Food and Nutrition Sciences
Vol.4 No.8A(2013), Article ID:35277,10 pages DOI:10.4236/fns.2013.48A011
Biophysical Characterization of Genistein in Its Natural Carrier Human Hemoglobin Using Spectroscopic and Computational Approaches
![]()
1Biophysics Division, Saha Institute of Nuclear Physics, Kolkata, India; 2Department of Chemistry, Tougaloo College, Tougaloo, USA; 3Saroj Mohan Institute of Technology, Hooghly, West Bengal, India; 4Department of Physics, Tougaloo College, Tougaloo, USA.
Email: †bsengupta@tougaloo.edu, †bsgupta.tougaloo@gmail.com, †pradeepsinp@yahoo.co.in, †pradeepk.sengupta@saha.ac.in,
†sandipanchakraborty.13@gmail.com
Copyright © 2013 Biswapathik Pahari et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received April 8th, 2013; revised May 9th, 2013; accepted May 17th, 2013
Keywords: Natural Drug Carrier; Fluorescence; Circular Dichroism; Molecular Dynamics; Docking
ABSTRACT
Steady state and time resolved fluorescence spectroscopy, combined with molecular dynamics simulation, have been used to explore the interactions of a therapeutically important bioflavonoid, genistein, with normal human hemoglobin (HbA). Binding constants estimated from the fluorescence studies were K = (3.5 ± 0.32) ´ 104 M−1 for genistein. Specific interactions with HbA were confirmed from flavonoid-induced fluorescence quenching of the tryptophan in the protein HbA. The mechanism of this quenching involves both static and dynamic components as indicated by: (a) increase in the values of Stern-Volmer quenching constants with temperatures, (b)  /
/![]() is slightly > 1 (where
is slightly > 1 (where 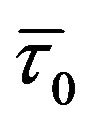 and
and ![]() are the unquenched and quenched tryptophan fluorescence lifetimes (averaged) respectively). Molecular docking and dynamic simulations reveal that genistein binds between the subunits of HbA, ~18 Å away from the closest heme group of chain α1, emphasizing the fact that the drug does not interfere with oxygen binding site of HbA.
are the unquenched and quenched tryptophan fluorescence lifetimes (averaged) respectively). Molecular docking and dynamic simulations reveal that genistein binds between the subunits of HbA, ~18 Å away from the closest heme group of chain α1, emphasizing the fact that the drug does not interfere with oxygen binding site of HbA.
1. Introduction
Flavonoids are polyphenolic compounds which are ubiquitous in plants of higher genera [1]. Flavonoids have varied uses as therapeutical agents in health disparities, including tumor, cancer, AIDS, diabetes, neurodegenerative and cardiac disorders [1-6]. The isoflavone genistein (structure shown in Scheme 1) has dual action as a topoisomerase inhibitor and a tyrosine kinase inhibitor and induces cell differentiation [7,8]. Tyrosine phosphorylation plays important roles in cell proliferation and cell transformation and so tyrosine kinase-specific inhibitors might be used as anticancer agents [9,10]. Genistein, has been shown to be a specific inhibitor of the EGF stimulated tyrosine kinase activity in cultured A431 cells [7].
Genistein induces DNA strand breakage through inhibiting topoisomerase II and induces a mature phenotype in the human myeloid HL-205 and erythroid K-562-J leukemia cells [7,8]. Although consumption of the flavonoids has been associated with the prevention of several degenerative diseases [1-6], their bioavailability is often poor probably due to their low aqueous solubility and possible interaction with plasma proteins [11]. Erythrocytes (red blood cells) have been used as drug delivery systems, due to their biocompatibility, biodegradability, usability and easy loading capability [12,13]. For designing and improving an erythrocyte based drug delivery system, the binding interactions of drugs with the main component of erythrocytes, hemoglobin (HbA), have to be fully understood. Hemoglobin is the most abundant blood protein and consists of two α and two β subunits which are noncovalently associated within erythrocytes as a 64.5 kDa tetramer [14,15]. In this article we have
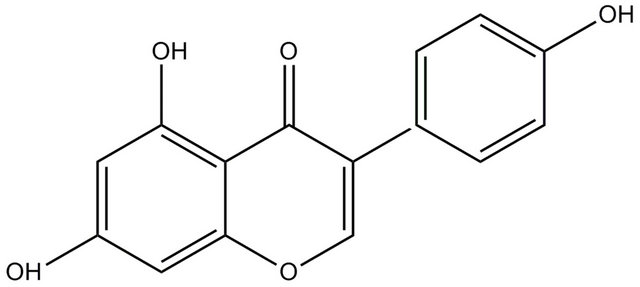
Scheme 1. Structure of the isoflavone; genistein (5,7-Dihydroxy-3-(4-hydroxyphenyl) chromen-4-one).
explored the interaction of genistein with human HbA using the intrinsic fluorescence of HbA and molecular dynamics approaches.
2. Materials and Methods
2.1. Sample Preparation
Lyophilized powder of human hemoglobin, genistein, and phosphate buffer were purchased from Sigma Chemicals, USA. Solvents used were of spectroscopic grade and obtained from Sigma-Aldrich. Purity of genistein was further confirmed by thin layer chromatography which showed only one spot under UV light. Absorption and fluorescence spectroscopic measurements were performed with genistein concentrations of 1 × 10−5 M. HbA was dissolved in pH 7.4 phosphate buffer solution (1 × 10−2 M) and the HbA stock solution (2 × 10−4 M) was kept in the dark at 277 K. The protein concentration was determined spectrophotometrically using the molar extinction coefficient of HbA at 276 nm (120,808 M−1·cm−1) [16,17]. For fluorescence quenching studies, the HbA concentration was kept constant at 10−5 M. Varying aliquots of concentrated methanolic solution of genistein were added to obtain final concentrations ranging from 0 - 2.5 × 10−5 M. The concentrations of methanol were always kept <1% (by volume) in all samples.
