Agricultural Sciences
Vol.5 No.10(2014), Article
ID:49299,9
pages
DOI:10.4236/as.2014.510095
Molecular Analysis of the Alpha-Tubulin Gene from the Microsporidium, Endoreticulatus sp. Zhenjiang, Isolated from Bombyx mori
Shengyan Xiao1, Zhongyuan Shen1,2*, Xudong Tang1,2, Li Xu1,2, Xuliang Fu1, Yajie Yue1, Nan Li1, Wei Wang1
1Jiangsu University of Science and Technology, Zhenjiang, China
2Sericultural Research Institute, Chinese Academy of Agricultural Sciences, Zhenjiang, China
Email: *szysri@163.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 19 June 2014; revised 22 July 2014; accepted 13 August 2014
ABSTRACT
We isolated a microsporidium from the silkworm, Bombyx mori, and classified it as Endoreticulatus sp. Zhenjiang based on morphological characteristics and phylogenetic analyses of ribosomal sequences. This microsporidium causes silkworm pebrine, although its original host and mode of transmission are unknown. To better understand its distribution and transmission mode, it is essential to have species specific molecular markers. Towards this goal, we characterized the alpha tubulin gene from Endoreticulatus sp. Zhenjiang in this study. The full-length alpha-tubulin cDNA from Endoreticulatus sp. Zhenjiang was cloned and sequenced (GenBank ID: KJ784483) using the rapid amplification of cDNA ends (RACE) protocol. The alpha-tubulin cDNA is 1382 bp long with an open reading frame spanning 1320 bp and consisting of a short 20-bp 5’-untranslated region (5’-UTR) and a 42-bp 3’-UTR with a stop codon and a poly (A) tail. This alpha-tubulin cDNA encodes a deduced polypeptide with 439 amino acids, including a complete tubulin domain and a tubulin C domain. This protein has an estimated isoelectric point of 5.1 and a predicted molecular weight of 48.6 kDa.
Keywords:Microsporidium, RACE, Tubulin Gene, Silkworm, Pebrine

1. Introduction
Microsporidia are unicellular eukaryotes and obligate intracellular parasites with the ability to infect a broad spectrum of hosts from protists to mammals, including humans and have worldwide distribution [1] [2] . All microsporidia identified to date belong to 160 genera and 1300 described species [3] [4] . Nosema bombycis is the first described microsporidium, which prevailed in Europe, America, and Asia during the mid-19th century. It is recognized as the pathogen of silkworm pebrine and still causes heavy economic losses in silk-producing countries such as China [5] . Besides Nosema, several microsporidian genera, such as Pleistophora, Thelohania, Vairimorpha, and Endoreticulatus have been found to infect silkworm. The genus Endoreticulatus clearly differs from N. bombycis in spore morphology and size, ultrastructure, and life cycle [6] . We have identified an Endoreticulatus microsporidium, which was isolated from silkworm in Zhenjiang City, Jiangsu Province, China. The fresh spores were oval, 2.9 ± 0.2 μm in length and 1.2 ± 0.2 μm in width. The organization of the rRNA gene was 5’-LSU-ITS-SSU-IGS-5S-3’, which is reverse compared to the organization of most microsporidian rRNA regions. This is the first Endoreticulatus microsporidium with a reversed rRNA gene arrangement [7] .
It is known that tubulin genes could offer useful markers to study phylogenesis [8] [9] , because the tubulin proteins are structural proteins with relatively conserved amino acid sequence between species. The tubulin gene family consists of six distinct, but highly conserved subfamilies that possess alpha-, beta-, gamma-, delta-, epsilon-, and zeta-tubulins, each with specific conserved sequences and are widely distributed among eukaryotes [9] [10] . Heterodimers of alphaand beta-tubulin proteins are the major components of microtubules, which are the central components of eukaryotic cilia, flagella, mitotic spindles and the cytoskeleton. Among the tubulin subfamilies, microsporidia have only alpha-, beta-, and gamma-tubulin genes. To understand the alpha-tubulin genes of microsporidia, we cloned and analyzed the complete cDNA sequence of the alpha-tubulin gene from Endoreticulatus sp. Zhenjiang. Molecular analysis revealed that the alpha-tubulin gene of Endoreticulatus sp. Zhenjiang had a shorter 5’-untranslated region (5’-UTR). In phylogenetic analysis, Endoreticulatus sp. Zhenjiang and Endoreticulatus sp. CHW-2004 Taiwan formed a separate clade supported by high bootstrap values. Based on the cDNA sequence, we designed specific primers to differentiate Endoreticulatus sp. Zhenjiang from other microsporidia.
2. Materials and Methods
2.1. Materials and Reagents
The microsporidia, Endoreticulatus sp. Zhenjiang, Nosema bombycis, Nosema philosamiae, Nosema sp. HA, Nosema sp. PA, Nosema antheraeae and silkworm p50 strain of Bombyx mori were maintained in the Sericulture Research Institute at the Chinese Academy of Agricultural Sciences.
SMART™ RACE cDNA Amplification Kit and Advantage 2 PCR Kit were purchased from Clontech Laboratories, Inc., Japan. RNAiso Plus, DNase I, rTaq enzyme, ExTaq enzyme, pMD18-T vectors, T4 DNA ligase and DNA size markers were purchased from Takara (Japan). All PCR primers were synthesized and DNA sequencing were performed by Shanghai Sangong Biological Engineering Technology & Services Co., Ltd. at shanghai in China. All other chemicals used were analytical grade.
