Journal of Biophysical Chemistry
Vol.2 No.4(2011), Article ID:8433,4 pages DOI:10.4236/jbpc.2011.24052
Binding of Hoechst with nucleic acids usingfluorescence spectroscopy
Institute of Cell Biophysics of Russian Academy of Sciences, Pushchino, Moscow Region, Russia. nvekshin@rambler.ru
Received 2 August 2011; revised 14 September 2011; accepted 27 September 2011.
Keywords: Hoechst; Fluorescence; Quenching; DNA; t-RNA; Oligonucleotide; Actinomycin
ABSTRACT
It has been shown that polarity of environment around Hoechst 33342 is almost unchanged while sorption of this fluorescent dye on a surface of the hairpin oligonucleotide HP1, t-RNA and DNA. At small concentrations, this dye, adsorbed on the surface of DNA, RNA or HP1, does not show any specificity to certain nucleotides. In the case of unwound sites of DNA or HP1, it can bind inside, but without the intercalation stacking with nucleotides. The energy transfer from nucleotide chromophores to Hoechst is absent due to their remoteness and also “bad” (non-stacking) orientation. The mutual fluorescence quenching of Hoechst by actinomycin D (AMD) and, vice versa, of 7-amino-actinomycin D (7AAMD) by Hoechst in DNA and HP1 is observed. It is due to dynamic deactivation and mutual replacing in binding sites.
1. INTRODUCTION
Many heterocyclic antibiotics, for instance, such as natural actinomycin D (AMD), its fluorescent derivative 7-amino-actinomycin D (7AAMD) [1-5] and synthetic fluorescent probe Hoechst 33342 or 33258 [5-10] at low concentrations form non-stacking complexes, stabilizing the structure of DNA. It leads to inhibition of the RNA polymerase reaction, suppression of protein synthesis and cell division. That is why these heterocyclic compounds exhibit anti-tumor activity (in contrast to stacking-intercalating dyes like ethidium bromide—the most powerful carcinogen). It is important to emphasize that these compounds at high (non-physiological) concentrations form the intercalating stacking complexes, inducing damages, breaks and disruptions of DNA [5]. Non-intercalating binding of various ligands with DNA was early observed in [11].
Full understanding of the molecular mechanisms of action of heterocyclic antibiotics at low concentrations on DNA and RNA can optimize the use of these compounds and their analogues as medical drugs.
Hoechst dyes 33342 and 33258 are closely related bisbenzimides. Hoechst 33258 is 2’-(4-hydroxyphenyl)-5- [5-(4-methylpiperazine-1-yl) benzimidazo-2-yl]-benzimidazole. Hoechst 33342 contains additional ethyl group. This group renders Hoechst 33342 a bit more hydrophobic than Hoechst 33258. Both dyes have the absorption band at 350 nm and the emission band in the “blue” region. They are non-intercalating fluorescent probes for non-covalent labeling of DNA. They are used in fluorescence spectroscopy, microscopy and flow cytometry to detect the nuclear and mitochondrial DNA [5-10].
For instance, the intensity and lifetime of fluorescence of nuclei, stained by Hoechst 33258, were measured by microscopy in [8]. The fluorescence intensity-based analysis of Hoechst 33258 and 7AAAD-stained images indicated the increased number and larger size of the DNA condensation centers in the G2/M-phases (in comparing to G0/1-phases).
It should be mentioned here that accumulation of the dye inside a cell leads to a big local concentration of the dye, i.e. to a high dye/nucleotide ratio.
Molecular oligonucleotide structures like d(CGC- [e6G]AATTCGCG in complexes with Hoechst 33258 and Hoechst 33342 were studied in work [7]. It was shown that the oligonucleotide, containing O6-ethyl-G-C base pairs, is stabilized by Hoechst. The dye binds in minor groove of B-structure. It should be noted that this job was carried out at a high dye / nucleotide ratio.