2.2. Spectroscopic Measurements
Steady state absorption spectra were recorded with Shimadzu UV2550 and Cecil model 7500 spectrophotometers. Steady state fluorescence measurements were carried out with Shimadzu RF5301 (equipped with a Fisher temperature controlled accessory) and Varian Cary Eclipse spectrofluorometers. A quartz cuvette of 1 cm path length was used in all experiments. Time resolved fluorescence decay measurements were performed using Jobin-Yvon nanosecond time correlated single photon counting (TCSPC) setup. Fluorescence decay measurements were performed using a nanosecond time correlated single photon counting setup with 295 nm excitation source (nanoLED-295) having pulse FWHM ~760 ps. An emission monochromator was used to block the scattered light and isolate the emission. Data analyses were performed using DAS6 Fluorescence Decay Analysis Software, provided with the TCSPC instrument and were fitted with a multi exponential decay function, 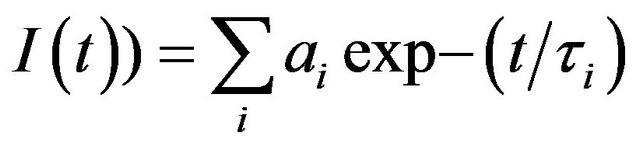 where ai and τi represent the amplitudes and decay times respectively of the individual components for multi-exponential decay profiles. The goodness of fit was estimated by using reduced χ2 (namely
where ai and τi represent the amplitudes and decay times respectively of the individual components for multi-exponential decay profiles. The goodness of fit was estimated by using reduced χ2 (namely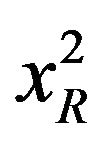 ) values as well as Durbin-Watson parameters (DW). A fit is considered acceptable for a given set of observed data and chosen function, when the
) values as well as Durbin-Watson parameters (DW). A fit is considered acceptable for a given set of observed data and chosen function, when the 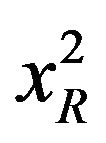 value is in the range 0.8 - 1.2 and the DW value is greater than 1.7, 1.75 and 1.8 for a single, double and triple exponential fit respectively [18]. Average lifetime is calculated using the equation,
value is in the range 0.8 - 1.2 and the DW value is greater than 1.7, 1.75 and 1.8 for a single, double and triple exponential fit respectively [18]. Average lifetime is calculated using the equation, 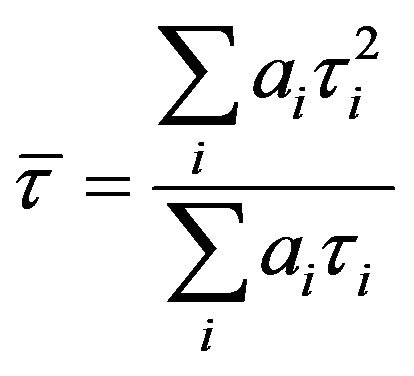 where ai and τi
where ai and τi
represent the amplitude and decay time respectively of the individual components for multi-exponential decay profiles. Circular dichroism spectra were acquired with Biologic Science Instruments (France) spectropolarimeter, using a rectangular cuvette with 1 mm path length. The scan rate was 60 - 100 nm/min, and three/five consecutive spectra were averaged to produce the final spectrum. All spectral measurements were performed at ambient temperature. The highest concentration of HbA for fluorescence decay, steady state, and circular dichroism experiments were kept at 10 µM in order to avoid scattering and related artifacts [6].
2.3. Docking Study
AutoDock4 [19] was employed to gain an insight into the genistein binding with HbA. 3-D atomic coordinates of HbA were obtained from the Brookhaven Protein Data Bank (PDB ID 2D60) and prepared for docking. Hemoglobin was considered as a tetramer. All hetero atoms were deleted and non-polar hydrogens were merged. The Kollman united-atom charge model was applied to the protein. Particular attention was given to the parameterization of the porphyrin rings. Partial atomic charges for the porphyrin ring were assigned using the GasteigerMarsili method while the valence state of the Iron (Fe) was added manually. Atomic solvation parameters and fragmental volumes were added to the protein. Grid maps used by the empirical free-energy scoring function in AutoDock were generated. A grid box of 100 × 100 × 100 grid points in size with a grid-point spacing of 0.375 Å was considered for docking. The map was centered such that it covered the entire protein including all possible binding sites.
The 3-D structure of genistein was built using the HYPERCHEM 7.5 [20] molecular builder module and optimized using the AM1 semi-empirical method to an RMS convergence of 0.001 kcal/(Ǻ mol) with the PolakRibiere conjugate gradient algorithm implemented in the HYPERCHEM 7.5 package. Rotatable bonds were assigned for the ligand and partial atomic charges were calculated using the Gasteiger-Marsili method after merging non-polar hydrogens. 100 docking runs were performed and for each run, a maximum of 2,500,000 GA operations were performed on a single population of 150 individuals. The weights for crossover, mutation and elitism were default parameters of 0.8, 0.02 and 1 respectively.
2.4. Molecular Dynamics of the HbA-Genistein Complex
MD simulations on free hemoglobin and its complex with genistein were performed using GROMACS [21,22] with OPLS all atom force-field [23]. Genistein parameters were developed according to the OPLS forcefield defined atomic groups. The partial atomic charges were initially assigned according to the OPLS force field using chemical-group analogy method and then readjusted to keep the charge neutrality of the atomic groups to make the ligand charge neutral. Ligand parameter were tested by comparing the GROMACS optimized structure with 1) the parameter-independent QM optimized structure of genistein in vacuum, and 2) the QM/MM optimized structures of genistein in explicit water. CPMD [24] was used for QM calculations and QM/MM calculations were carried-out using GROMACS-CPMD [25]. For QM/MM, the genistein molecule was considered in the QM sub-system and the water was considered in MM sub-system and their interaction between the subsystems was described by the QM/MM Hamiltonian [25].
The crystal structure coordinates of HbA obtained from the protein data bank (PDB ID: 2D60) and used in the docking study, were considered for the simulation of free HbA. For HbA-genistein complex, the lowest energy docked complex obtained from the docking study was used for MD simulation. The OPLS parameters for the heme prosthetic group were taken from the previously published parameter set [26]. The coupling of heme with His-92 of HbA through Fe-N bonding was facilitated through “specbond” option implemented in GROMACS. The HbA structure was primarily subjected to molecular dynamics simulation using GROMACS to check the planarity of the porphyrin ring and proper positioning of the distal histidine residue of hemoglobin that coordinates to the Iron (Fe) of the heme moiety, an essential structural feature for the biological functioning of HbA. Then both ligand bound/unbound structures of HbA were subjected to in vacuo minimization using steepest descent algorithm. Each structure was soaked in a water box containing SPC water molecules such that all the protein atoms were at a distance equal or greater than 1 nm from the box edges. Then the system was further minimized using 500 steps of steepest descent algorithm in a water box. Then each minimized system was subjected to 100 ps of position restrained dynamics at 300 K where proteins were kept fixed by adding restraining forces, but water molecules were allowed to move. Final production simulations were performed in the isothermal-isobaric (NPT) ensemble at 300 K, by coupling to an external bath with a coupling constant of 0.1 ps using Berendsen methods. Pressure was kept constant (1 bar) by using the time-constant for pressure coupling set to 1 ps. The LINCS [27] algorithm was used to constrain bond lengths, allowing the use of 2 fs time steps. Van der Waals and Coulomb interactions were truncated at 1.2 nm. Conformations generated during MD simulation were stored at every 5 ps. All the analyses were carried out using the available trajectory analysis tools of GROMACS packages.
