American Journal of Plant Sciences
Vol.3 No.2(2012), Article ID:17574,7 pages DOI:10.4236/ajps.2012.32031
Enzymes of Phenylpropanoid Metabolism Involved in Strengthening the Structural Barrier for Providing Genotype and Stage Dependent Resistance to Karnal Bunt in Wheat
![]()
1Department of Molecular Biology & Genetic Engineering, Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, India; 2Defence Institute of Bio-Energy Research, Haldwani, India.
Email: *purwar_shalini@rediffmail.com
Received May 25th, 2011; revised July 1st, 2011; accepted July 15th, 2011
Keywords: Karnal Blunt; Peroxidase; Phenylalanine Ammonia Lyase; Phenylpropanoid Metabolism; Polyphenol Oxidase; Tilletia Indica; Wheat
ABSTRACT
The role of lignifications and enzymes involved in the phenylpropanoid (PP) biosynthesis i.e. phenylalanine ammonia lyase (PAL), Peroxidase (POD), Polyphenol oxidase (PPO) in providing resistance to Karnal Bunt (KB) during different developmental stages of resistant (HD-29) and susceptible genotype (WH-542) and its recombinant inbred lines (RILs) of wheat were investigated. The enzymes of PP pathway were expressed constitutively in both the susceptible and resistant genotype. However, the activity was higher in all the developmental stages of resistant genotype and its RILs, indicating that this genotype has a significant higher basal level of these enzymes as compared to the susceptible line and could be used as marker(s) to define KB resistance. The activity of PAL and POD was significantly higher in WSv stage (Z = 16) while the specific activity of PPO was higher in WS3 (Z = 77) stage as compared to the other physiological stages in both the genotypes. In resistant genotype the lignin content increased two-fold and three-fold at WS2 and WS3 stage, respectively, while in susceptible genotype no significant increase in lignin content was observed. The pathway might be associated with the enhancement of structural defense barrier due to lignifications of cell wall as evident from the enhanced synthesis of lignin in all the stages of resistant genotype. Our results clearly indicate the possible role of enzymes of PP metabolism provides genotype and stage dependant structural barrier resistance in wheat against KB.
1. Introduction
Karnal bunt (KB) caused by Tilletia indica (Syn. Neovossia indica), a semi-biotroph fungus occurs sporadically and may assume epidemic proportions in certain areas incurring qualitative and quantitative losses in wheat production. Although wheat genotype with varying levels of resistance to KB have been identified among Indian, Chinese and Brazilian varieties of wheat [1] yet plant breeders have so far not been able to develop resistance in any of the cultivars of the aestivum group. Since long it has been recognized that infection of KB pathogen occurs during heading and florets that are infected locally [2,3] by air borne and water borne sporidia. Many aspect of KB resistance is still unknown and the proper understanding to KB resistance and differential immunity at biochemical and physiological level would help to devise strategies for control of the disease. Therefore, elucidation of the biochemical basis of resistance and stage specific immunity against KB is needed to be examined for analyzing the role of enzymes of phenylpropanoid metabolism which provides structural barriers in conferring the resistance against KB in wheat.
Phenylpropanoids (PPs) belongs to the largest group of secondary metabolites (including lignin, flavonoids, phytoalexins, tannins etc.) produced by plants in response to biotic and abiotic stresses [4-6]. There are several PPsbased mechanisms of defense against pathogens, for example, construction of structural lignin containing barriers preventing the pathogen penetration into the plant tissues. Another mechanism is the use of phytoalexin and scopoletin, which could act as broad-range antibiotics. Additionally, scopoletin being an efficient peroxidase substrate may act as scavenger of reactive oxygen species and thus prevent, or reduce, oxidative damage to infected plant cells [7]. PPs exert direct antimicrobial activity and also serve in signaling and chemotaxis to both pathogenic and symbiotic microorganisms. It is thought that the molecular basis for the protective action PPs in plants is their antioxidant and free radical scavenging properties. Plant defense has three main aspects; accumulating PR protein [8], lignifying and cross linking the cell walls of tissues that are distant from the HR sites. During compatible and incompatible interaction between plants and pathogen, the shikimic acid pathway is involved in plant defense [9]. It has received considerable attention due to the synthesis of secondary metabolites such as lignin, phenolics and phytoalexin, which add mechanical rigidity and strength to the cell wall and provide barriers to infection of pathogen. In these protection processes, key enzymes of phenylpropanoid pathway such as Phenylalanine ammonia lyase (PAL), Peroxidase (POD) and Polyphenoloxidase (PPO), which are involved in the biosynthesis of lignin metabolites that provides structural barriers against fungus [10].
PAL is the entry-point enzyme in the phenylpropanoid biosynthesis pathway and its presence has been demonstrated in pathogen infected plants [11]. Studies of several different species of plants show that PAL activity increased with the biotic and abiotic stress [12]. The phenolic compounds produced in plants due to pathogen attack are often converted into more reactive species by PPO and POD [13]. Peroxidases (PODs) are associated with the active defense reactions in higher plants in response to foreign organisms [14]. They are involved in the oxidation of phenolic compounds in cell wall, suberization and lignifications of host plant cells during the defense reaction against pathogenic agents [15]. PPO catalyzing the oxygen-dependent oxidation of phenols to quinones is ubiquitous among angiosperms and is assumed to be active in plant defense against pests and pathogens [16].
The present study illuminate possible role of phenylpropanoid metabolism in impart stage specific and differential immunity as well as their role in providing structural barrier against KB pathogen has been demonstrated by comparing the basal levels of enzyme activeties like PAL, POD and PPO that are involved in tissue lignifications in developing wheat spikes and different recombinant inbred lines.
