American Journal of Molecular Biology
Vol.2 No.3(2012), Article ID:20973,10 pages DOI:10.4236/ajmb.2012.23029
Identification of ebs1, lsm6 and nup159 as suppressors of spt10 effects at ADH2 in Saccharomyces cerevisiae suggests post-transcriptional defects affect mRNA synthesis*
![]()
Department of Molecular, Cellular, and Biomedical Sciences, University of New Hampshire, Durham, NH, USA
Email: #cldenis@unh.edu
Received 11 January 2012; revised 27 April 2012; accepted 14 May 2012
Keywords: CCR4-NOT Complex; Transcription; mRNA Degradation; mRNA Export
ABSTRACT
Suppression of the effects of an spt10 mutation on ADH2 expression is a phenotype shared by a small number of genes whose protein products are either components of the CCR4-NOT complex required for mRNA deadenylation and degradation (CCR4, CAF1, NOT4) or have been shown to interact with the complex (DBF2, SRB9, SRB10). In this work, we conducted a screen for additional suppressors of spt10 at ADH2 to identify new factors related to CCR4 function. In addition to reisolating ccr4 and caf1 alleles, three previously unidentified suppressors of spt10 were obtained: ebs1, lsm6, and nup159. These three genes are known or presumed to affect mRNA export or degradation. Mutations in EBS1, LSM6 and NUP- 159 not only suppressed spt10-induced ADH2 expression but also, like ccr4 and caf1 defects, reduced the ability of ADH2 to derepress. None of these defects affected the expression of CCR4-NOT complex components or the formation of the CCR4-NOT complex. The reduced ADH2 expression was also not the result of increased degradation of ADH2 mRNA, as the lsm6 and nup159 alleles, like that of a ccr4 deletion, actually slowed ADH2 degradation. Our results indicate that alterations in factors that slow mRNA degradation or affect mRNA transport may also interfere with the synthesis of mRNA and suggest an integration of such events in gene expression.
1. INTRODUCTION
The CCR4 protein is involved in several processes controlling the metabolism of mRNA in the cell: mRNA initiation, elongation, and degradation. CCR4 was originally identified as a mutation that suppressed the increased ADH2 expression on glucose caused by a spt10 allele, cre1-1 [1]. SPT10 is known to function in part by controlling histone gene expression, which in turn can alter chromatin structure and gene expression at sensitive loci [2]. The interaction of CCR4 with SPT10 is not limited to the ADH2 promoter, as ccr4 also suppressed spt10 at the his4-912d locus [3]. CCR4 is also required for maximal expression of ADH2 and other nonfermentative genes [1] and appears involved in other processes such as cell wall integrity, amino acid metabolism, cell cycle progression, and DNA repair [4-8]. The effects of CCR4 at ADH2 and presumably many of these other processes occur at the transcriptional level implicating CCR4 in the control of mRNA initiation.
CCR4 functions in the context of two CCR4-NOT complexes, 1.0 and 1.9 MDa in size. The 1.0 MDa complex contains CCR4, CAF1, the five NOT proteins (NOT1- NOT5) and three other proteins, CAF40, CAF130 and BTT1 [6,9,10]. In addition to CCR4, mutations in both CAF1 and NOT4 suppress the effects of an spt10 mutation at ADH2 and affect the derepression of ADH2, suggesting that they also are involved in the activation of transcription [6,11].
The NOT1-5 proteins, however, are clearly involved in the repression of transcription [12,13], initially identified by their binding and restricting TFIID access to noncanonical TATAA sequences [14]. In addition, the CCR4- NOT proteins have been suggested to play a role in mRNA elongation [15]. Recent studies indicate that the CCR4-NOT complex is responsible for restarting transcription after transcriptional arrest has occurred [16].
One defined biochemical function for CCR4 is that it is the catalytic component of the cytoplasmic deadenylase complex [18-20]. CCR4 mRNA poly (A) deadenylation is the first step in targeting the mRNA for degradation and caf1 or not4 defects also lead to defects in deadenylation [19,20]. A ccr4 defect, therefore, in stabilizing mRNA would augment mRNA levels. This effect would be opposite in nature to the ccr4 suppression of spt10 effects at ADH2 or the requirement of CCR4 for ADH2 and other genes’ syntheses. The CCR4 effect on ADH2 RNA accumulation implies that it operates by mechanisms other than or in addition to controlling mRNA degradation.