3. Results and Discussions
3.1. Steady State and Time Resolved Fluorescence Results
Figures 1(a) and (b) present the fluorescence emission and excitation spectra and Figure 1(c) shows the absorption spectra of hemoglobin with increasing concentration of genistein. It is evident from Figure 1(a) that addition of genistein induces significant changes in the emission intensity of tryptophan in human hemoglobin with no major change in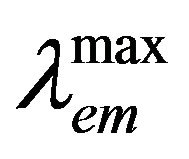 . The quenching of tryptophan fluorescence with increasing concentrations of the flavonoid genistein, suggest genistein is binding at a region in HbA which is close to the tryptophan residue. The excitation wavelength used for this is 280 nm where unbound genistein has little absorbance (see the blue absorption profile in Figure 1(c). Figures 1(b) and (c) show the fluorescence excitation and absorbance of HbA (10 µM) with (0 µM ―, 5 µM ..., 10 µM - - -, 15 µM - . -, 20 µM - . . -) genistein in 10 mM phosphate buffer at pH 7. There is no appreciable change in the
. The quenching of tryptophan fluorescence with increasing concentrations of the flavonoid genistein, suggest genistein is binding at a region in HbA which is close to the tryptophan residue. The excitation wavelength used for this is 280 nm where unbound genistein has little absorbance (see the blue absorption profile in Figure 1(c). Figures 1(b) and (c) show the fluorescence excitation and absorbance of HbA (10 µM) with (0 µM ―, 5 µM ..., 10 µM - - -, 15 µM - . -, 20 µM - . . -) genistein in 10 mM phosphate buffer at pH 7. There is no appreciable change in the 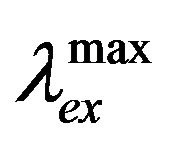 in Figure 1(b). However, significant difference in the absorption profiles between unbound and conjugated HbA was observed confirming the binding of genistein with HbA. It is pertinent to mention that fluorescence excitation spectra look at the excited state of the chromophore tryptophan, whereas absorption spectroscopy studies the ground state characteristics of the whole HbA along with tryptophan and genistein.
in Figure 1(b). However, significant difference in the absorption profiles between unbound and conjugated HbA was observed confirming the binding of genistein with HbA. It is pertinent to mention that fluorescence excitation spectra look at the excited state of the chromophore tryptophan, whereas absorption spectroscopy studies the ground state characteristics of the whole HbA along with tryptophan and genistein.
Quenching studies of protein tryptophan fluorescence
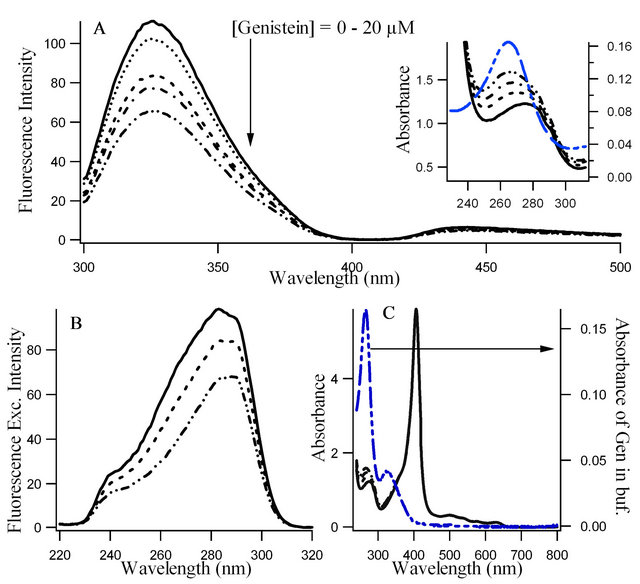
Figure 1. Fluorescence emission spectra (λex = 280 nm), (b) Fluorescence excitation spectra (λem = 340 nm) and (c) Absorption spectra, of hemoglobin (10 μM) in presence of increasing concentrations of genistein (0 µM ―, 5 µM ..., 10 µM - - -, 15 µM - . -, 20 µM - . . -). The absorption of unbound genistein in buffer (blue) is shown in Figure 1(c) using the right axis. Furthermore, in this figure the absorption of tryptophan at 280 nm as well as the Soret (~406 nm) and heme (> 500 nm) bands of HbA protein are displayed. Figure 1(a) inset highlights the absorption profiles of Figure 1(c) in the UV region to indicate the changes in the absorption spectra of tryptophan of HbA upon binding of genistein.
by ligands is a convenient means for exploring ligandprotein interactions [5,6]. The ratio F0/F for each addition of genistein are plotted against the genistein concentrations (see Figure 2(a)), where F0 and F are the fluorescence intensities of Trp in HbA in the absence and presence of genistein, respectively. It is observed that the ratio F0/F increases linearly with the genistein concentration and a linear regression equation following Stern-Volmer relation is obtained. Figure 2(a) presents corresponding Stern-Volmer plots based on the equation.
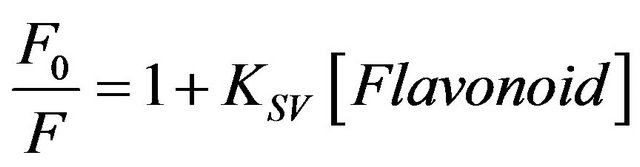 (1)
(1)
where KSV is the Stern-Volmer quenching constant for the quenching of HbA tryptophan fluorescence by flavonoid [16]. The KSV values obtained are ca. 3.21 ± 0.002 ´ 104 M−1 for genistein which indicate that HbA tryptophan fluorescence is efficiently quenched by the flavonoid. To understand the fluorescence quenching better, temperature dependent studies were performed. Figure 2(b) presents the variation of KSV with increasing temperature where significant dependence between the two is observed within the experimental error. A linear SternVolmer plot (Figure 2(a)) can be expected to arise from collisional and/or static quenching [5]. The increase in KSV with rise in temperature clearly indicates the influence of collisional or dynamic factor in the fluorescence quenching mechanism. To clarify this aspect, time resolved studies are performed on HbA at different concentrations of genistein which is shown in Figure 1(c) and also discussed later.