2. Material and Methods
2.1. Plant Material
In the present study two parent genotypes of bread wheat (Triticum aestivum), one highly resistant HD-29 (designnated as PR) and another highly susceptible WH-542 (designated as PS) based on their pathogenicity testing under field conditions against KB, and their respective recombinant inbred lines (RILs) were used [17]. A total of 8 resistant RILs designated as (R1 - R8) and 7 susceptible RILs designated as (S1 - S7) were used in the present investigation. The seeds of these genotypes were collected from the Crop Research Centre, G. B. Pant University of Agriculture and Technology, Pantnagar (UK) and Department of Plant Breeding, Punjab Agricultural University, Ludhiana (Punjab).
2.2. Selection of Different Stages of Developing Wheat Spike
Based on the degree of susceptibility to pathogen invasion, different stages of developing wheat spikes were selected for evaluation of enzymes involved in phenylpropanoid metabolism. Different growth stages of wheat plants were described according to Zadok’s scale and spikes of resistant and susceptible parents were selected for investigating the molecular basis of stage dependent immunity are as follows: Vegetative stem flag leaf stage (WSv, Z = 16,) as control stage, boot stage (WS1, Z = 45), ear head just peeping out at the tip or from the center (WS2, Z = 58) more prone stage and post-anthesis stage (WS3, Z = 77).
2.3. Determination of Enzymes of Phenylpropanoid Metabolism
The phenylalanine ammonia lyase (PAL), peroxides (POD) and polyphenoloxidase (PPO) enzymes activity were determined in all the selected stages of developing spike of both resistant (PR) and susceptible (PS) parents and their RILS of wheat against KB.
Phenylalanine ammonia lyase (PAL): PAL activity was measured according to the method of [18] Dickerson et al. (1984) with slight modifications. The reaction mixture contained 0.1 M Tris-HCl buffer (pH 8.5), 1 mM 2-marcaptoethanol, 15 mM L-phenylalanine, 0.1 g soluble polyvenylpolypyrrolidone (PVP) and enzyme extract. The reaction mixture was incubated at 30˚C for 15 min. The reaction was terminated by the addition of 6 N HCl and the absorbance of reaction mixture was measured at 290 nm. One enzyme unit represented the amount of enzyme that produces one nmole of cinnamic acid min–1·g·FW–1.
Peroxidase Enzyme Assay (POD): POD activity was determined by measuring the appearance of pink/brown colour resulting from guaiacol oxidation in the presence of hydrogen peroxide according to the method of [19] Zieslin and Ben-Zaken (1993). The reaction mixture containing 50 µl of 0.02 M Guaiacol, 0.5 ml of 0.38 M H2O2 and 2.0 ml of 0.2 M sodium phosphate buffer (pH 5.8). 50 µg of protein extract were added in 3.0 ml disposable cuvette and the absorbance was measured after every 1 min for 5 min at 470 nm. Enzyme unit can be defined as an increase in absorbance by 1.0 at 470 nm·min–1 at room 25˚C and specific activity of POD was expressed as unit mg·protein–1.
Polyphenoloxidase enzyme (PPO): PPO activity was determined according to the method of [20] Chunhua et al. (2001). The reaction mixture (1 ml) containing 50µg of protein extract and 10 mM phosphate buffer pH 7.0. Each sample was aerated for 2 min in a small test tube followed by the addition of catechol as the substrate at a final concentration of 20 nM. PPO activity was expressed as the change in one unit of absorbance at 420 nm·min–1·g·FW–1 of sample.
2.4. Estimation of Lignin
The lignin quantification was done by derivatization with thioglycolic acid. Thioglycolic acid displaces lignin from its normal covalent attachment to the cell wall and enables it to be extractable from the cell wall by alkali. Acidification of alkaline extract causes precipitation of lignin thioglycolic acid (LTGA). Leaf and different stages of wheat spikes were homogenized in methanol and centrifuged at 10,000 rpm for 5 minutes. The resulting alcohol insoluble residue (AIR) was dried (24 h at 60˚C) and used for lignin determination as described [21] Barber and Ride (1988). The absorbance of the solution containing 10 mg/ml of LTGA prepared in 0.5 N NaOH was measured at 280 nm.
Protein estimation was done according to the method of [22] Bradford (1976). Each biochemical analysis was performed in triplicate. The maximum difference among the three values was less than 5% of the mean.
3. Results and Discussion
The functions of phenylpropanoid (PP) compounds in plant defense range from preformed or inducible physical and chemical barriers against infection to signal molecules involved in local and systemic signaling for defense gene induction. In present manuscripts we report the possible role of enzymes of PP metabolism in imparting stage specific immunity and providing structural barrier against KB pathogen. In order to ascertain the role of the basal expression levels of different enzymes involved in phenylpropanoid metabolism like phenyl ammonium lyase (PAL), peroxidase (POD) and polyphenyl oxidase (PPO) were analyzed using enzymatic assay at different physiological stages of wheat spikes of resistant and susceptible genotypes and their recombinant inbred lines (RILs).