2.2. Total RNA Isolation and DNA Extraction
First, the fourth instar B. mori larvae were allowed to feed on mulberry leaves artificially infected with Endoreticulatus sp. Zhenjiang spores (107 spores/larvae). Then, microsporidial spores of Endoreticulatus sp. Zhenjiang were isolated from infected silkworm for morphological analyses and maintained in the Sericulture Research Institute at the Chinese Academy of Agricultural Sciences.
Total RNA was isolated from the midguts of silkworm infected with microsporidia using RNAiso Plus. The total RNA was to deal with DNase I, insuring the absence of contaminating DNA. Quality of the total RNA was determined by 260/280 absorbance ratio as well as by running an aliquot on an agarose gel. The RNA were then stored at −80˚C until further use. Genomic DNA was extracted from the microsporidia as described by Dong et al. [11] .
2.3. Cloning the Alpha-Tubulin cDNA from Endoreticulatus sp. Zhenjiang
The full-length alpha-tubulin cDNA sequence of Endoreticulatus sp. Zhenjiang was obtained using the SMART™ RACE cDNA Amplification Kit (Clontech). Gene specific primers for the 5’-RACE and 3’-RACE were designed based on the conserved regions in the known microsporidia alpha-tubulin genes in the National Center for Biotechnology Information (NCBI) database.
To amplify the 5’-end of the alpha-tubulin cDNA from Endoreticulatus sp. Zhenjiang, the following primers were used: Universal Primer Mix, UPM: 5’-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3’ and GSP1: 5’-GAGAATATGCCACAAGGGGAAAGTG-3’. The 3’-end of the Endoreticulatus sp. Zhenjiang alpha-tubulin cDNA was amplified using the primers GSP2: 5’-CGAGATAAGAAGGATTGCTGAGAACTGC-3’- and UPM: 5’-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3’. The amplified products were sequenced and subsequently, the full-length cDNA was obtained by overlapping the two sequences.
2.4. Analysis of Sequence Data
Alpha-tubulin gene sequences from Endoreticulatus sp. Zhenjiang were analyzed by BLASTn in the microsporidian genome database (Microsporidia DB: http://microsporidiadb.org/micro/) [12] . The nucleotide sequences obtained in the present study and those for microsporidian species compared, including those for 15 alpha-tubulin genes were aligned with the Clustal X 1.83 software [13] . Phylogenetic trees were constructed with nucleotide sequences of the protein-coding regions using MEGA5 [14] [15] . The evolutionary history was inferred from the maximum likelihood (ML) and neighbor joining (NJ) methods [9] . One thousand bootstrap replications were performed to test the robustness of the estimated phylogenetic trees [7] .
2.5. Species Specific Amplification
Based on the sequence alignment, species specific primers were designed from the Endoreticulatus sp. Zhenjiang alpha-tubulin gene sequence region that varied from the other microsporidia. These primer sequences are: Spe-Forward—GGAGCCAGGAGTGATCAACGAGG and Spe-Reverse—AAGAACAGTCGGCCTCTTCTCG. PCR amplifications were performed using the following parameters: denaturation at 95˚C for 5 min; followed by 35 cycles of 95˚C for 30 s, 55˚C for 30 s, 72˚C for 1 min and 30 s, and then a final extension at 72˚C for 10 min [16] . The following genomic DNA were used as templates in the PCR amplifications: Endoreticulatus sp. Zhenjiang, N. bombycis, N. philosamiae, Nosema sp. HA, Nosema sp. PA, N. antheraeae. Ten samples of each microsporidan species were tested to insure the reliability of the experimental results. PCR products were analyzed on 1.0% agarose gels.
3. Results
3.1. cDNA Cloning and Sequence Analysis
The full length alpha-tubulin gene sequence was obtained using the RACE protocol, and deposited in GenBank (KJ784483). The sequence is 1382 bp long with a 20-bp 5’-UTR, and a 42-bp 3’-UTR. The 3’-UTR has a stop codon and a poly (A) tail, but there is no predicted polyadenylation signal sequence (AATAAA, Figure 1(A)). The open reading frame (ORF) of the Endoreticulatus sp. Zhenjiang alpha-tubulin cDNA was 1320 bp with a 52.1% GC content. The putative polypeptide has 439 amino acids with a calculated molecular weight of about 48.6 kDa, an isoelectric point of 5.1 and without signal peptide. A complete tubulin domain (46 - 242) and a complete tubulin C domain (244 - 389) (Figure 1(B)) were detected using the SMART analysis software (http://smart.embl-heidelberg.de/). BLAST homology analysis and multiple sequence alignments showed that Endoreticulatus sp. Zhenjiang alpha-tubulin gene sequence shares 99.4% nucleotide similarity and 99.5% amino acid sequence similarity with Endoreticulatus sp. CHW-2004 Taiwan (Table 2).