Ensemble and single-molecule fluorescence measurements of Hoechst 33258 with calf thymus DNA at various [Hoechst 33258]/[DNA bp] ratios were performed in work [9] to elucidate the mechanism of their interaction. Upon binding of the dye to this DNA at the [Hoechst 33258]/[DNA bp] ratio = 0.05, the relative fluorescence quantum yield of the dye was increased from 0.02 to 0.58. The fluorescence decay was fitted by mono-exponential model with lifetime of 3.6 ns. It was concluded that the dye at low [Hoechst 33258]/[DNA bp] ratios binds in minor groove of DNA. At higher ratios, the dye partially intercalates in G:C sequences, reflected by the lifetime component to 1.5 - 2 ns. However, it should be emphasized here that preparations of calf thymus DNA usually contain large amount of breaks and damaged chains. Therefore, the easy penetration of Hoechst into these flexible sites (not into the rigid double helix) in such samples could occur.
While binding to cellular DNA, the Hoechst dye can block the DNA replication during cell division. Consequently, Hoechst at low concentrations could be applied, in principal, as anti-bacterial and anti-tumor drug. At high concentrations, Hoechst leads to disruption of the DNA macromolecule [10].
Since Hoechst is capable to bind with various types of nucleotide-contained biopolymers of cells, it is very important to compare its interaction with different biopolymers in solution. In the present work, the fluorescence study on mechanism of interaction of Hoechst 33342 with native lambda DNA, yeast t-RNA and synthetic hairpin oligonucleotide HP1 in aqueous solution is carried out at very low (sub-micromolar) concentrations of the dye. Also, AMD and 7AAMD are applied for this aim.
2. EXPERIMENTAL PROCEDURES
2.1. Materials
Lambda DNA (“Invitrogen”), yeast t-RNA (“GibcoBRL, Life Technilogies”), hairpin oligonucleotide HP1 (“Midland Certified Reagent Company”), AMD (“Serva”) 7AAMD (“Fluka”) and Hoechst 33342 tri-hydrochloride (“Sigma”) were used. Structural formula of HP1, containing only one guanine residue, is presented below.

In aqueous solution, the HP1 oligonucleotide exists as a hairpin structure with the formation of nine base pairs.
Concentrations of HP1, RNA and DNA in experiments were near 10 mM (of nucleotides). Concentration of Hoechst and 7AAMD in the most cases was 0.5 mM, AMD - 1.3 mM.
2.2. Instrumentation
Fluorescence spectra (uncorrected) of Hoechst 33342 and 7AAMD in the 20 mM Tris buffer with 1 mM EDTA (pH 7.7) with or without DNA, RNA or HP1 were recorded in 1-cm quartz cuvettes at 20˚C. The excited state lifetime of Hoechst 33342 and 7AAMD was measured by phase method with the “SLM-4800” spectrofluorometer in special mirror cuvettes [12], increased the fluorescence intensity by many times.
3. RESULTS AND DISCUSSION
3.1. Binding of Hoechst with Transport RNA
Unlike AMD and 7AAMD, which are unable to communicate with t-RNA [3,5], the dye Hoechst 33342 binds to t-RNA extremely effective. This leads to dramatically increasing in its fluorescence intensity, comparing the emission of the dye in the buffer without t-RNA (Figure 1). The life-time of Hoechst at binding was enhanced from 0.4 ns to 4.4 ns. The observed increase in the lifetime and intensity of Hoechst emission upon the binding is due to a fixation of the dye on the rigid t-RNA surface (a free Hoechst molecule is flexible and has a lot of vibrations).
If to normalize the both spectra at maximum, no any sufficient shift between them is observed (not shown in figure). Lack of any considerable shift in the emission spectrum of Hoechst suggests that the polarity of the environment around the dye is almost unchanged. The Hoechst/t-RNA complex contains a number of water molecules. It can be concluded also that no multiple strong interactions, including multiple hydrogen bond formation of the dye with nucleotide chromophores, participate in the complex formation.
Thus, there is no doubt that Hoechst, unlike of AMD and 7AAMD, well sticks on the surface of t-RNA due to hydrophobic interactions. Moreover, it can be concluded that this fluorescent dye is not specific for DNA only, but has an affinity to various polynucleotide chains.