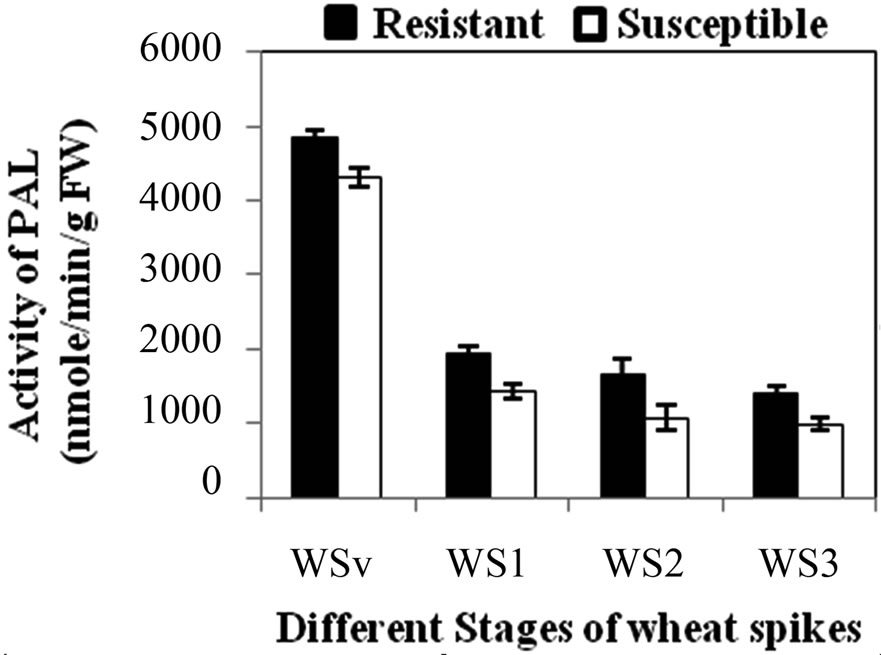
PAL activity was significantly higher in all the developmental stages of resistant genotype (hd-29) than the susceptible genotype (Wh-542). PAL activity showed decreasing trend from WSv stages to WS3 stages, in resistant and susceptible genotype it decreases from 4851 to 1397 U and 4306 to 991 U, respectively (Figure 1(a)). When comparing PAL activity in WS2 stage of resistant RILs, R7 showed lowest activity and R1, R3 and R5 showed similar activity while others resistant RILs (R2, R4, R6 and R8) showed moderate PAL activity as compared to parent resistant genotype (PR) (Figure 1(b)). In case of susceptible RILs, S5 showed lowest and S1 and S4 showed similar activity while others susceptible RILs (S2, S3, S6 and S7) showed moderate PAL activity as compared to parent susceptible genotype (PS) (Figure 1(c)). PAL activity is high in resistance genotype and its RILs mainly in R5, associated with enhancement of lignifications and also in acceleration of activities of enzymes involved in PP pathway. In general, KB pathogen shows susceptibility at WS2 stage [3] due to lack of physical barrier and accumulation of compounds such as lignin and phenol compounds that are toxic to infection that have been implicated in the resistance responses [23]. Recent studies have shown that initial stages of resistant and later stages of susceptible wheat spikes strengthen the cell wall structure resulting in restriction of pathogen invasion in plant tissues [24]. Hence, the WS2 stage of wheat spike of susceptible genotype is more prone to KB infection as compared to resistant genotype. During earlier stages, higher level of expression of defense enzymes and accumulation of chemical at vegetative leaf stages prevented fungal mycelial colonization in leaves. Similar findings were reported in mature tobacco plants where differential expression of PAL was observed in the four major organs (leaves, stems, roots and flowers) [25]. Activity of PAL and POD enzymes showed enhanced in pepper roots during interactions with Arbuscular mycorrhizal fungi and/or Verticillium dahlia [26].