3.2. Phylogenetic Trees Constructed from Alpha-Tubulin Gene Sequence
The ML and NJ phylogenies inferred from alpha-tubulin sequence alignments are based on the nucleotide alignment of 15 microsporidian sequences (Table 1). The ML and NJ methods revealed that in all phylogenetic trees, Endoreticulatus sp. Zhenjiang was within the same clade as Endoreticulatus sp. CHW-2004 Taiwan with high bootstrap support (100%), while N. bombycis, N. philosamiae, N. spodopterae, N. plutellae, and Nosema sp. PX1 formed a separate clade. It is evident that Endoreticulatus sp. Zhenjiang was distantly related to Nosema species (Figure 2). This was further supported by genetic distances, i.e. the pairwise distance of alpha-tubulin
Table 1 . GenBank accession numbers of alpha-tubulin gene from Endoreticulatus sp. Zhenjiang and other species used in this study.
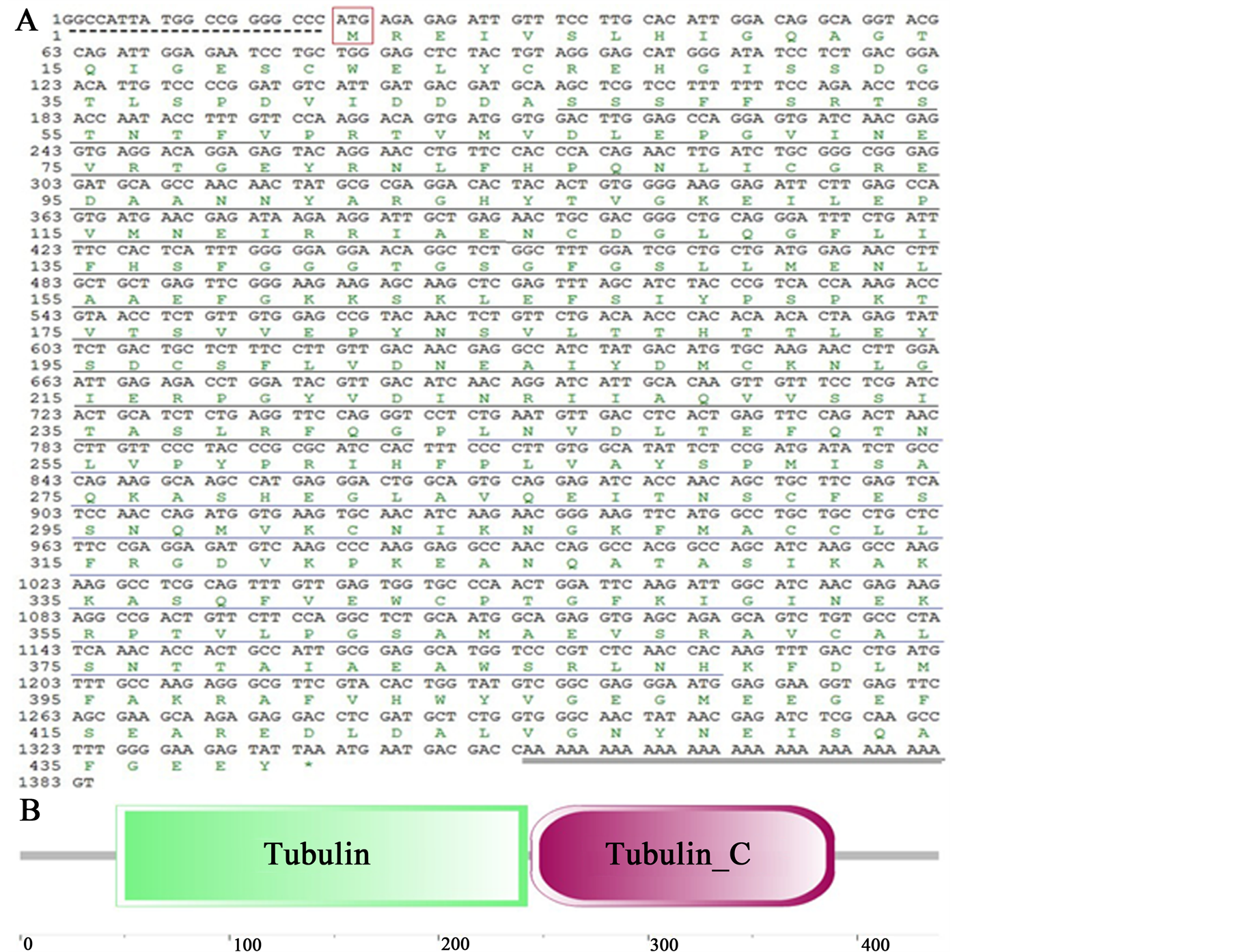
Figure 1. Nucleotide sequence, deduced amino acid sequence, and conserved domains of Endoreticulatus sp. Zhenjiang alpha-tubulin. (A) Nucleotide and deduced amino acid sequence of alpha-tubulin cDNA; (B) Schematic domain structure of alpha-tubulin. The start codon is boxed in red. 5’-terminal untranslated region is marked with a dotted underline. Stop codon is denoted by an asterisk (*). The polyadenylation tail is marked with double underlines. The tubulin domain is underlined in black, and tubulin_C domain is underlined in blue.