Fluorescence lifetime measurements were used to obtain further confirmation regarding the nature of the fluorescence quenching, as is discussed above in Figures 1 and 2. Lifetime measurements of HbA tryptophan fluorescence both in presence and absence of the flavonoid (see Table 1) have been performed. We observed that in presence of genistein, a decrease in ![]() occurred, which should be the case if dynamic quenching was the sole responsible factor in fluorescence quenching. However, this change in
occurred, which should be the case if dynamic quenching was the sole responsible factor in fluorescence quenching. However, this change in ![]() is very little. Thus the fact that the average lifetimes (
is very little. Thus the fact that the average lifetimes (![]() ) computed from the decay parameters does not change appreciably with increase in genistein concentrations (see Table 1 and Figure 2(c)), corroborate the conclusion drawn from the steady state fluorescence data that both static and dynamic mechanisms are responsible for the observed fluorescence quenching, implying that flavonoids indeed bind to hemoglobin. Since the intrinsic fluorescence of human hemoglobin originates
) computed from the decay parameters does not change appreciably with increase in genistein concentrations (see Table 1 and Figure 2(c)), corroborate the conclusion drawn from the steady state fluorescence data that both static and dynamic mechanisms are responsible for the observed fluorescence quenching, implying that flavonoids indeed bind to hemoglobin. Since the intrinsic fluorescence of human hemoglobin originates
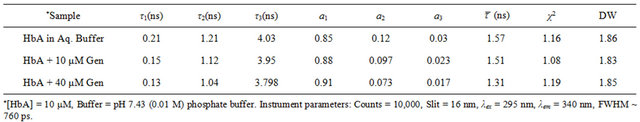
Table 1. Fluorescence decay parameters of HbA (tryptophan) in buffer and in presence of genistein.
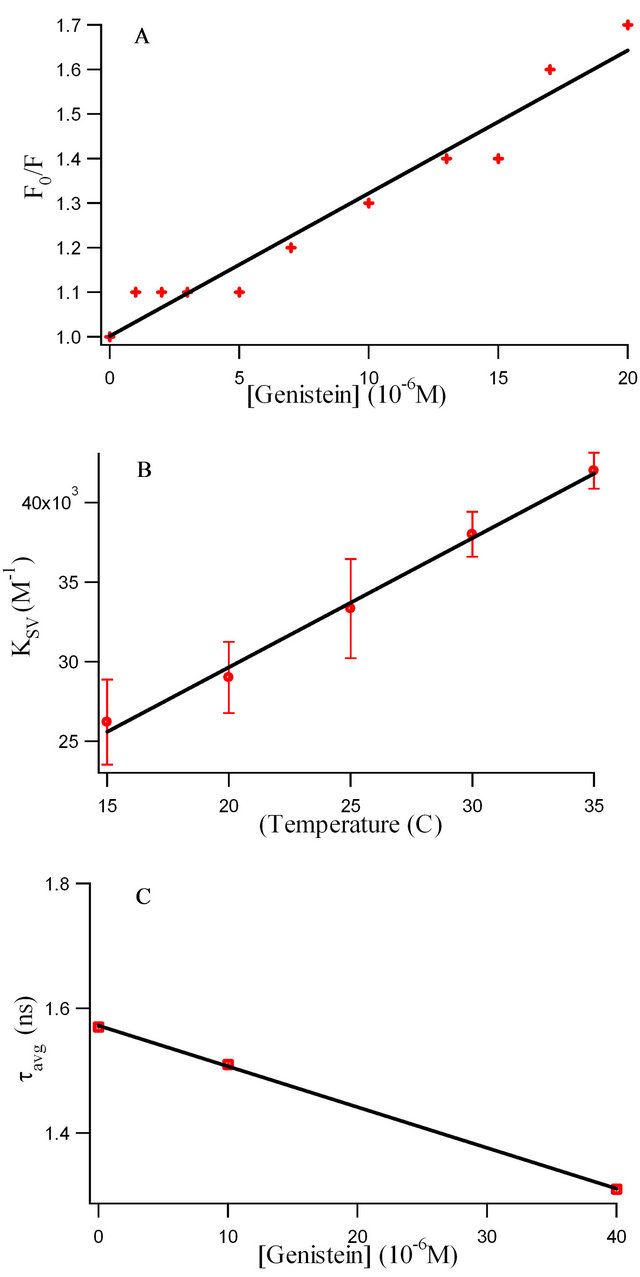
Figure 2. (a) Stern-Volmer plots of the HbA tryptophan fluorescence quenching with increasing genistein concentration at 25˚C. B: Variation of Ksv with temperatures. Each data point indicates the average of three experiments; error bars indicate standard deviations. C: The plot of 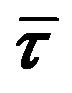 with increase in genistein concentrations.
with increase in genistein concentrations.
primarily from β-37 tryptophan, it seems reasonable to infer that the β-37 tryptophan residue is presumably at or near the binding site of the flavonoids. The apparent binding constant “K” and the number of binding site(s) “n” were estimated from fluorescence titration studies, using the plot of Log(Fo − F)/F vs. Log(1/([Dt] − (Fo − F))[Pt]/F0)) [5] (Figure 3) which is based on the equation:
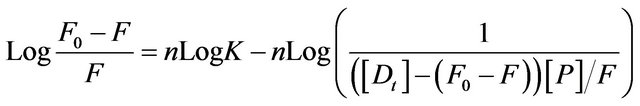 (2)
(2)
where F0 and F are the fluorescence intensity of HbA in absence and presence of flavonoid (D) respectively, [Dt] is the total flavonoid concentration and [Pt] is the total protein concentration, n is the number of binding sites and K is the binding constant.
Table 2 shows the spectroscopic parameters as well as binding constants “K” and number of binding sites “n” for the binding of genistein with HbA. From Figure 3, we observed a K value of (3.5 ± 0.32) ´ 104 and n ~ 1.
The binding forces contributing to interactions of flavonoids with proteins often include a van der Waals interaction, hydrophobic force, electrostatic interactions, hydrogen bond, etc.
The free energy change (ΔG0) of reaction provides the main line of evidence for confirming the binding force. The thermodynamic parameter is evaluated using the following equation:
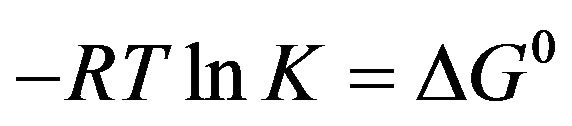 (3)
(3)
where K and R are the binding constant and gas constant, respectively which is provided in Table 2. The spontaneity of the binding of genistein with HbA is evident from negative value of ΔG0.