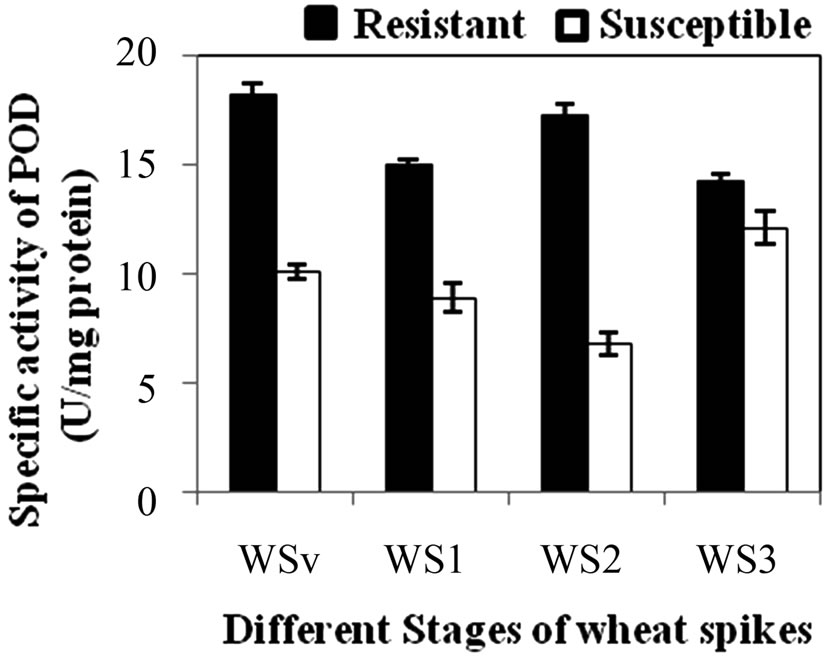
POD specific activity was significantly dominant in all developmental stages of the resistant genotype than susceptible genotype. The specific activity of POD was significantly higher (p ≤ 0.05) in leaves (WSv) and WS2 stage than the WS1 and WS3 stages. The specific activity of POD was found maximum in resistant and susceptible genotype at WSv (18.3 U·mg·protein–1) and WS2 (12.1 U·mg·protein–1) stage, respectively (Figure 2(a)). The developmental stages results indicated the enhanced expression of POD at the WS2 stage, which reinforces its role in stage dependent immunity against Karnal bunt. At this stage (S2), POD specific activity was also checked in RILs of both resistant and susceptible genotype. Among resistant RILs, R7 showed lowest activity and R3 and R5 showed similar activity while others resistant RILs (R1, R2, R4, R6 and R8) showed moderate POD activity as
 (a)
(a)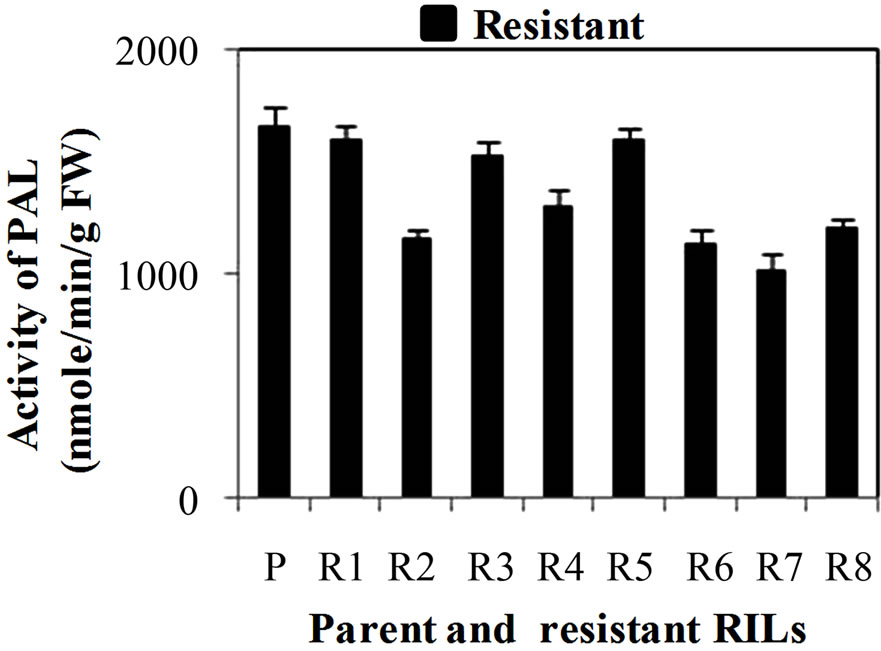 (b)
(b)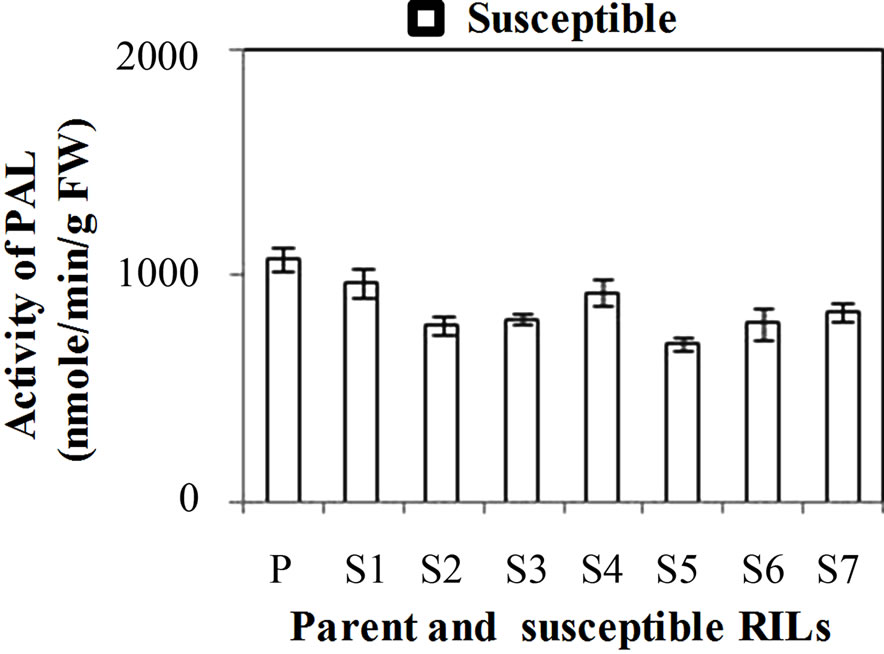 (c)
(c)
Figure 1. (a) Changes in activity of Phenylalanine ammonia lyase (PAL) (nmole/min/g FW) at different stages of developing wheat spikes of Resistant (HD-29) and Susceptible (WH-542) genotypes. Vegetative stem flag leaf stage (WSv, Z = 16,) as control stage, boot stage (WS1, Z = 45), ear head just peeping out at the tip or from the center (WS2, Z = 58) more prone stage and post-anthesis stage (WS3, Z = 77); (b) Activity of PAL at WS2 stage in parent and resistant recombinant inbred lines (RILs); (c) Activity of PAL at WS2 stage in parent and susceptible RILs. (Error bars are standard deviation).