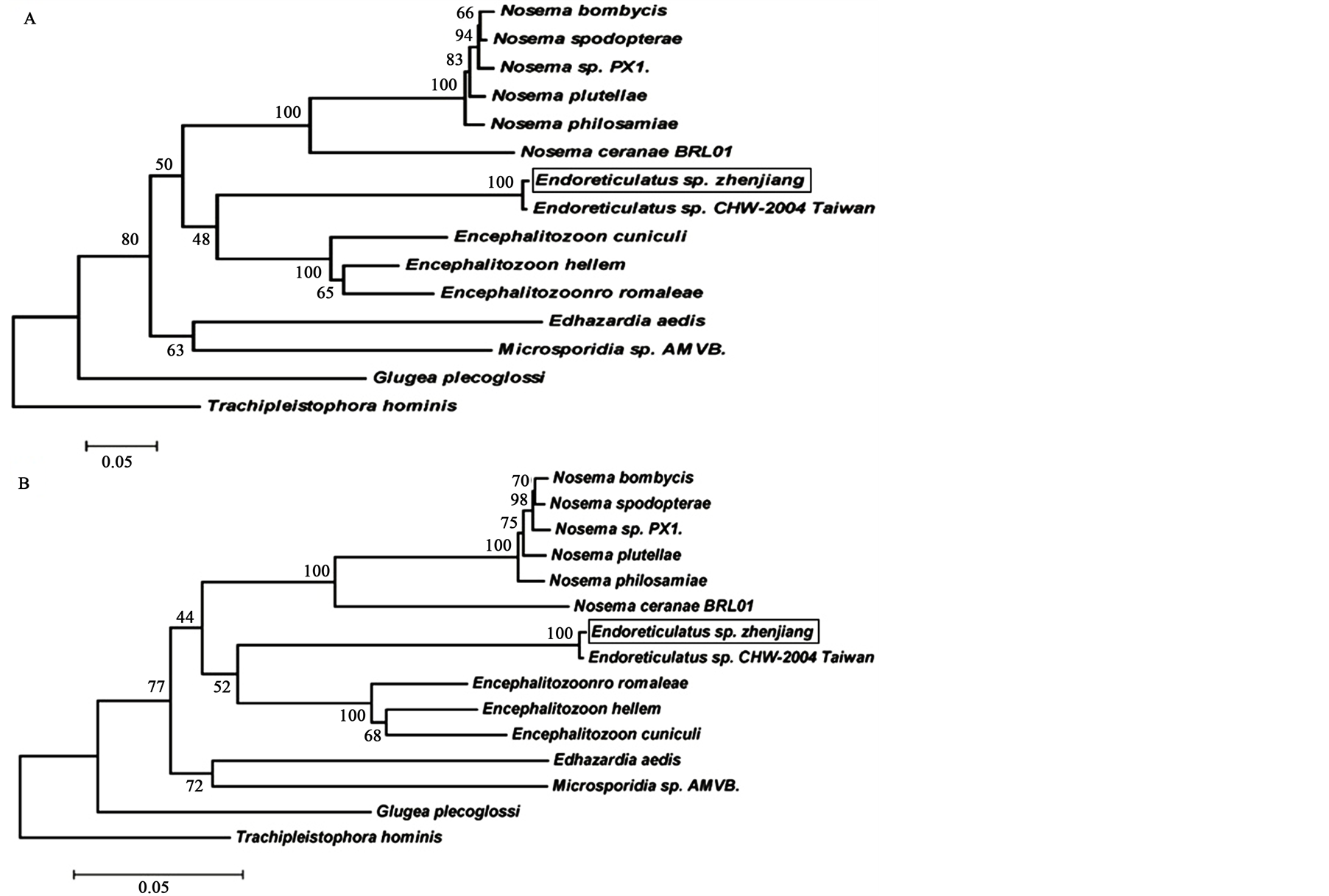
Figure 2. Nucleotide sequence, deduced amino acid sequence, and conserved domains of Endoreticulatus sp. Zhenjiang alpha-tubulin. (A) Nucleotide and deduced amino acid sequence of alpha-tubulin cDNA; (B) Schematic domain structure of alpha-tubulin. The start codon is boxed in red. 5’-terminal untranslated region is marked with a dotted underline. Stop codon is denoted by an asterisk (*). The polyadenylation tail is marked with double underlines. The tubulin domain is underlined in black, and tubulin_C domain is underlined in blue.
nucleotide sequences between Endoreticulatus sp. Zhenjiang, N. bombycis, N. philosamiae, N. spodopterae, N. plutellae, and Nosema sp. PX1 varied from 0.482 to 0.529 (Table 2). These results are consistent with our previous report [7] .
3.3. Species Specific Diagnosis by PCR
Molecular diagnosis by PCR is often used for species identification and offers superior sensitivity and specificity when compared to microscopy. PCR amplification of a specific alpha-tubulin gene sequence was performed using gene specific primers (Spe-F/Spe-R) to distinguish Endoreticulatus sp. Zhenjiang from N. bombycis, N. philosamiae, N. spodopterae, N. plutellae, and Nosema sp. PX1 by PCR. The results showed that the expected fragment was amplified only from the genomic DNA of Endoreticulatus sp. Zhenjiang (Figure 3).