3.2. Binding of Hoechst with Hairpin Oligonucleotide HP1
When the binding of Hoechst to HP1 occurs, a sharp increase in its fluorescence intensity arises (Figure 2). However, this strong enhancement of the intensity is not accompanied by any sufficient shift of the emission spectrum, i.e. the situation is the same as in the case of t-RNA. It is radically different from that was described for spectral shifts of 7AAMD, when it binds inside HP1. Lack of any considerable shift in the emission spectrum of Hoechst suggests that the polarity of the environment around the dye is almost unchanged. Sorption of Hoechst on the surface of oligonucleotide HP1 does not affect on the form of emission spectrum. This is not too surprising, since the HP1 oligonucleotide has polar sites on the surface and, moreover, contains bound water.
The life-time of Hoechst at binding was increased from 0.4 ns to 4 ns. The increase in the lifetime and in-
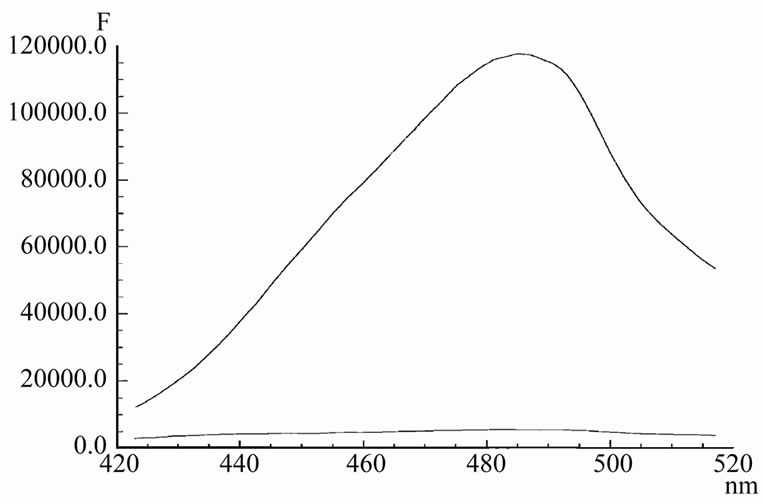
Figure 1. The emission spectrum of free Hoechst (lower curve) and Hoechst in the presence of t-RNA (upper curve) in 20 mM Tris with 1 mM EDTA, pH 7.7. Concentration of Hoechst was 0.5 mM. Excitation was 380 nm.
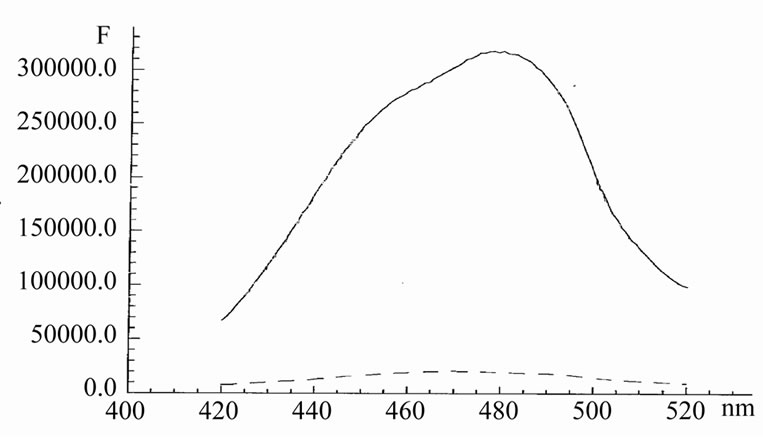
Figure 2. The emission spectrum of Hoechst in the presence of oligonucleotides HP1 (upper curve) and without it (lower curve) in 20 mM Tris with 1 mM EDTA, pH = 7.7. Concentration of Hoechst was 0.5 mM. Excitation was at 380 nm.
tensity of Hoechst emission upon the binding is due to a fixation of the dye on the rigid HP1 surface.
