achieve metal dependent ionization of the prenyl diphosphate substrates (N,D)DXX(S,T)XXXE. The modification of these two GPP-interacting residues tested in our experiments gave similar results, leading to a loss of GES function. These results indicate that conformational changes in the processing of the GPP substrate are needed for GES function. It is likely that any mutation of these residues affects the conformation, and either induces the release of GPP out of the catalytic region before it is transformed into a monoterpenol, or even prevents its access to the catalytic region. These observations are corroborated by the fact that any disturbance of the substrate stability in the catalytic pocket (i.e. GPP from yeast FPPS) [13,23] results in a partial or total loss of enzymatic function. One could thus infer that the loss of GES specificity by GPP destabilization would not result in a shift towards a geraniol/linalool monoterpenol synthase function, but rather in a loss of activity.
DM6 is located at the very bottom of the pocket, and affects an amino-acid pointing to a region complementtary to the area tested in DM1 and DM2. This drastic M→P change observed in P. frutescens (R)-LIS, and recreated in DM6, had the potential to affect the nearby helix and thus to produce constraints on the pocket. However, it resulted in a GES activity very similar to that observed with the wild type GES protein (Table 2). This area is therefore likely to play no, or only a minor, role in the GES activity and specificity.
DM8 is on a loop at the very bottom of the pocket site, and is therefore located outside the helix parts of the catalytic site. It is likely that this residue forms part of the entrance of the GES pore. Amino acids located on a loop are not considered to be critical in monoterpene synthase activity, but they have been shown to be important in diterpene synthases [24]. DM8 was tested because the L→M modification could destabilize the neighboring metallic cations from the catalytic pocket. This might affect the enzymatic reaction via substrate wobbling at the main catalytic site, and release different reaction products. However, quite expectedly, this minor modification did not affect GES efficiency or specificity, indicating that its presence in O. basilicum (R)-LIS may be fortuitous.
DM9 corresponds to a single amino-acidic difference observed between O. basilicum GES and LIS, at the end of the protein sequence, but within the catalytic site. DM9 had no effect on GES protein activity. This was very surprising, since changing the negatively charged D into a positively charged H at this position, facing SARM and in contact with Mg2+, was expected to substantially affect the enzyme activity. This outcome, together with the results observed with DM3, DM4 and DM8, suggests that these residues, which are involved in the coordination of the divalent cations, act strongly and that the link is very difficult to disturb with a single directed mutagenesis.
DM10 is also at the end of the protein sequence and corresponds to a single amino-acid difference, observed between P. frutescens GES and LIS. Albeit being next to the amino-acidic position described in DM9, in P. frutescens this H→N mutation acts on the carbonate part of the substrate. In DM10, this area was modified to make this amino acid and the previous one look similar to the amino acid observed in P. frutescens. Strain K197G DM10 exhibited a monoterpenol profile, intermediate between the profile observed with the untransformed erg20K197G yeast strain, and that corresponding to the same strain expressing the wild-type GES (Table 2). This area therefore participates in the formation of the catalytic site. More interestingly, the enzyme with DM10 was altered in its efficiency in behaving as GES, but maintained its entire specificity, with no shift of production towards other terpenes. This type of behavior is clearly different to that observed by Kampranis et al. [11] for S. fruticosa sabinene and 1,8 cineole synthases.
4. CONCLUSION
The data from this study provides initial insight into the location and composition of the GES catalytic site. This catalytic site has the shape of a pocket surrounded by the cyan amino-acids from Figure 2(A) and (B) that were mutated in our study. It is possible to draw an imaginary line leaving F355 and D507 on the left side and L277, A381 and L442 on the right side from the catalytic site. All introduced mutations affecting the enzymatic efficiency were located on the right half of the catalytic site, as shown in Figure 2, suggesting that the most critical residues for the enzymatic function are located in this area. Conversely, none of the mutations located on its left half resulted in a reduced enzymatic efficiency. The residues from the right half of the catalytic site are involved in holding the carbon chain of GPP, whereas the residues from the left half of the catalytic site are involved in interactions with the 3 Mg2+, which holds the pyrophosphate O− charges. Destabilization of the Mg2+ - GPP link was impossible to achieve, contrary to our observations of the yeast farnesyl diphosphate synthase site, where single amino-acidic mutations in K197 or K254 residues could induce a release of intermediate GPP out of the catalytic site [13,23]. Furthermore, none of the GES→LIS modifications we tested was able to induce the formation of new products, contrary to our observations of sabinene or 1,8 cineole synthase [11]. We expected that at least one of the tested DM would be able to induce not only a change in the amount of detected geraniol, but also a modification in the monoterpenol’s specificity, with the detection of linalool or other monoterpenols. This was clearly not the case, suggesting that the enzymatic specificity of GES is more difficult to modify than its enzymatic efficiency. The transformation from the intermediate carbocation into products other than geraniol might therefore be due to a coordinated interaction produced by several critical amino-acids, such as the amino-acid observed in the present study, or could be induced by specific action from an area which has not yet be identified by 3D predictions and functionally directed mutagenesis experiments. Further biochemical and structure-function studies are needed, to determine the specificities of the GES catalytic site with greater accuracy.
5. ACKNOWLEDGEMENTS
The authors thank professor Eran Pichersky for providing the O. basilicum GES cDNA.