4. Discussion
Microsporidia are fungi-related obligate intracellular parasites with a highly reduced and compact genome, which affect microsporidian transcription and gene expression regulation [17] -[23] . For example, overlapping transcription, a highly unusual phenomenon among eukaryotes, has been observed in some microsporidian species. In the microsporidian Antonospora locustae, a locust parasite, and the mammalian parasite, Encephalitozoon cuniculi, high frequencies of multi-gene transcripts with short 5’-UTRs have been observed in the spore stage [24] -[26] . In this study, we report the full-length alpha-tubulin cDNA sequences of Endoreticulatus sp. Zhenjiang using the RACE protocol. Molecular analysis indicated that the 5’-UTR of the Endoreticulatus sp. Zhenjiang alpha tubulin was only 20 bp which was much shorter than in Saccharomyces cerevisiae. In addition,
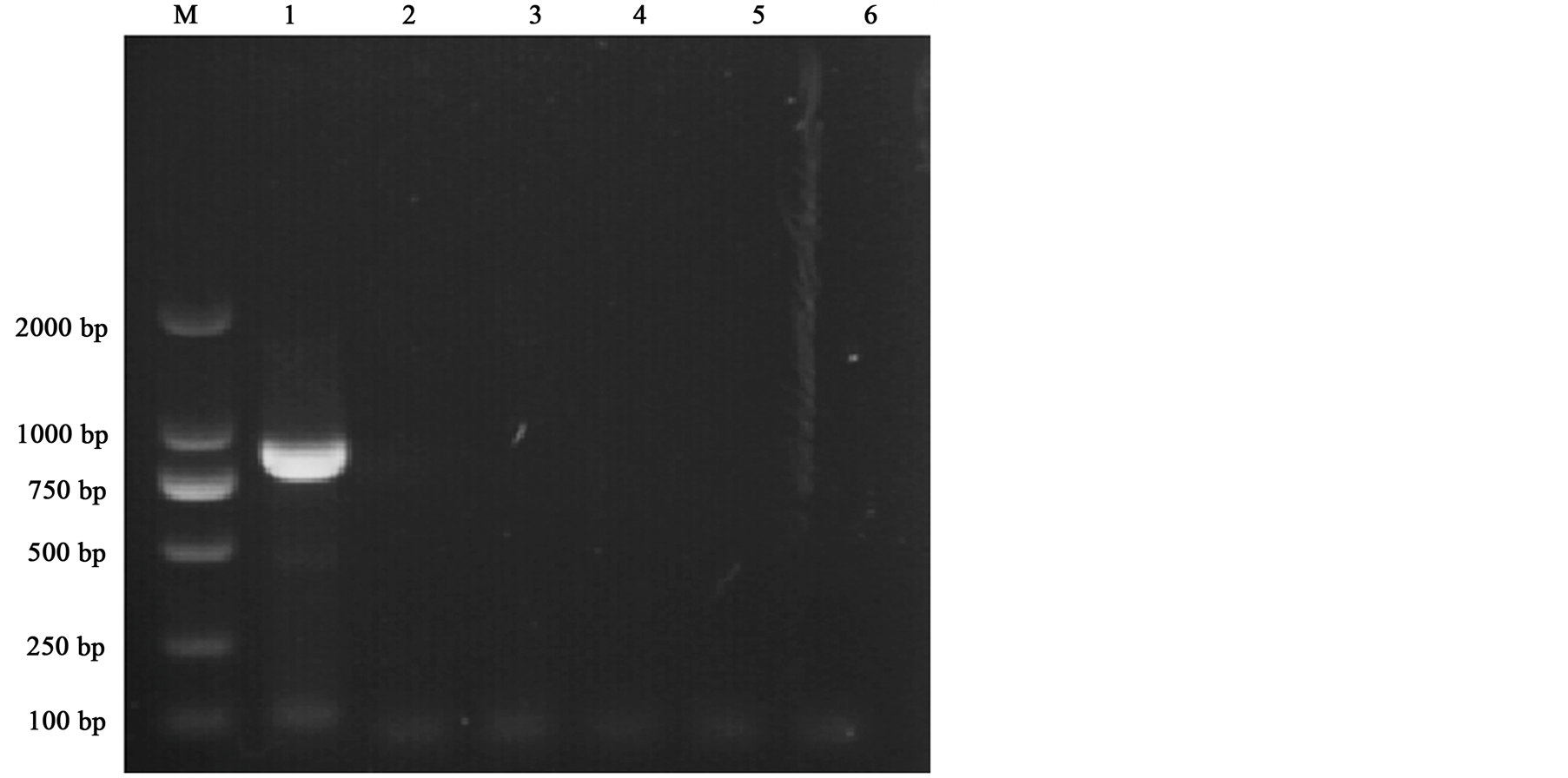
Figure 3. Species specific diagnosis by PCR. Lanes 1-6: Endoreticulatus sp. Zhenjiang, Nosema bombycis, Nosema philosamiae, Nosema sp. HA, Nosema sp. PA, and Nosema antheraeae.
Table 2. Comparison of microsporidian alpha-tubulin nucleotide sequences: percentage of identity (top diagonal) and pairwise distance (bottom diagonal).
the 3’-UTR was also short with 42 bp that includes 29 dA residues without a polyadenylation signal. All these features are likely due to the high compaction of microsporidia genome, which also has a gene transcription pattern that is different from other microsporidia [27] .