 (a)
(a)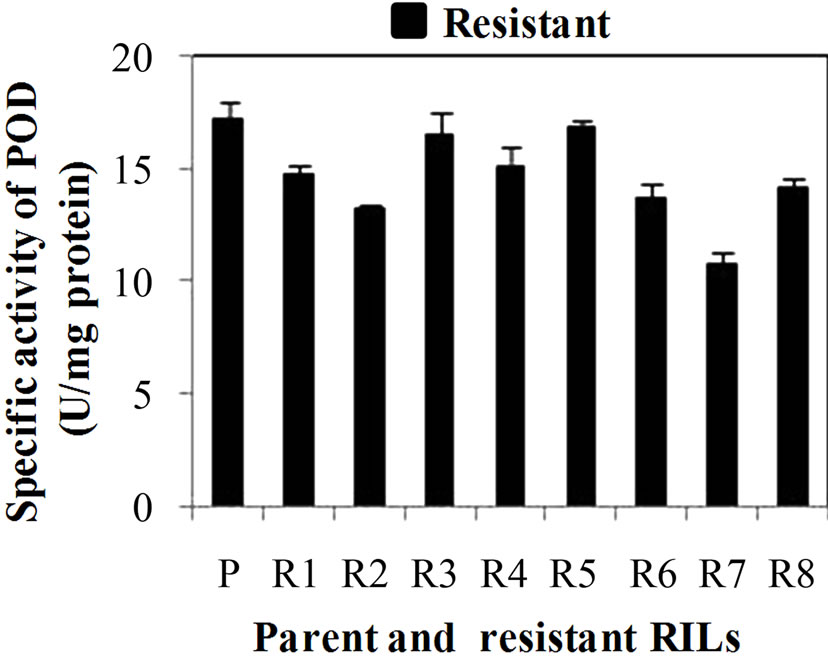 (b)
(b)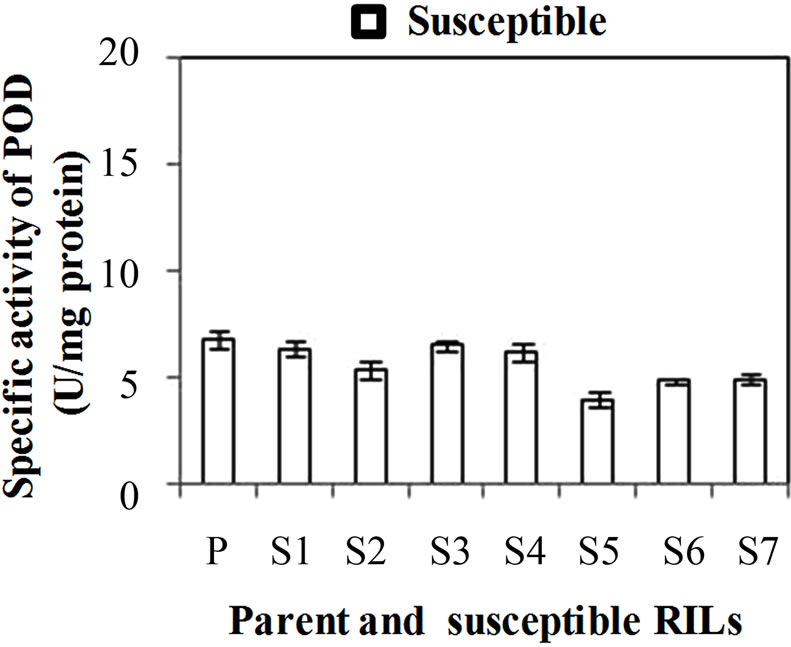 (c)
(c)
Figure 2. (a) Changes in specific activity of Peroxidase (POD) (U/mg protein) at different stages of developing wheat spikes of Resistant (HD-29) and Susceptible (WH-542) genotypes. Vegetative stem flag leaf stage (WSv, Z = 16,) as control stage, boot stage (WS1, Z = 45), ear head just peeping out at the tip or from the center (WS2, Z = 58) more prone stage and post-anthesis stage (WS3, Z = 77); (b) Specific activity of POD at WS2 stage in parent and resistant recombinant inbred lines (RILs); (c) Specific activity of POD at WS2 stage in parent and susceptible RILs. (Error bars are standard deviation).
compared to parent resistant genotype (PR) (Figure 2(b)). In case of susceptible RILs, maximum specific activity of POD was observed in S1 and lowest in S5 in comparison to other susceptible RILs. S3 and S4 showed similar activity while others susceptible RILs (S2, S6 and S7) showed moderate POD activity as compared to parent susceptible genotype (PS) (Figure 1(c)). The basal activity of POD was high in resistant genotype and its RILs especially in R5 than susceptible genotype and its RILs. The oxidative enzymes such as POD and PPO, which catalyze the formation of lignin and other oxidative phenols, contribute to the formation of defense barriers for reinforcing the cell structure [15]. These enzymes have been correlated with defense against pathogens in several plants, including tomato [27] and wheat [10]. POD catalyses conversion of cinnamyl alcohol in to lignin by oxidative polymerization indicating enhanced lignifications of cell walls in developing spikes of wheat. POD activity is frequently increases in plants infected by pathogens and the level of its activity is clearly correlated with the resistance phenomena such as lignin production [19].
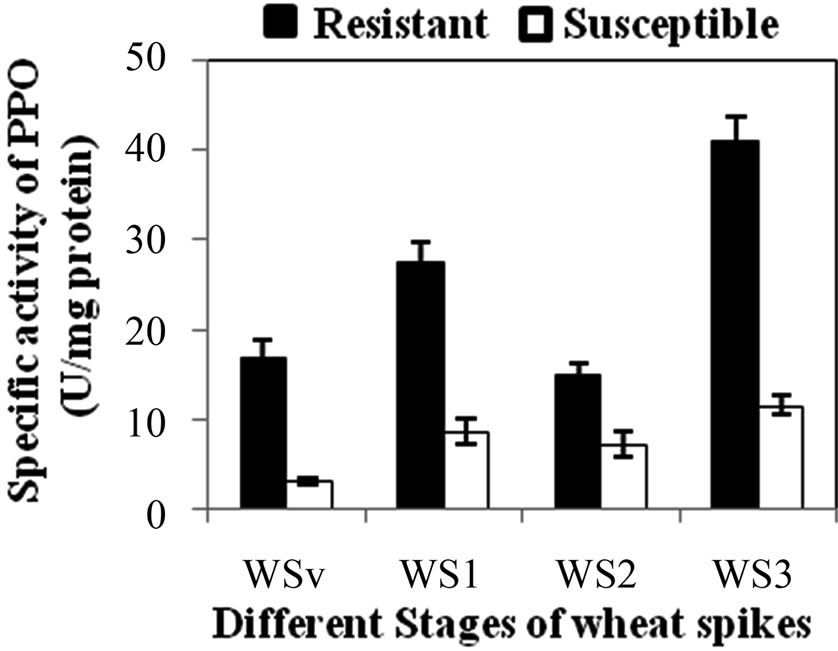
PPO specific activity was significantly higher in all the developmental stages of resistant genotype than the susceptible genotype. Specific activity of PPO was observed maximum in WS3 stage of resistant and susceptible genotype as 41 and 11.5 U·mg·protein–1, respectively (Figure 3(a)). PPO specific activity was also checked in WS2 stages of both resistant and susceptible RILs. Among resistant RILs, R7 showed lowest activity and R1and R5 showed similar activity while others resistant RILs (R2-R4, R6 and R8) showed moderate PPO activity as compared to parent resistant genotype (PR) (Figure 3(b)). In case of susceptible RILs, maximum and minimum PPO activity was observed in S1 and S3 stage, respecttively, in comparison to other susceptible RILs. On other hand, S3 and S4 showed similar activity while others susceptible RILs (S2, S6 and S7) showed moderate PPO activity as compared to parent susceptible genotype (PS) (Figure 3(c)). PPO activity was higher in resistant genotype and its RILs showed that enhanced PPO during maturation of the flower and was not induced in mature leaves even under stress conditions. PPO catalyzing the oxygen-dependent oxidation of phenol to anions is ubiquitous in the plant system and is assumed to be involved in plant defense against pest and pathogens. Li L and Steffens J. C. [16] reported that the over-expression of PPO in transgenic tomato results in enhanced disease resistance.