3.3. Binding of Hoechst with DNA
Preparations of DNA from phage lambda do not contain a large amount of breakages and damages (in contrast to preparations of thymus DNA). Binding of Hoechst with lambda DNA also leads to increase in the fluorescence intensity (Figure 3). The fluorescence intensity of Hoechst here was in ~ 1.5 - 2 times less than in the case of HP1 or t-RNA. It means that Hoechst interacts with the double helix not such strong as with single chains or hairpin structures. The life-time of Hoechst at binding with DNA was increased from 0.4 ns to only 3 ns.
Furthermore, there was a shift (albeit it was small) to shorter wavelengths of the emission spectrum. This means that Hoechst not only sticks on the surface of DNA, but also partly penetrates inside. The binding occurs especially easily into the unwound sites, but hardly into dou-
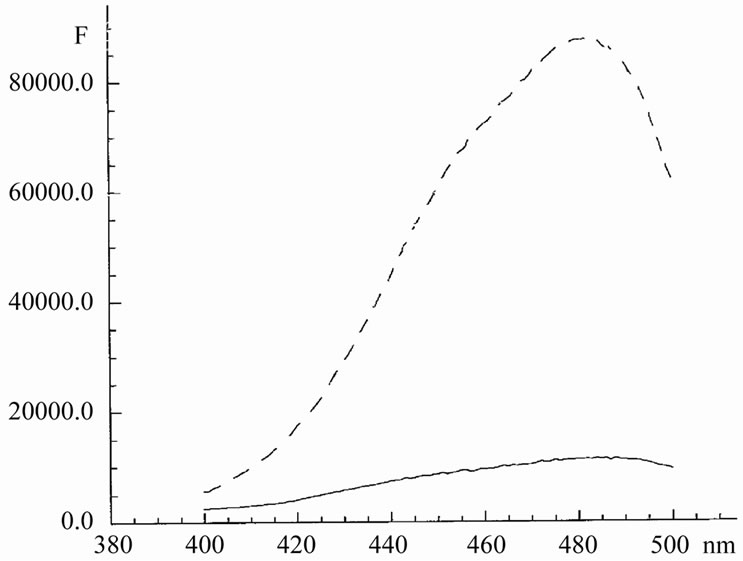
Figure 3. The emission spectrum of Hoechst in the presence of lambda DNA (upper curve) and without it (lower curve) in 20 mM Tris with 1 mM EDTA, pH 7.7. Concentration of Hoechst was 0.5 mM. Excitation was at 380 nm.
ble helix. This observation helps us to explain the fact, established by the authors [10], that Hoechst at some conditions may induce the condensation of DNA.
3.4. Lack of Energy Transfer from Nucleotides to Hoechst
In principal, the energy transfer (not by the inductiveresonance Forster’s mechanism, but by the exchange Dexter’s one) from nucleotides to Hoechst could take place. However, in reality, it is not found.
Indeed, as it seen form Figure 4, there is no any nucleotide band at 260 nm in the excitation spectrum of the Hoechst/DNA complex. The same picture was observed in the case of hairpin HP1 instead of DNA. The band at 275 nm at the excitation spectrum is the own shoulder of Hoechst. This shoulder is clearly visible in the solution of free dye, without DNA or HP1. The ratio of the intensities of the bands at 275 nm and 360 nm, as seen from Figure 4, is virtually identical for Hoechst in water and for Hoechst in DNA. Thus, we came to conclusion about the absence of energy transfer from nucleotides to Hoechst. This is result of spatial remoteness of Hoechst from nucleotide chromophores and also of their mutual “bad” non-stacking orientation.
Since the excitation spectrum upon binding of Hoechst with HP1 or DNA remains practically unchanged, it means the weakness of interactions of the dye with nucleotides. This result again confirms that Hoechst binds mainly at the surface of DNA. Only a small portion of the dye population binds inside.