REFERENCES
- Withers, S.T. and Keasling, J.D. (2007) Biosynthesis and engineering of isoprenoid small molecules. Applied Microbiology and Biotechnology, 73, 980-990. doi:10.1007/s00253-006-0593-1
- Hayashi, K., Kawaide, H., Notomi, M., Sakigi, Y., Matsuo, A. and Nosaki, H. (2006) Identification and functional analysis of bifunctional ent-kaurene synthase from the moss Physcomitrella patens. FEBS Letters, 580, 6175- 6181. doi:10.1016/j.febslet.2006.10.018
- Dittmar, K. and Liberles, D. (2010) Evolution after gene duplication. Wiley-Blackwell, New York.
- Trapp, S.C. and R.B. (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics, 158, 811-832.
- Martin, D.M., Aubourg, S., Schouwey, M.B., Daviet, L., Schalk, M., Toub, O., Lund, S.T. and Bohlmann, J. (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biology, 10, 226. doi:10.1186/1471-2229-10-226
- Dickschat, J.S. (2011) Isoprenoids in three-dimensional space: The stereochemistry of terpene biosynthesis. Natural Product Reports, 28, 1917-1936. doi:10.1039/c1np00063b
- Mukherjee, M. and Datta, A.K. (2007) The basils—A review. Plant Arches, 7, 473-483.
- Morales, M.R. and Simon, J.E. (1997) “Sweet Dani”: A new culinary and ornamental lemon basil. HortScience, 32, 148-149.
- Iijima, Y., Gang, D.R., Fridman, E., Lewinsohn, E. and Pichersky, E. (2004) Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiology, 134, 370-379. doi:10.1104/pp.103.032946
- Iijima, Y., Davidovich-Rikanati, R., Fridman, E., Gang, D., Bar, E., Lewinsohn, E. and Pichersky, E. (2004) The biochemical and molecular basis for the divergent patterns in the biosynthesis of terpenes and phenylpropenes in the peltate glands of three cultivars of basil. Plant Physiology, 136, 3724-3736. doi:10.1104/pp.104.051318
- Kampranis, S.C., Ioannidis, D., Purvis, A., Mahrez, W., Ninga, N., Katerelos, N.A., Ansour, S., Dunwell, J.M., Degenhardt, J., Makris, A.M., Goodenough, P.W. and Johnson, C.B. (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: Structural insights into the evolution of terpene synthase function. Plant Cell, 19, 1994-2005. doi:10.1105/tpc.106.047779
- Masumoto, N., Korin, M. and Ito, M. (2010) Geraniol and linalool synthases from wild species of perilla. Phytochemistry, 71, 1068-1075. doi:10.1016/j.phytochem.2010.04.006
- Fischer, M.J.C., Meyer, S., Claudel, P., Bergdoll, M. and Karst, F. (2011) Metabolic engineering of monoterpene synthesis in yeast. Biotechnology and Bioengineering, 108, 1883-1892. doi:10.1002/bit.23129
- Sagot, I., Bonneu, M., Balguerie, A. and Aigle, M. (1999) Imaging fluorescence resonance energy transfer between two green fluorescent proteins in living yeast. FEBS Letters, 447, 53-57. doi:10.1016/S0014-5793(99)00258-6
- Coelho, E., Perestrelo, R., Neng, N.R., Câmara, J.S., Coimbra, M.A., Nogueira, J.M.F. and Rocha, S.M. (2008) Optimisation of stir bar sorptive extraction and liquid desorption combined with large volume injection-gas chromatography-quadrupole mass spectrometry for the determination of volatile compounds in wines. Analytica Chimica Acta, 624, 79-89. doi:10.1016/j.aca.2008.06.032
- Sali, A. and Blundell, T.L. (1993) Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology, 234, 779-815. doi:10.1006/jmbi.1993.1626
- Hyatt, D.C., Youn, B.Y., Zhao, Y.X., Santhamma, B., Coates, R.M., Croteau, R.B. and Kang, C.H. (2007) Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proceedings of National Academy of Sciences USA, 104, 5360-5365. doi:10.1073/pnas.0700915104
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876-4882. doi:10.1093/nar/25.24.4876
- DeLano, W.L. (2002) The PyMOL molecular graphics system on World Wide Web. http://www.pymol.org
- Chen, F., Tholl, D., Bohlmann, J. and Pichersky, E. (2011) he family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant Journal, 66, 212- 229. doi:10.1111/j.1365-313X.2011.04520.x
- Köksal, M., Jin, Y., Coates, R.M., Croteau, R. and Christianson, D.W. (2011) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature, 469, 116-120. doi:10.1038/nature09628
- Hillwig, M.L., Xu, M., Toyomasu, T., Tiernan, M.S., Wei, G., Cui, G., Huang, L. and Peters, R.J. (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant Journal, 68, 1051-1060. doi:10.1111/j.1365-313X.2011.04756.x
- Fischer, M.J.C., Meyer, S., Claudel, P., Bergdoll, M. and Karst, F. (2011) Identification of a lysine residue important for the catalytic activity of yeast farnesyl diphosphate synthase. Protein Journal, 30, 334-339. doi:10.1007/s10930-011-9336-y
- Zhou, K., Gao, Y., Hoy, J.A., Mann, F.M., Honzatko, R.B. and Peters, R.B. (2012) Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. Journal of Biological Chemistry, 287, 6840-6850. doi:10.1074/jbc.M111.337592
NOTES
*Corresponding author.