The 5’-UTR and 3’-UTR of eukaryotic mRNA play important roles in the post-transcriptional gene expression regulation. Particularly, the 3’-UTR guides the mRNA 3’ end processing with the end processing signal. The 3’-UTR not only controls stability, degradation rate of mRNA and gives assistance to identify specific codons, but also controls translation time, sites, initiation, and efficiency [28] . The spatial and temporal regulation of eukaryotic translation initiation is largely achieved through the 5’-UTR [29] . Highly expressed genes such as housekeeping genes tend to have shorter 5’-UTRs with simple structures, but genes that need strict regulation such as growth factors and proteins associated with cell proliferation, differentiation, and apoptosis, have much longer 5’-UTRs with complex structures [30] [31] .
Traditional, taxonomical species classification of microsporidia is mainly based on ultrastructural and ecological features. For example, the differential diagnosis of Nosema species was based on subtle differences in overlapping characteristics such as spore size, number of nuclei per cell, type of cell division, microsporidium host relationships, and primary site of infection. However, many microsporidia are considered to have a wide host range, complicated life cycles, and parasite host relationships. Therefore, the majority of the previously published classifications differ significantly and the taxonomy of microsporidia, particularly the species identification still remains controversial. Molecular markers using the tubulin protein and rRNA sequence data in combination with phylogenetic analysis can be effective tools to redefine the phylogenetic relationship between microsporidia [7] [27] . The alphaand beta-tubulin proteins are the most abundant proteins in eukaryotic cells and their evolution may have paralleled the nucleus. Data on recent phylogenies based on alphaand beta-tubulin protein-coding genes have enabled to carry out more robust phylogenetic analyses [32] -[36] . In this study, phylogenetic trees were constructed from Endoreticulatus sp. Zhenjiang alpha-tubulin sequence and 14 other microsporidian sequences. We found that Endoreticulatus sp. Zhenjiang was within the same clade as Endoreticulatus sp. CHW-2004 Taiwan, but it was distantly related to other microsporidia, which was consistent with our previous results based on SSU rRNA subunit phylogenetic tree [7] . This also explains the credibility of the tubulin gene for phylogenetic analysis.
5. Conclusion
A number of methods have been explored to effectively detect microsporidia and range from the earliest visual identification to microscopic examination and serological and molecular detection techniques [37] -[39] . Currently, molecular diagnostic methods have made great progress due to higher specificity and sensitivity in the detection of microsporidia [40] [41] . Although replacing microscopy with more sensitive and specific nucleic acid based methods is hampered by the higher cost of cloning and sequencing, multiplexing the detection of more than one parasite in a single test has been found to be very effective and would decrease the cost of the test without the need for multiple PCR equipment, which is expensive. In this study, we designed gene specific primers to amplify the Endoreticulatus sp. Zhenjiang alpha-tubulin gene from its genomic DNA. These primers and the conditions used, specifically were amplified only from the genomic DNA of Endoreticulatus sp. Zhenjiang but not from N. bombycis, N. philosamiae, Nosema sp. HA, Nosema sp. PA or N. antheraeae. Therefore, this study reported that the specific identification of Endoreticulatus sp. Zhenjiang among the other microsporidia can be a more reliable detection method to improve the epidemiology of the disease.
Acknowledgements
This work was supported by the earmarked fund for Modern Agro-industry Technology Research System. We are grateful to everyone who generously provided us with access to the software used in this work. We also thank all our partners and laboratory members for their kind help and criticism.
References
- Li, Y., et al. (2009) Identification of a Novel Spore Wall Protein (SWP26) from Microsporidia Nosema bombycis. International Journal for Parasitology, 39, 391-398. http://dx.doi.org/10.1016/j.ijpara.2008.08.011
- Didier, E.S., Snowden, K.F. and Shadduck, J.A. (1998) Biology of Microsporidian Species Infecting Mammals. Advances in Parasitology, 40, 283-320. http://dx.doi.org/10.1016/S0065-308X(08)60125-6
- Corradi, N. and Keeling, P.J. (2009) Microsporidia: A Journey through Radical Taxonomical Revisions. Fungal Biology Reviews, 23, 1-8. http://dx.doi.org/10.1016/j.fbr.2009.05.001
- Corradi, N., Haag, K.L., Pombert, J.F., Ebert, D. and Keeling, P.J. (2009) Draft Genome Sequence of the Daphnia Pathogen Octosporeabayeri: Insights into the Gene Content of a Large Microsporidian Genome and a Model for Host-Parasite Interactions. Genome Biology, 10, R106. http://dx.doi.org/10.1186/gb-2009-10-10-r106
- Cai, S., Lu, X., Qiu, H., Li, M. and Feng, Z. (2011) Identification of a Nosema bombycis (Microsporidia) Spore Wall Protein Corresponding to Spore Phagocytosis. Parasitology, 138, 1102-1109.http://dx.doi.org/10.1017/S0031182011000801
- Wan, Y., et al. (1995) Separation and Research of Pathogenic Microsporidia SCM (Endoreticulatus. sp), Bombyx mori. Acta Sericologic Sinica, 21, 168-172.