3.5. Displacement of Hoechst from HP1 and DNA by Actinomycin D
AMD is a typical representative among actinomycins,
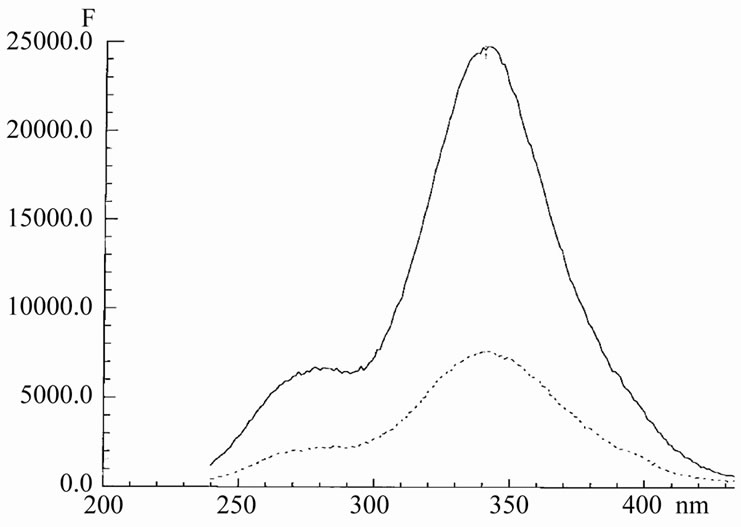
Figure 4. The excitation spectrum of Hoechst at the presence of 6 µM lambda DNA (upper curve) and without DNA (lower curve) in 20 mM Tris with 1 mM EDTA, pH 7.7. Concentration of Hoechst was 0.5 mM. Emission was at 460 nm.
consisting of a flat phenoxazone chromophore (4,6-dimethyl-2-amino-phenoxazone-3-one-1,9-dicarboxylic acid) and two identical penta-peptide-lacton groups, two amino groups of which are acylated by two carboxyl chromophores.
Even small amounts of AMD, added to complexes Hoechst/HP1 and Hoechst/DNA, induced a sharp decrease in the intensity of Hoechst fluorescence (Figure 5 and Figure 6). In the case of DNA, the maximum of emission shifts from 478 nm to 485 nm. The life-time of Hoechst was decreased here from 3 ns to 2 ns.
It is interestingly that in the case of DNA, the quenching effect was expressed in a much greater extent. It means that the contact between Hoechst by AMD in lambda DNA is realized in much degree that in the case of HP1.
There are three possible reasons for reducing in the fluorescence intensity of Hoechst by AMD: energy transfer, dynamic deactivation and displacement. Effects of trivial screening and reabsorption also give here some small contribution.
3.6. Displacement of 7AAMD in HP1 by Hoechst
In the hairpin oligonucleotide HP1, the 7AAMD molecule is linked strongly enough. But addition an excess amounts of Hoechst 33342 to this complex induces strong decrease in fluorescence intensity of 7AAMD (Figure 7).
The observed fluorescence quenching can not be explained by resonance energy transfer, because the 7- AAMD emission spectrum does not overlap with the absorption band of Hoechst. Also, the effects of screening and reabsorption are too small here. The life-time of 7AAMD was decreased by Hoechst from 2 ns to 1.5 ns.
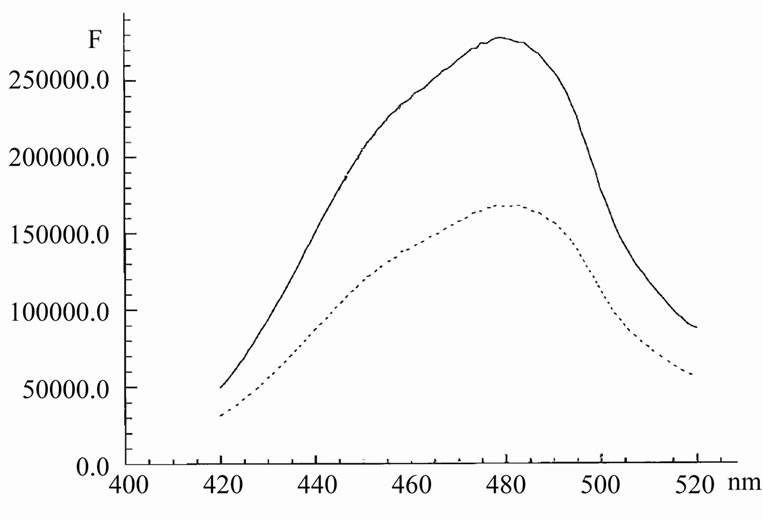
Figure 5. The emission spectrum of Hoechst in the presence of hairpin oligonucleotide HP1 initially (upper curve) and after adding of 1.3 mM AMD (lower curve) in 20 mM Tris with 1 mM EDTA, pH 7.7. Concentration of Hoechst was 0.5 mM. Excitation was 380 nm.