In order to study the correlation between activation of enzymes and lignin production at different developmental stages of resistant and susceptible genotype lignin estimation was done. Lignin quantity was found much higher in all the developmental stages of resistant genotype
 (a)
(a)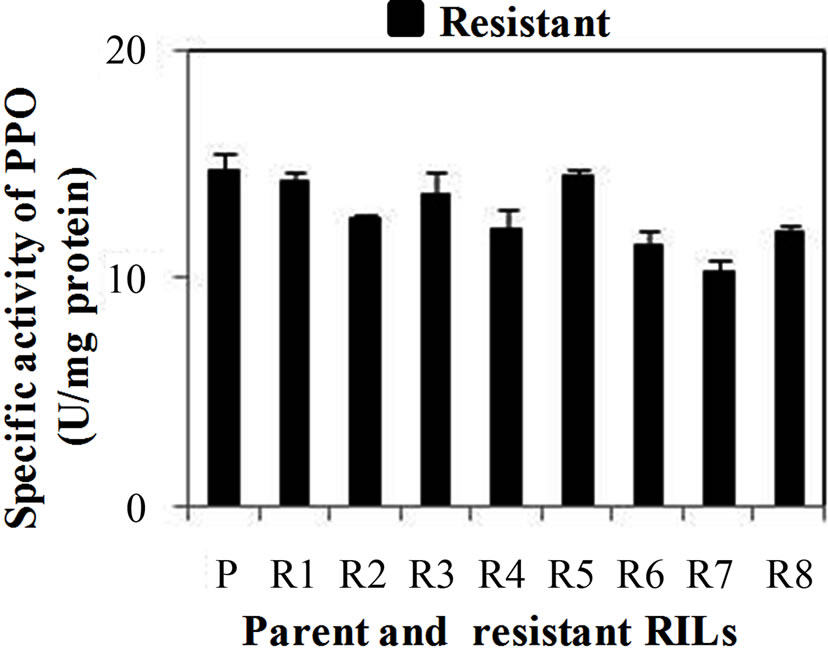 (b)
(b)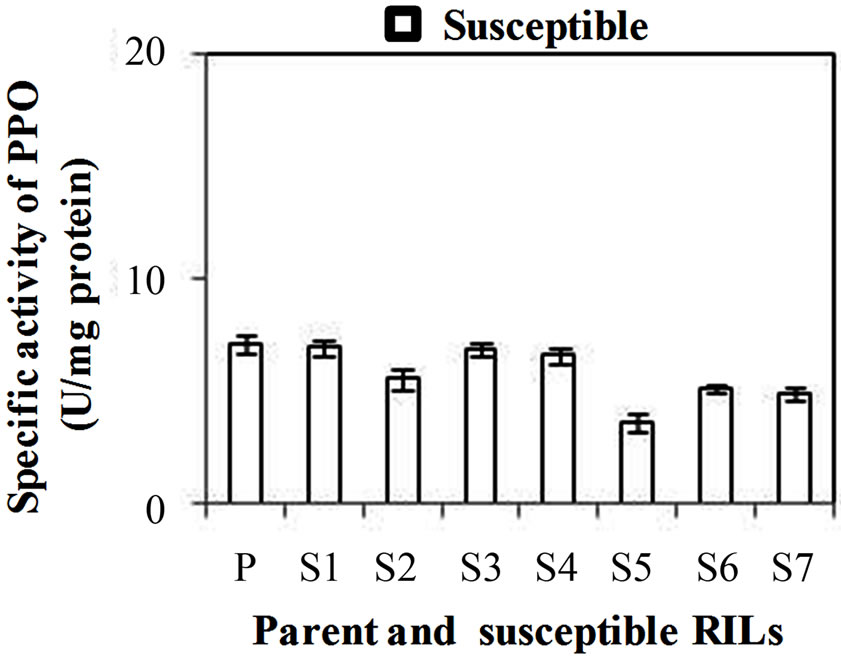 (c)
(c)
Figure 3. (a) Changes in specific activity of Polyphenol oxidase (PPO) (U/mg protein) at different stages of developing wheat spikes of Resistant (HD-29) and Susceptible (WH-542) genotypes. Vegetative stem flag leaf stage (WSv, Z = 16,) as control stage, boot stage (WS1, Z = 45), ear head just peeping out at the tip or from the center (WS2, Z= 58) more prone stage and post-anthesis stage (WS3, Z = 77); (b) Specific activity of PPO at WS2 stage in parent and resistant recombinant inbred lines (RILs); (c) Specific activity of PPO at WS2 stage in parent and susceptible RILs. (Error bars are standard deviation).