- Xu, X., et al. (2012) Phylogenetic Characterization of a Microsporidium (Endoreticulatus sp. Zhenjiang) Isolated from the Silkworm, Bombyx mori. Parasitology Research, 110, 815-819.http://dx.doi.org/10.1007/s00436-011-2560-8
- Keeling, P.J. (2003) Congruent Evidence from Alpha-Tubulin and Beta-Tubulin Gene Phylogenies for a Zygomycete Origin of Microsporidia. Fungal Genetics and Biology, 38, 298-309. http://dx.doi.org/10.1016/S1087-1845(02)00537-6
- Zhu, F., Shen, Z., Xu, L. and Guo, X. (2013) Molecular Characteristics of the Alphaand Beta-Tubulin Genes of Nosemaphilosamiae. Folia Parasitologica, 60, 411-415. http://dx.doi.org/10.14411/fp.2013.043
- McKean, P. G., Vaughan, S. and Gull, K. (2001) The Extended Tubulin Superfamily. Journal of Cell Science, 114, 2723-2733.
- Dong, S., Shen, Z., Xu, L. and Zhu, F. (2010) Sequence and Phylogenetic Analysis of SSU rRNA Gene of Five Microsporidia. Current Microbiology, 60, 30-37. http://dx.doi.org/10.1007/s00284-009-9495-7
- Aurrecoechea, C., Barreto, A., Brestelli, J., Brunk, B.P., Caler, E.V., Fischer, S., et al. (2011) AmoebaDB and MicrosporidiaDB: Functional Genomic Resources for Amoebozoa and Microsporidia Species. Nucleic Acids Research, 39, D612-D619. http://dx.doi.org/10.1093/nar/gkq1006
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F. and Higgins, D.G. (1997) The CLUSTAL_X Windows Interface: Flexible Strategies for Multiple Sequence Alignment Aided by Quality Analysis Tools. Nucleic Acids Research, 25, 4876-4882. http://dx.doi.org/gka797
- Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731-2739. http://dx.doi.org/10.1093/molbev/msr121
- Yanagawa, N., Tamura, G., Oizumi, H., Endoh, M. and Motoyama, T. (2011) MAGE Expressions Mediated by Demethylation of MAGE Promoters Induce Progression of Non-Small Cell Lung Cancer. Anticancer Research, 31, 171-175.
- Gao, K., Deng, X., Qian, H., Wu, P., Qin, G., Liu, T., et al. (2012) Novel Protein of IBP from Silkworm, Bombyx mori, Involved in Cytoplasmic Polyhedrosis Virus Infection. Journal of Invertebrate Pathology, 110, 83-91. http://dx.doi.org/10.1016/j.jip.2012.02.011
- Katinka, M.D., Duprat, S., Cornillot, E., Méténier, G., Thomarat, F., Prensier, G., et al. (2001) Genome Sequence and Gene Compaction of The Eukaryote Parasite Encephalitozoon cuniculi. Nature, 414, 450-453. http://dx.doi.org/10.1038/35106579
- Peyretaillade, E., El-Alaoui, H., Diogon, M., Polonais, V., Parisot, N., Biron, D.G., et al. (2011) Extreme Reduction and Compaction of Microsporidian Genomes. Research in Microbiology, 162, 598-606. http://dx.doi.org/10.1016/j.resmic.2011.03.004
- Cuomo, C.A., Desjardins, C.A., Bakowski, M.A., Goldberg, J., Ma, A.T., Becnel, J.J., et al. (2012) Microsporidian Genome Analysis Reveals Evolutionary Strategies for Obligate Intracellular Growth. Genome Research, 22, 2478-2488. http://dx.doi.org/10.1101/gr.142802.112
- Keeling, P.J. and Fast, N.M. (2002) MICROSPORIDIA: Biology and Evolution of Highly Reduced Intracellular Parasites. Annual Review of Microbiology, 56, 93-116. http://dx.doi.org/10.1146/annurev.micro.56.012302.160854
- Keeling, P.J., Fast, N.M., Law, J.S., Williams, B.A. and Slamovits, C.H. (2005) Comparative Genomics of Microsporidia. Folia Parasitologica, 52, 8-14. http://dx.doi.org/10.14411/fp.2005.002
- Waller, R.F., Jabbour, C., Chan, N.C., Celik, N., Likic, V.A., Mulhern, T.D., et al. (2009) Evidence of a Reduced and Modified Mitochondrial Protein Import Apparatus in Microsporidian Mitosomes. Eukaryotic Cell, 8, 19-26. http://dx.doi.org/10.1128/EC.00313-08
- Corradi, N. and Slamovits, C.H. (2011) The Intriguing Nature of Microsporidian Genomes. Briefings in Functional Genomics, 10, 115-124. http://dx.doi.org/10.1093/bfgp/elq032
- Corradi, N., Gangaeva, A. and Keeling, P.J. (2008) Comparative Profiling of Overlapping Transcription in the Compacted Genomes of Microsporidia Antonospora locustae and Encephalitozoon cuniculi. Genomics, 91, 388-393. http://dx.doi.org/10.1016/j.ygeno.2007.12.006
- Williams, B.A., Slamovits, C.H., Patron, N.J., Fast, N.M. and Keeling, P.J. (2005) A High Frequency of Overlapping Gene Expression in Compacted Eukaryotic Genomes. Proceedings of the National Academy of Sciences of the United States of America, 102, 10936-10941. http://dx.doi.org/10.1073/pnas.0501321102
- Grisdale, C.J. and Fast, N.M. (2011) Patterns of 5’ Untranslated Region Length Distribution in Encephalitozoon cuniculi: Implications for Gene Regulation and Potential Links between Transcription and Splicing. Journal of Eukaryotic Microbiology, 58, 68-74. http://dx.doi.org/10.1111/j.1550-7408.2010.00523.x
- Wang, J., Huang, K., Mao, X., Zhao, Y. and Lu, C. (2001) Small Subunit Ribosomal RNA (ssurRNA) Gene of Microsporidia. Acta Biochiimica et Biophysica Sinica, 33, 229-232.