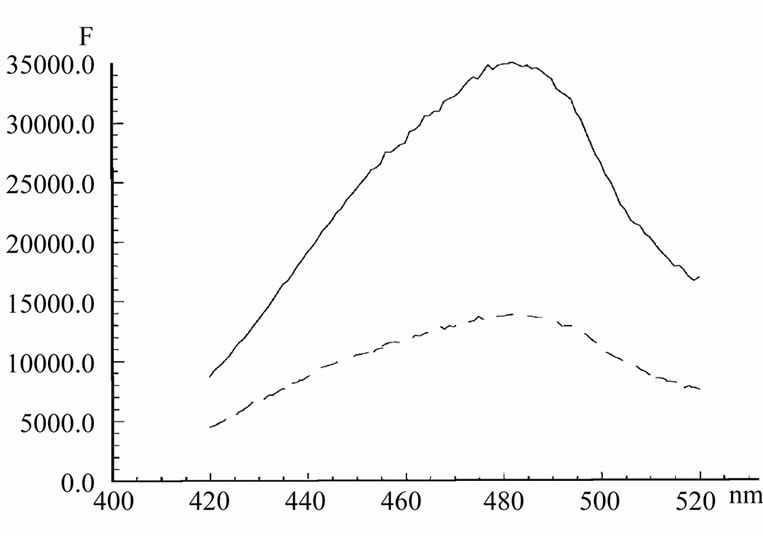
Figure 6. The emission spectrum of Hoechst in the presence of lambda DNA initially (upper curve) and after adding of 1.3 mM AMD (lower curve) in 20 mM Tris with 1 mM EDTA, pH 7.7. Concentration of Hoechst was 0.5 mM. Excitation was 380 nm.
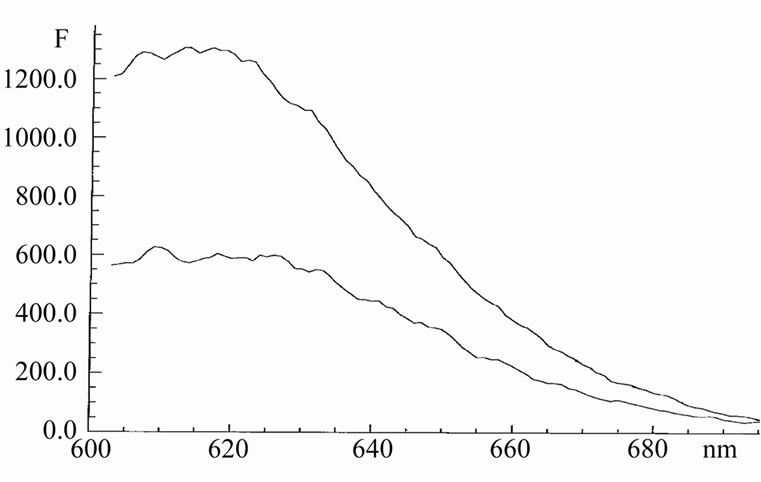
Figure 7. The emission spectrum of 7AAMD in oligonucleotides HP1 initially (upper curve) and after adding 6 mM Hoechst (lower curve) in 20 mM Tris and 1 mM EDTA, pH 7.7. Concentration of 7AAMD was 0.5 mM. Excitation was 570 nm.