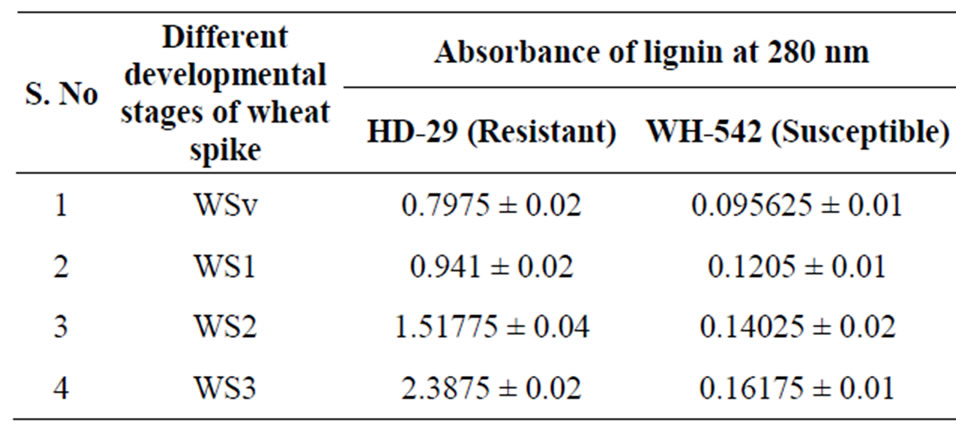
Table 1. Lignin quantification at different developmental stages of resistant genotype (HD-29) and susceptible genotype (WH-542) of wheat.
then the susceptible genotype. In developing wheat spike of resistant genotype the quantity of lignin increased two-fold and three-fold at WS2 and WS3 stage, respectively, while in case of susceptible genotype slight increase in lignin content was observed, that was not signifycant (Table 1). Lignin is one of the most abundant biopolymers, which provides resistance to plants against pathogens and makes the cell wall more resistant to pathogen attack. The deposition of lignin has been implicated as a defense response in wheat genotypes resistant to several diseases [21]. Nonetheless, increased synthesis of lignin, as well as structural modifications of lignin itself, represent another important defence mechanism, in order to protect plasmalemma from the ROS injury, thus preventing membrane damages due to lipid peroxidation [28].
In conclusion, analysis of the above data suggests that high tolerance of resistant genotype is a result of involvement of multi component signaling leading to genotype and stage dependent immunity against KB. Our results gives evidences in favor of the role of phenylpropanoid (PP) pathways is important in defining the KB resistance as WS2 stage and the period just before the onset of anthesis is most prone to get infected and higher expression of PP pathways enzymes during this period definitely bears influence on disease severity [4]. Although, plant defense responses are invariably complex multicomponent process in nature, it is not easy to define which components are both necessary and sufficient to confer disease resistance. Both the isolation of mutants selectively impaired in the production of specific phenylpropanoid end products and the production of phenocopies of such mutants by reverse genetics offer useful approaches to address the functionality of stress-induced phenylpropanoids. Therefore, the roles of PP pathways in enticing plant defense response can’t be ruled out and probably play an important role in arresting the mycelial proliferation in host tissues after initial disease development. Such results may provide an insight and a possible tool to define and cure not only KB, but also other floret infecting diseases.
4. Acknowledgements
The authors are grateful to Department of Science and Technology New Delhi, for providing research fund to carrying out the present work.
REFERENCES
- G. Fuentes-Davila and S. Rajaram, “Sources of Resistance to Tilletia indica in Wheat,” Crop Protection, Vol. 13, No. 1, 1994, pp. 20-24. doi:10.1016/0261-2194(94)90131-7
- H. S. Dhaliwal, A. S. Randhawa, K. Chand and D. V. Singh, “Primary Infection and Further Development of Karnal Bunt of Wheat,” Indian Journal of Agricultural Sciences, Vol. 53, No. 4, 1983, pp. 239-244.
- M. Rana, C. Arora, B. Ram and A. Kumar, “Floret Specificity of Karnal Bunt Infection Due to Presence of Fungal Growth Promotory Activity in Wheat Spikes,” Journal of Plant Biology, Vol. 28, No. 3, 2001, pp. 283-290.
- R. A. Dixon, L. Achnine, P. Kota, C. J. Liu, M. S. S. Reddy and L. Wang, “The Phenylpropanoid Pathway and Plant Defence: A Genomics Perspective,” Molecular Plant Pathology, Vol. 3, No. 5, 2002, pp. 371-390. doi:10.1046/j.1364-3703.2002.00131.x
- L. G. Korkina, “Phenylpropanoids as Naturally Occurring Antioxidants: From Plant Defense to Human Health,” Cell and Molecular Biology, Vol. 53, 2007, pp. 15-25.
- T. Vogt, “Phenylpropanoid Biosynthesis,” Molecular Plant, Vol. 3, No. 1, 2010, pp. 2-20. doi:10.1093/mp/ssp106
- P. Bednarek, B. Schneider, A. Svatos, N. J. Oldham and K. Hahlbrock, “Structural Complexity, Differential Response to Infection, and Tissue Specificity of Indolic and Phenylpropanoid Secondary Mechanism in Arabidopsis roots,” Plant Physiology, Vol. 138, 2005, pp. 1058-1070. doi:10.1104/pp.104.057794
- N. P. Shetty, J. D. Jensen, A. Knudsen, C. Finnie, N. Geshi, A. Blennow, D. B. Collinge and H. J. L. Jørgensen, “Effects of β-1,3-Glucan from Septoria tritici on Structural Defence Responses in Wheat,” Journal of Experimental Botany, Vol. 60, No. 15, 2009, pp. 4287-4300. doi:10.1093/jxb/erp269
- I. E. Somssich and K. Hahlbrock, “Pathogen Defence in Plants—A Paradigm of Biological Complexity,” Trends in Plant Science, Vol. 3, 1998, pp. 86-90. doi:10.1016/S1360-1385(98)01199-6
- M. Mohammadi and H. Kazemi, “Changes in Peroxidase and Polyphenol Activity in Susceptible and Resistant Wheat Heads Inoculated with Fusarium graminearum and Induced Resistance,” Plant Science, Vol. 162, 2002, pp. 491-498. doi:10.1016/S0168-9452(01)00538-6
- K. M. Herrmann, “The Shikimate Pathway: Early Steps in the Biosynthesis of Aromatic Compounds,” Plant Cell, Vol. 7, 1995, pp. 907-919.