- Jiang, Y., Ling, Y. and Zhao, W. (2001) Progress in the Studies on Function of 3’ Untranslated Region on Post-Transcriptional Leval. Chinese Bulletin of Botany, 18, 3-10.
- Pickering, B.M. and Willis, A.E. (2005) The Implications of Structured 5’ Untranslated Regions on Translation and Disease. Seminars in Cell & Developmental Biology, 16, 39-47. http://dx.doi.org/10.1016/j.semcdb.2004.11.006
- Hughes, T.A. (2006) Regulation of Gene Expression by Alternative Untranslated Regions. Trends in Genetics, 22, 119-122. http://dx.doi.org/10.1016/j.tig.2006.01.001
- Fang, Y., Xue, J. and Tian, L. (2008) Regulation of Translation Initiation and Targeted Gene Therapy of Tumor. Chinese Journal of Cancer Biotherapy, 15, 396-400.
- Akiyoshi, D.E., Weiss, L.M., Feng, X., Williams, B.A.P., Keeling, P.J., Zhang, Q., et al. (2007) Analysis of the β-Tubulin Genes from Enterocytozoon bieneusi Isolates from a Human and Rhesus Macaque. Journal of Eukaryotic Microbiology, 54, 38-41. http://dx.doi.org/10.1111/j.1550-7408.2006.00140.x
- Keeling, P.J., Luker, M.A. and Palmer, J.D. (2000) Evidence from β-Tubulin Phylogeny That Microsporidia Evolved from within the Fungi. Molecular Biology and Evolution, 17, 23-31. http://dx.doi.org/10.1093/oxfordjournals.molbev.a026235
- Keeling, P.J. and Doolittle, W.F. (1996) Alpha-Tubulin from Early-Diverging Eukaryotic Lineages and the Evolution of the Tubulin Family. Molecular Biology and Evolution, 13, 1297-1305. http://dx.doi.org/10.1093/oxfordjournals.molbev.a025576
- Lee, R.C., Williams, B.A., Brown, A.M., Adamson, M.L., Keeling, P.J. and Fast, N.M. (2008) α- and β-Tubulin Phylogenies Support a Close Relationship between the Microsporidia Brachiola algerae and Antonospora locustae. Journal of Eukaryotic Microbiology, 55, 388-392. http://dx.doi.org/ 10.1111/j.1550-7408.2008.00348.x
- Haag, K.L., Larsson, J.I., Refardt, D. and Ebert, D. (2011) Cytological and Molecular Description of Hamiltosporidium tvaerminnensis gen. etsp. nov., a Microsporidianparasite of Daphnia magna, and Establishment of Hamiltosporidium magnivora comb.nov. Parasitology, 138, 447-462. http://dx.doi.org/10.1017/S0031182010001393
- Stine, S.W., Vladich, F.D., Pepper, I.L. and Gerba, C.P. (2005) Development of a Method for the Concentration and Recovery of Microsporidia from Tap Water. Journal of Environmental Science and Health, Part A: Toxic Hazardous Substances and Environmental Engineering, 40, 913-925. http://dx.doi.org/10.1081/ESE-200056105
- Kokoskin, E., Gyorkos, T.W., Camus, A., Cedilotte, L., Purtill, T. and Ward, B. (1994) Modified Technique for Efficient Detection of Microsporidia. Journal of Clinical Microbiology, 32, 1074-1075.
- Lee, S.H., Joung, M., Yoon, S., Choi, K., Park, W.Y. and Yu, J.R. (2010) Multiplex PCR Detection of Waterborne Intestinal Protozoa: Microsporidia, Cyclospora, and Cryptosporidium. Korean Journal of Parasitology, 48, 297-301. http://dx.doi.org/10.3347/kjp.2010.48.4.297
- Sanders, J.L. and Kent, M.L. (2011) Development of a Sensitive Assay for the Detection of Pseudoloma neurophilia in Laboratory Populations of the Zebrafish Danio rerio. Diseases of Aquatic Organisms, 96, 145-156. http://dx.doi.org/10.3354/dao02375
- Taniuchi, M., Verweij, J.J., Sethabutr, O., Bodhidatta, L., Garcia, L., Maro, A., et al. (2011) Multiplex Polymerase Chain Reaction Method to Detect Cyclospora, Cystoisospora, and Microsporidiain Stool Samples. Diagnostic Microbiology and Infectious Disease, 71, 386-390. http://dx.doi.org/10.1016/j.diagmicrobio.2011.08.012
NOTES

*Corresponding author.