Hence, contacting to the surface of oligonucleotide, Hoechst began in touch with 7AAMD that leads to direct deactivation of the exited state of 7AAMD or (and) forcing it out of the hairpin. The intensity of the 7AAMD emission has to be decreased in the both cases.
4. CONCLUSIONS
Thus, we can conclude that polarity of the environment around fluorescent dye Hoechst 33342 is almost unchanged while it sorption on the surface of HP1, tRNA or DNA. At small concentrations, Hoechst binds mainly at the surface of DNA, RNA or HP1. At small concentrations, it has no any specificity to certain nucleotides. In the case of unwound sites of DNA or hairpins, it binds inside, but without stacking with nucleotides. The energy transfer from nucleotides to Hoechst is absent due to remoteness of Hoechst from nucleotide chromophores and also of their “bad” non-stacking orientation. The mutual quenching of emission of Hoechst by actinomycin D (AMD) and, vice versa, of emission of 7-amino-actinomycin D (7AAMD) by Hoechst in DNA and HP1 is observed. It is due to two main reasons: dynamic deactivation and mutual replacing in binding sites.
REFERENCES
- Chen, F.M., Sha, F. and Chin, K.H. (2004) The nature of actinomycin D binding to d(AACCAXYG) sequence motif. Nucleic Acids Research, 32, 271-277. doi:10.1093/nar/gkh178
- Vekshin, N., Savintsev, I., Kovalev, A., Yelemessov, R. and Wadkins, R. (2001) Solvatochromism of the excitation and emission spectra of 7-amino-actinomycin D: Implications for drug recognition of DNA secondary structures. The Journal of Physical Chemistry: B, 105, 8461- 8467. doi:10.1021/jp011168p
- Vekshin, N.L. and Kovalev, A.E. (2006) Prompt nonstacking binding of actinomycin D to hairpin oligonucleotide HP1 and slow redistribution from HP1 to DNA. The Journal of Biochemistry, 140, 185-191. doi:10.1093/jb/mvj150
- Bitehtina, M.A. and Vekshin, N.L. (2008) 7-amino-actinomycin as a fluorescent probe to un-twisting and denaturation of DNA. Bioorganicheskaya Kchimiya, 34, 781-785.
- Vekshin, N.L. (2011) Biophysics of DNA-antibiotic complexes. Nova Press Inc., New York.
- Van Zant, G. and Fry, C.G. (1983) Hoechst 33342 of mouse bone marrow: Effects on colony-forming cells. Cytometry, 4, 40-46. doi:10.1002/cyto.990040106
- Sriram, M., Van der Marel, G.A., Roelen, H.L., Van Boom, J.H. and Wang, A.H. (1992) Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: Molecular structure of d(CGC- [e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342. The EMBO Journal, 11, 225-232.
- Murata, S., Herman, P. and Lakowicz, J.R. (2001) Texture analysis of fluorescence lifetime images of ATand GC-rich regions in nuclei. Journal of Histochemistry and Cytochemistry, 49, 1443-1452. doi:10.1177/002215540104901112
- Adhikary, A., Buschmann, V., Muller, Ch. and Sauer, M. (2003) Ensemble and single-molecule fluorescence spectroscopic study of the binding modes of the bis-benzimidazole derivative Hoechst 33258 with DNA. Nucleic Acids Research, 31, 2178-2186. doi:10.1093/nar/gkg308
- Saito, M., Kobayashi, M., Iwabuchini, S.I., Motita, Y., Takamura, Y. and Tamiya, E. (2004) DNA condensation monitoring after interaction with Hoechst 33258 by atomic force microscopy and fluorescence spectroscopy. The Journal of Biochemistry, 36, 813-823. doi:10.1093/jb/mvh191
- Zimmer, C. and Wahnert, U. (1986) Nonintercalating DNA-binding ligands: Specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Progress in Biophysics and Molecular Biology, 47, 31-112. doi:10.1016/0079-6107(86)90005-2
- Vekshin, N.L. (2009) Fluorescence spectroscopy of biomacromolecules. Photon-vek, Pushchino.