- B. Munch-Mani and A. J. Slusarenko, “Production of Salicylic Acid Precursors Is a Major Function of Phenylalanine Ammonia Lyase in Resistance of Arabidopsis to Peronospora parasitica,” Plant Cell, Vol. 8, 1996, pp. 203-212.
- R. Gomez-Vasquez, R. Day, H. Buschmann, S. Randles, J. R. Beeching and R. M. Cooper, “Phenylpropanoids, Phenylalanine Ammonia Lyase and Peroxidases in Elicitor Challenged Cassava (Manihot esculenta) Suspension Cells and Leaves,” Annals of Botany, Vol. 94, 2004, pp. 87-97. doi:10.1093/aob/mch107
- K. Kerby and S. Somerville, “Enhancement of Specific Intercellular Peroxidases Following Inoculation of Barley with Erysiphe graminis sp. Hordei,” Physiological and Molecular Plant Pathology, Vol. 35, 1989, pp. 323-337. doi:10.1016/0885-5765(89)90037-4
- K. G. Welinder, “Plant Peroxidases: Structure, Function Relationships,” In: C. Penel, T. Gaspar and H. Greppin, Eds., Plant Peroxidases 1980-1990, Topics and Detailed Literature on Molecular, Biochemical and Physiological Aspects, Université de Genève, Geneva, 1992, pp. 1-24.
- L. Li and J. C. Steffens, “Over-Expression of Polyphenol Oxidase in Transgenic Tomato Plants Results in Enhanced Bacterial Disease Resistance,” Planta, Vol. 215, No. 2, 2002, pp. 239-247. doi:10.1007/s00425-002-0750-4
- V. C. Gupta, J. P. Jaiswal, I. Sharma and A. Kumar, “Investigating the Role of Cystatinin Conferring Stage Dependent Resistance against Karnal Bunt of Wheat,” Food and Agricultural Immunology, Vol. 21, No. 1, 2010, pp. 65-79. doi:10.1080/09540100903427314
- D. P. Dickerson, S. F. Pascholati, A. E. Hagerman, L. G. Butler and R. L. Nicholson, “Phenylalanine AmmoniaLyase and Hydroxycinnamate: CoA Ligase in Maize Mesocotyls Inoculated with Helminthosporium maydis or Helminthosporium carbonum,” Physiological and Molecular Plant Pathology, Vol. 25, 1984, pp. 111-123. doi:10.1016/0048-4059(84)90050-X
- N. Zieslin and R. Ben-Zaken, “Peroxidase Activity and Presence of Phenolic Substances in Penduncles of Rose Flowers,” Plant Physiology and Biochemistry, Vol. 31, 1993, pp. 333-339.
- C. H. Shi, Y. Dai, X. Bingle, X. L. Xu, Y. S. Xie and Q. L. Liu, “The Purification and Spectral Properties of Polyphenol Oxidase I from Nicotiana tabacum,” Plant Molecular Biology Reporter, Vol. 19, 2001, pp. 381-385. doi:10.1007/BF02772840
- Barbar and Rider, “A Quantitative Assay of Induced Lignification in Wounded Leaves and Its Use to Survey Potential Elicitors of the Response,” Physiological and Molecular Plant Pathology, Vol. 32, No. 1, 1988, pp. 85- 197.
- M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding,” Analytical Biochemistry, Vol. 7, No. 72, 1976, pp. 248-254. doi:10.1016/0003-2697(76)90527-3
- B. G. Flavia and C. M. Jair, “Custodio DS and Goussaini MM (2005) Resistance Induction in Wheat Plants by Silicon and Aphid,” Scientia Agricola, Vol. 62, No. 6, 2005, pp. 547-551.
- C. Chen, R. R. Belanger, N. Benhamou and T. C. Pullitz, “Defense Enzymes Induced in Cucumber Roots by Treatment with Plant-Growth Promoting Rhizobacteria (PGPR),” Physiological and Molecular Plant Pathology, Vol. 56, 2000, pp. 13-23. doi:10.1006/pmpp.1999.0243
- T. Fukasawa-Akada, S. D. Kung and J. C. Watson, “Phenylalanine Ammonia Lyase Gene Structure, Expression and Evolution in Nicotiana,” Plant Molecular Biology, Vol. 30, 1996, pp. 711-722. doi:10.1007/BF00019006
- G. Idoia, A. Jone and G. Nieves, “Defence-Related Enzymes in Pepper Roots during Interactions with Arbuscular mycorrhizal Fungi and/or Verticillium dahliae,” Biological Control, Vol. 51, No. 3, 2006, pp. 293-310.
- Borden and Higgins, “Hydrogen Peroxidase Play a Critical Role in the Defense Response of Tomato to Clodosporicem Fulvum,” Plant Physiology and Biochemistry, Vol. 61, 2002, pp. 227-236.
- M. Iriti and F. Faoro, “Plant Defense and Human Nutrition: The Phenylpropanoids on the Menu,” Curr. Top. Nutrac. Res., Vol. 2, 2004, pp. 47-95.
Abbreviations
AIR: Alcoholic insoluble residue;
KB: Karnal blunt;
LTGA: Lignin thioglycolic acid;
POD: Peroxidase;
PAL: Phenylalanine ammonia lyase;
PPs: Phenylpropanoids;
PPO: Polyphenol oxidase;
PVP: Poly venyl polypyrrolidone;
RIL: Recombinant inbred lines.
NOTES
*Corresponding author.