3.2. Far Ultraviolet Circular Dichroism (CD) Spectroscopic Studies
To investigate the possible effect of the flavonoid on the secondary structure of HbA, we used far-UV CD spectroscopy. The CD spectrum of HbA in aqueous buffer (in the absence of flavonoid) has two characteristic peaks of
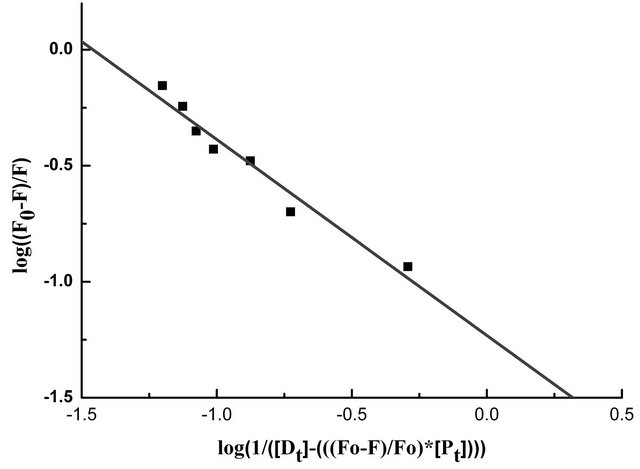
Figure 3. The plot of log(1/([Dt] − (F0 − F))[Pt]/F0) vs. log(F0 − F)/F0 of the fluorescence quenching data. The concentration of HbA, [Pt] = 10.0 × 10−6 M.

Table 2. Binding and thermodynamic parameters for Genistein-HbA interactions at 25˚C.
negative ellipticities at 208 nm and 222 nm indicating its predominantly α-helical secondary structure (Figure 4). As shown in the figure, the CD spectrum of the HbA remains essentially unchanged upon addition of different concentrations of flavonoids (hemoglobin concentration remaining fixed).
The quenching observed for HbA in presence of genistein must be due to some specific interaction that increases the local concentrations of the flavonoids around the tryptophan residue(s) in hemoglobin. The significant dependence of KSV on temperature indicates that the observed fluorescence quenching is influenced by both static and dynamic factors. However the variation of ![]() with increasing genistein concentrations is minor. (N.B. - For static quenching the complexed fluorophores are nonfluorescent, and the only observed fluorescence arises from the uncomplexed fraction. Therefore, the average lifetime of the uncomplexed fluorophores (
with increasing genistein concentrations is minor. (N.B. - For static quenching the complexed fluorophores are nonfluorescent, and the only observed fluorescence arises from the uncomplexed fraction. Therefore, the average lifetime of the uncomplexed fluorophores (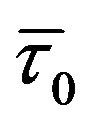 ) remains unchanged and consequently
) remains unchanged and consequently 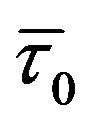 /
/![]() = 1 (
= 1 (![]() is the average lifetime in the presence of quencher). By contrast, for dynamic quenching
is the average lifetime in the presence of quencher). By contrast, for dynamic quenching 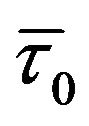 /
/![]() = F0/F > 1 (where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively) [16]. It should be noted that although there is a dramatic increase (~60%) in KSV with increase in temperature from 15˚C to 35˚C as is shown in Figure 2(b) confirming the existence of both static and dynamic factors in fluorescence quenching, the decrease in
= F0/F > 1 (where F0 and F are the fluorescence intensities in the absence and presence of quencher, respectively) [16]. It should be noted that although there is a dramatic increase (~60%) in KSV with increase in temperature from 15˚C to 35˚C as is shown in Figure 2(b) confirming the existence of both static and dynamic factors in fluorescence quenching, the decrease in ![]() with increase in genistein concentrations is only by ~16.56% (as shown in Figure 2(c)). This small change in
with increase in genistein concentrations is only by ~16.56% (as shown in Figure 2(c)). This small change in ![]() reflects a local structural change in the microenvironment of tryptophan of HbA, with no appreciable global change in the protein structure, as is evident from the CD spectra in Figure 4.
reflects a local structural change in the microenvironment of tryptophan of HbA, with no appreciable global change in the protein structure, as is evident from the CD spectra in Figure 4.
3.3. Molecular Dynamics Studies
To explore the effect of genistein binding on the structural stability of HbA, we have analyzed the dynamic structural properties (RMSD and radius of gyration, Rg) of both the ligand free and bound HbA obtained from MD simulation and shown in Figure 5.
The RMSD of the backbone Cα atom of the simulated protein over the time is a reliable parameter to analyze the stability of the system. As evident from Figure 5(a), during the first 3 ns of the simulation, both the systems undergo structural readjustments according to its environments, and monotonically reach the equilibrium state characterized by a stable RMSD profile. A closer look at each trajectory obtained from the MD simulation reveals that the HbA remains stable throughout the simulation for both ligand free and bound HbA. Also, genistein binding does not induce any structural perturbations in HbA. Observed RMSD for free and genistein bound HbA are highly similar, 0.59 ± 0.05 nm and 0.58 ± 0.04 nm respectively during the last 7 ns of simulation. On the other hand, Rg provides a good measure of the overall structural dimension of the protein. HbA is a globular protein with four chains. The calculated Rg value for free HbA is 2.439 ± 0.02 nm, while that of genistein bound HbA is 2.43 ± 0.011 nm. Again the high similarity values of free and ligand bound HbA signifies no such structural unfolding induced by the binding of the ligand, genistein. This agrees well the CD spectra observed in Figure 4.
The interaction energy components between HbA and genistein were analyzed in MD simulation during last 7 ns of the simulation and shown in Figure 6. LennardJones (LJ) interactions between the ligand and the pro-
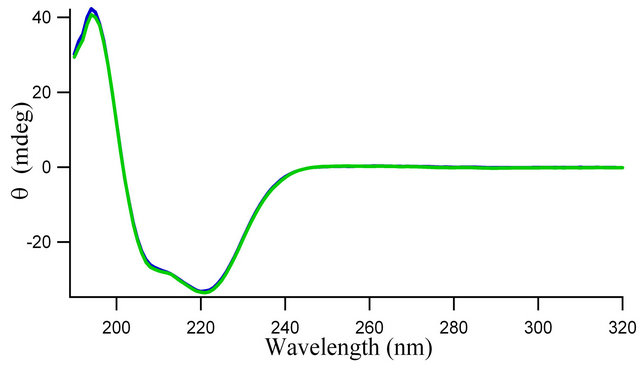
Figure 4. Circular dichroism spectra of 3.0 × 10−6 M hemoglobin (HbA) in absence (blue) and presence (green) of 10.0 × 10−6 M genistein in 0.01 M phosphate buffer, pH 7.4.

Figure 5. Variation of (a) RMSD (nm) and (b) Radius of gyration (Rg, nm) with simulation time for HbA (black) and HbA with bound genistein (red) obtained from molecular dynamics simulation.
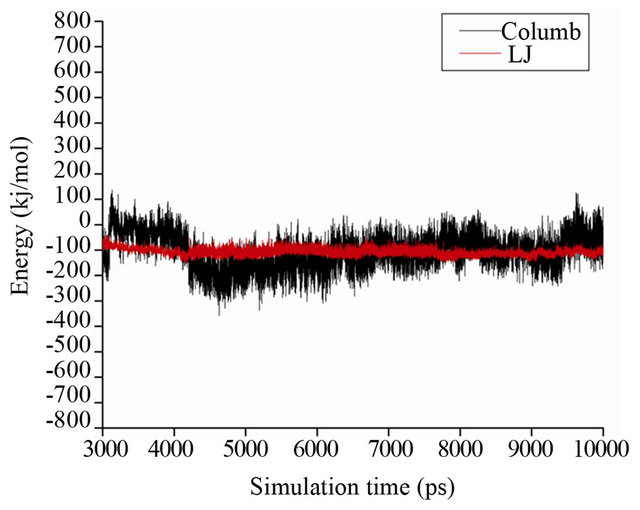
Figure 6. Analysis of different energetic component between HbA and genistein during molecular dynamics simulation. Black and red line represents Columb and van der Waals (Leanard-Jones, LJ) interactions between protein and ligand.
tein are more stable compared to the Coulombic interactions. The average LJ interaction energy obtained from MD simulation is found to be −107.9 ± 13.7 KJ/mole while Columbic interactions are found to be −104.7 ± 67.1 KJ/mole averaged over the last 7 ns of the simulation. The high standard deviation of average Coulombic interactions indicates less specificity of the Coulomb interactions in the binding of genistein with HbA while the stable LJ interactions pattern signifies its contribution to the binding process. Figure 7(a) reveals the backbone trace of the MD average structure of HbA (light color) and HbA-genistein complex (dark color). As evident from the figure, the tetrameric structure of HbA constituted by two α chains and two β chains is highly preserved during simulation and the structure of HbA in presence and absence of genistein is highly similar with calculated RMSD of 0.4 nm over the entire protein backbone. Also the helical nature of each chain is unperturbed during the simulation for both the systems.
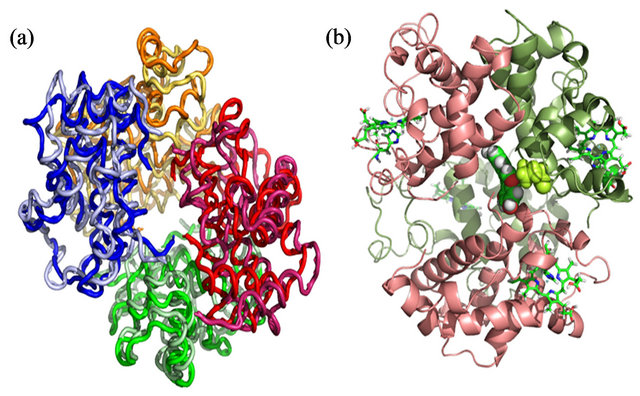
Figure 7. (a) Backbone trace of the MD average structure of HbA (light color) and HbA-genistein complex (dark color). All the four chains have been colored differently. (b) Ribbon representation of HbA-genistein complex. Two α chains and two β chains are colored pink and green accordingly. Four heme groups are represented in sick representation, while genistein and Trp-37 are represented in CPK representation. Trp-37 is represented in green CPK mode.
Figure 7(b) reveals the details of the genistein binding with HbA in atomistic details. The lowest energy docked conformation and the MD average structure starting from the lowest energy docked complex reveals very similar binding mode where genistein readily enters the central cavity formed by four subunits of HbA, namely α1, α2, β1, β2 (shown in pink and deep green respectively). It is to be mentioned that this binding site is similar to the binding site of another bio-flavonoid daidzein within HbA [8]. Particularly, genistein strongly binds at the interface of α2 and β2 domains, very close to the Trp-37, which is evident from the figure (Figure 7(b)). There is a strong van der Waals overlap between genistein and Trp-37. This further confirms that the nearby Trp-37 serves as good optical probe for monitoring genistein binding to HbA. Genistein docked ~5 Å apart from Trp-37 in β2 chain and the average distance between them remains stable during the dynamics, as evident from Figure 8(a). This observation is supported by the fact that the fluorescence of β-37 Trp is exploited to sense the ligand binding to that pocket, as is shown in Figures 1-3.
Another important aspect, we have characterized from our MD simulation is the distances between genistein and Fe atom of the porphyrin moiety. It is to be mentioned that the heme group plays a determining role in the biological functioning of HbA. We have calculated the minimum distances between genistein and the Fe atom of the four heme groups during MD simulation and the results are shown in Figure 8(b). Genistein binds between the subunits of HbA, which is sufficiently away from the heme. The closest distance of genistein to any of the four heme group is ~18 Å (heme group of α1 and β2), emphasizing the fact that the ligand does not interfere with oxygen binding and biological functioning of HbA.
In the tetrameric form of normal adult hemoglobin, the binding of oxygen is a cooperative process. Binding of oxygen to heme group in one chain induces conformational changes which are transmitted to the remaining three monomers in the tetramer, where it induces a similar conformational change in the other heme sites such that binding of oxygen to these sites becomes easier. Thus the inter-domain contacts are highly important to modulate the biological functioning of HbA. We have characterized the mean inter-chain contacts of both free HbA and genistein bound HbA obtained from MD simulation and shown in Figure 9. As evident from the figure,
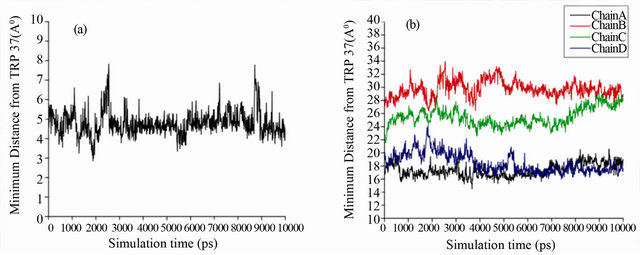
Figure 8. (a) Minimum distance between Trp-37 and genistein during MD simulation. (b) Distances between four Fe atoms in four chains and genistein during MD simulation.
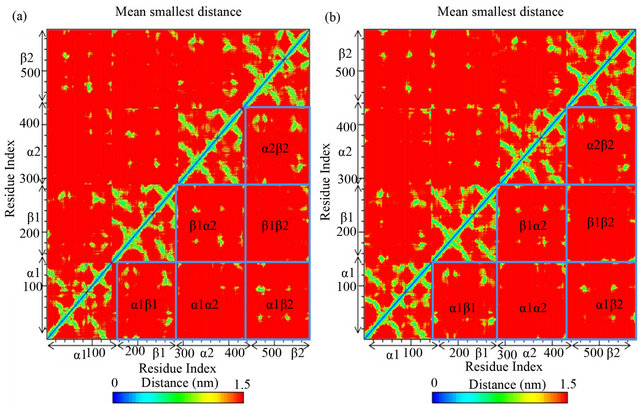
Figure 9. Mean contact distances between residues observed during MD simulation. (a) HbA only; (b) HbA-genistein complex.
genistein binding does not influence the interchain contacts significantly. Only slight variations are observed in the interdomain contacts of α1 and β1 chain and also in between α1 and β2. But, the overall essential contacts between any two chains remain unperturbed upon genistein binding to HbA.
4. Concluding Remarks
Since the pharmacological actions of flavonoids in vivo are closely related to their binding with cellular targets including proteins, the investigation on binding of flavonoids with proteins is very significant. HbA has been found to play an important role in the distribution and bioavailability of flavonoids [5,6]. Besides, flavonoids are emerging as potentially useful drugs for cardiac and neurodegenerative disorders which involves oxidative stress [1-6,28-30]. With this scenario in mind we have explored the interactions of the isoflavonoid genistein with HbA. We have fruitfully exploited the intrinsic protein tryptophan fluorescence for this study. Molecular modeling studies have been further employed to provide detailed insights at atomistic level regarding the flavonoid recognition by HbA. The rationale of this study is that the small flavonoid molecules can easily penetrate the erythrocyte membrane and reaches inside the cells where hemoglobin is present in high concentrations.
In summary, our present study using steady state and time resolved fluorescence and circular dichroism spectroscopic and molecular modeling approaches, shows how genistein interacts with HbA. The spectroscopic data suggest that the flavonoid bind with hemoglobin and quench its fluorescence. Molecular docking studies reveal that the position of Trp-37 (β chain) residue, the emitting fluorophore in HbA is present just ~5 Ǻ away from the binding cavity. This approach can easily be applied to other flavonoid derivatives, which would open the door to new avenues for the “screening and design” of the most suitable flavonoid derivatives from among numerous structural variants of this new generation of rapidly emerging therapeutic drugs of immense importance in modern medicine.
5. Acknowledgements
BSG is thankful for the travel award from AAAS WIRC @ MSIs/NSF which provided the travel and part of the research supply support for this international collaboration, as is presented in the present study. BSG also likes to acknowledge the financial and research supports from NIH/NCMHHD/RIMI grant # 1P20MD002725; HBCUUP Grant-NSF ID: 0811638; JHS program with the contract # N01-HC-95172; HHMI grant # 52007562 at Tougaloo College. PKB and S. Chakraborty thank the financial support from MS-INBRE (#USM-GR04015-05-9; NIH/NCRR #P20RR016476). PKS acknowledges CSIR, India for the award of an Emeritus Scientist grant (no. 21(0864) 11/ EMR-II) and Prof. Milan K. Sanyal, Director, Saha Institute of Nuclear Physics, for providing the opportunity to continue his scientific endeavors at this institute and for extending necessary facilities. We thank Prof. Abhijit Chakrabarti of Structural Genomics Division, and Prof. Samita Basu of Chemical Sciences Division of SINP for letting us use the Circular Dichroism spectrometer and TCSPC fluorescence lifetime facility respectively.
REFERENCES
- St. Rusznyák and A. Szent-Györgyi, “Vitamin P: Flavonols as Vitamins,” Nature, Vol. 138, No. 3479, 1936, p. 27.
- S. Chaudhuri, A. Banerjee, K. Basu, B. Sengupta and P. K. Sengupta, “Interaction of Flavonoids with Red Blood Cell Membrane Lipids and Proteins: Antioxidant and Antihemolytic Effects,” International Journal of Biological Macromolecule, Vol. 41, No. 1, 2007, pp. 42-48. doi:10.1016/j.ijbiomac.2006.12.003
- B. Sengupta, T. Uematsu, P. Jacobsson and J. Swenson, “Exploring the Antioxidant Property of Bioflavonoid Quercetin in Preventing DNA Glycation: A Calorimetric and Spectroscopic Study,” Biochemical and Biophysical Research Communications, Vol. 339, No. 1, 2006, pp. 355- 361. doi:10.1016/j.bbrc.2005.11.019
- B. Sengupta and J. Swenson, “Properties of Normal and Glycated Human Hemoglobin in Presence and Absence of Antioxidant,” Biochemical and Biophysical Research Communications, Vol. 334, No. 3, 2005, pp. 954-959. doi:10.1016/j.bbrc.2005.06.181
- S. Chakraborty, S. Chaudhuri, B. Pahari, J. Taylor, P. K. Sengupta and B. Sengupta, “A Critical Study on the Interactions of Hesperitin with Human Hemoglobin: Fluorescence Spectroscopic and Molecular Modeling Approach,” Journal of Luminescence, Vol. 132, No. 6, 2012, pp. 1522- 1528. doi:10.1016/j.jlumin.2012.01.021
- G. Rusak, H. O. Gutzeit and J. L. Müller, “Structurally Related Flavonoids with Antioxidative Properties Differentially Affect Cell Cycle Progression and Apoptosis of Human Acuteleukemia Cells,” Nutrition Research, Vol. 25, No. 2, 2005, pp. 143-155. doi:10.1016/j.nutres.2004.12.003
- A. Constantinou, K. Kiguchi and E. Huberman, “Induction of Differentiation and DNA Strand Breakage in Human HL-60 and K-562 Leukemia Cells by Genistein,” Cancer Research, Vol. 50, No. 9, 1990, pp. 2618-2624.
- D. A. Frank and A. C. Sartorelli, “Alterations in Tyrosine Phosphorylation during the Granulocytic Maturation of HL-60 Leukemia Cells,” Cancer Research, Vol. 48, No. 1, 1988, pp. 52-58.
- T. Akiyama, J. Ishida, S. Nakagawa, H. Ogawara, S.-I. Watanabe, N. Itoh, M. Shibuya and Y. Fukami, “Genistein, a Specific Inhibitor of Tyrosine-Specific Protein Kinases,” Journal of Biological Chemistry, Vol. 262, No. 12. 1987, pp. 5592-5595.
- K. Kiguchi, A. I. Constantinou and E. Huberman, “Genistein-Induced Cell Differentiation and Protein-Linked DNA Strand Breakage in Human Melanoma Cells,” Cancer Communications, Vol. 2, No. 8, 1990, pp. 271-277.
- A. Bolli, M. Marino, G. Rimbach, G. Fanali, M. Fasano and P. Ascenzi, “Flavonoid Binding to Human Serum Albumin,” Biochemical and Biophysical Research Communications, Vol. 398, No. 3, 2010, pp. 444-449. doi:10.1016/j.bbrc.2010.06.096
- F. K. Alanzi, G. E.-D. I. Harisa, A. Maqboul, M. A. Hamid, S. H. Neau and I. A. Alsarra, “Biochemically Altered Human Erythrocytes as a Carrier for Targeted Delivery of Primaquine: An in Vitro Study,” Archives of Pharmacal Research, Vol. 34, No. 4, 2011, pp. 563-571. doi:10.1007/s12272-011-0406-7
- M. Hamidi, H. Tajerzadeh, A. R. Dehpour, M. R. Rouini and S. Ejtemaee-Mehr, “In Vitro Characterization of Intact Human Erythrocytes Loaded by Enalaprilat,” Drug Delivery, Vol. 8, No. 4, 2001, pp. 223-230. doi:10.1080/107175401317245903
- M. L. Doyle, J. M. Holt and G. K. Ackers, “Effects of NaCl on the Linkages between O2 Binding and Subunit Assembly in Human Hemoglobin: Titration of the Quaternary Enhancement Effect,” Biophysical Chemistry, Vol. 64, No. 1-3, 1997, pp. 271-287. doi:10.1016/S0301-4622(96)02235-1
- X. Yang, J. Chou, G. Sun, H. Yang and T. Lu, “Synchronous Fluorescence Spectra of Hemoglobin: A Study of Aggregation States in Aqueous Solutions,” Microchemical Journal, Vol. 60, No. 3, 1998, pp. 210-216. doi:10.1006/mchj.1998.1648
- J. R. Lakowicz, “Principles of Fluorescence Spectroscopy,” 3rd Edition, Springer-Verlag, New York, 2006.
- W. B. Gratzer, “Medical Research Council Labs,” Holly Hill, London.
- D. Sahoo, P. Bhattacharya and S. Chakravorti, “Quest for Mode of Binding of 2-(4-(dimethylamino) Styryl)-1-Methylpyridinium Iodide with Calf Thymus DNA,” Journal of Physical Chemistry B, Vol. 114, No. 5, 2010, pp. 2044- 2050. doi:10.1021/jp910766q
- G. M. Morris, D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew and A. J. Olson, “Automated Docking Using a Lamarkian Genetic Algorithm and an Empirical Binding Free Energy Function,” Journal of Computational Chemistry, Vol. 19, No. 14, 1998, pp. 1639-1662. doi:10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
- Hyperchem, Hypercube, Inc., 2002.
- D. V. D. Spoel, B. Hess, G. Groenhof, A. E. Mark and H. J. Berendsen, “GROMACS: Fast, Flexible, and Free,” Journal of Computational Chemistry, Vol. 26, No. 16, 2005, pp. 1701-1718. doi:10.1002/jcc.20291
- E. Lindahl Erik, B. Hess and D. V. D. Spoel, “GROMACS 3.0: A Package for Molecular Simulation and Trajectory Analysis,” Journal of Molecular Modelling, Vol. 7, No. 8, 2001, pp. 306-317.
- W. L. Jorgensen and J. Tirado-Rives, “The OPLS Force Field for Proteins. Energy Minimizations for Crystals of Cyclic Peptides and Crambin,” Journal of the American Chemical Society, Vol. 110, No. 6, 1988, pp. 1657-1666. doi:10.1021/ja00214a001
- R. Car and M. Parrinello, “Unified Approach for Molecular Dynamics and Density-Functional Theory,” Physical Review Letters, Vol. 55, No. 22, 1985, pp. 2471-2474. doi:10.1103/PhysRevLett.55.2471
- P. K. Biswas and V. Gogonea, “A Regularized and Renormalized Electrostatic Coupling Hamiltonian for Hybrid Quantum-Mechanical—Molecular-Mechanical Calculations,” Journal of Chemical Physics, Vol. 123, No. 16, 2005, pp. 164114-164122. doi:10.1063/1.2064907
- V. Gogonea, J. M. Shy and P. K. Biswas, “Electronic Structure, Ionization Potential, and Electron Affinity of the Enzyme Cofactor (6R)-5,6,7,8-Tetrahydrobiopterin in the Gas Phase, Solution, and Protein Environments,” Journal of Physical Chemistry B, Vol. 110, No. 45, 2006, pp 22861- 22871. doi:10.1021/jp061653q
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, “LINCS: A Linear Constraint Solver for Molecular Simulations,” Journal of Computational Chemistry, Vol. 18, No. 12, 1997, pp. 1463-1472. doi:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H
- B. Pahari, B. Sengupta, S. Chakraborty, B. Thomas, D. McGowan and P. K. Sengupta, “Contrasting Binding of Fisetin and Daidzein in γ-Cyclodextrin Nanocavity,” Journal of Photochemistry and Photobiology B: Biology, Vol. 118, No. 1, 2013, pp. 33-41. doi:10.1016/j.jphotobiol.2012.10.010
- B. Pahari, S. Chakraborty, S. Chaudhuri, B. Sengupta and P. K. Sengupta, “Binding and Antioxidant Properties of Therapeutically Important Plant Flavonoids in Biomembranes: Insights from Spectroscopic and Quantum Chemical Studies,” Chemistry and Physics of Lipids, Vol. 165, No. 4, 2012, pp. 488-496. doi:10.1016/j.chemphyslip.2011.10.006
- B. Sengupta, B. Pahari, L. Blackmon and P. K. Sengupta, “Prospect of Bioflavonoid Fisetin as a Quadruplex DNA Ligand: A Biophysical Approach,” PloS One, Vol. 8, No. 6, 2013, pp. 1-11.
NOTES
*Contributed equally to the paper.
#Permanent address: Gandhi Centenary B. T. College, Habra, India.
†Corresponding authors.