To gain further insight into the mechanisms of how CCR4 activates gene expression, we have reconducted the screen for factors that can suppress spt10-enhanced expression at ADH2. The only known suppressors of spt10 effects are defects in the CCR4, CAF1 and NOT4 proteins, all components of the CCR4-NOT complex, and the DBF2, SRB9 and SRB10 proteins, which are known to associate with the CCR4-NOT complexes [5, 21]. The SRB9 and SRB10 proteins have been shown to be part of mediator, the transcriptional activator complex [21,22]. Hence, the ability to suppress spt10-enchanced ADH2 expression reflects a common factor related to CCR4 function, and possibly to a role not directly involving effects on mRNA degradation. Utilizing this screen for suppressors of spt10, we have identified three new genes which when mutated are capable of suppressing spt10: NUP159, LSM6 and EBS1. These three factors, like CCR4, CAF1 and NOT4 are known or presumed to be involved in controlling mRNA export or degradation. These results suggest that defects in factors involved in post-transcriptional regulation of mRNA metabolism may result in corresponding decreases in transcriptional initiation. The CCR4 requirement at ADH2 may, therefore, result from indirect effects involving the integration of mRNA synthesis with the subsequent nuclear export and degradation of mRNA.
2. MATERIALS AND METHODS
2.1. Yeast Strains and Growth Medium
Yeast strains are listed in Table 1. Strains were grown on YEP medium (1% yeast extract/2% Bacto-peptone) supplemented with 2% glucose and 2% agar (YD plates). Allyl alcohol was added to YD plates in the concentration indicated in the test. ADH assays were conducted as previously described [6].
2.2. Analysis of spt10 Suppressors
Standard genetic techniques were used to identify complementation groups. Yeast transformations were conducted with either an YCp50-LEU2, YEpl3, or YEp24 based genomic library. Western analysis was conducted as described [6] using antibody specific to CCR4-NOT protein components [23].
2.3. RNA Analysis
S1 nuclease protection assays were conducted to quantitate ADH2 mRNA levels as described [17,24]. The rates of ADH2 mRNA half-lives were determined following growth of yeast on YEP medium containing 2% glycerol/ 2% ethanol and shifting to YEP medium containing 8% glucose as previously indicated [17].

Table 1. Yeast strains.
3. RESULTS
3.1. A Screen for Suppressors of spt10 at ADH2
In order to identify other factors that are functionally related to the CCR4-NOT proteins, we conducted a search for additional suppressors of the spt10 effects at the ADH2 locus using a strain carrying the defective adh1-1 allele. The increased expression of ADH2 caused by spt10 results in an inability of strains to grow in the presence of ally1 alcohol. Ally1 alcohol can be converted by ADH enzymes to acrolein that kills the cells. While ccr4 alleles were previously identified by such a screen [1], in order to identify new suppressors we used a modified selection process. In the previous screen, mutations that allowed growth on ally1 alcohol-containing plates were then tested for sensitivity to the respiratory inhibitor Antimycin A; complete loss of both ADH I and ADH II enzyme activity in the cell would require the utilizetion of oxidative phosphorylation rendering such cells Antimycin A sensitive. This secondary requirement resulted in the elimination of nearly 90% of the ally1 alcohol revertants [1]. Since there is a narrow range of ADH enzyme activities that allows growth on both ally1 alcohol and resistance to antimycin A we were intent on identifying new suppressors of spt10 that resulted in ADH II activity that fell into this range.
Strain 553-4b, containing a spt10 mutant allele called cre1-1, was chosen for this screen since the cre1-1 allele, unlike an spt10 deletion, does not result in phenotypes typically associated with ccr4 or not defects: these phenotypes include temperature sensitivity, slow growth, and sensitivity to caffeine. Approximately 1 × 108 cells of strain 553-4b were spread onto YD plates containing 10 mM ally1 alcohol. 198 ally1 alcohol resistant colonies were selected for analysis. Complementation analysis, following mating each of these revertants to strain 43- 2B-M1 carrying a defective adh2 allele, identified 99 of these revertants as containing mutations in the ADH2 gene (data not shown).
Since ccr4 and defects in other CCR4-NOT complex components are sensitive to caffeine and a number of other factors such as high temperature and elevated salt concentration, the remaining 99 revertants were tested for sensitivity to these conditions. Sixteen of the 99 showed sensitivity to one or more of the following conditions: 8 mM caffeine, 37˚C, 16˚C, 0.75 M MgCl2, or 0.2 M LiC1 (Table 2; data not shown) and were chosen for further analysis.
3.2. Identification of Known Suppressors of spt10
To test if any of these 16 revertants generated in this screen were ccr4 mutations, Western analysis was conducted on each strain to determine if the CCR4 protein was absent. Extracts from only one revertant, 553-4b-1- 65, lacked detectable CCR4 protein (data not shown; see also Figure 1). In addition, genetic complementation analysis was conducted by crossing a ccr4-containing strain to all the revertants. In this case, only revertant 553-4b-1-65 failed to complement a ccr4 allele. Similarly, a caf1 strain, 1005-2-3c-s, was crossed to all revertants to identify those that contained caf1 mutations. Re
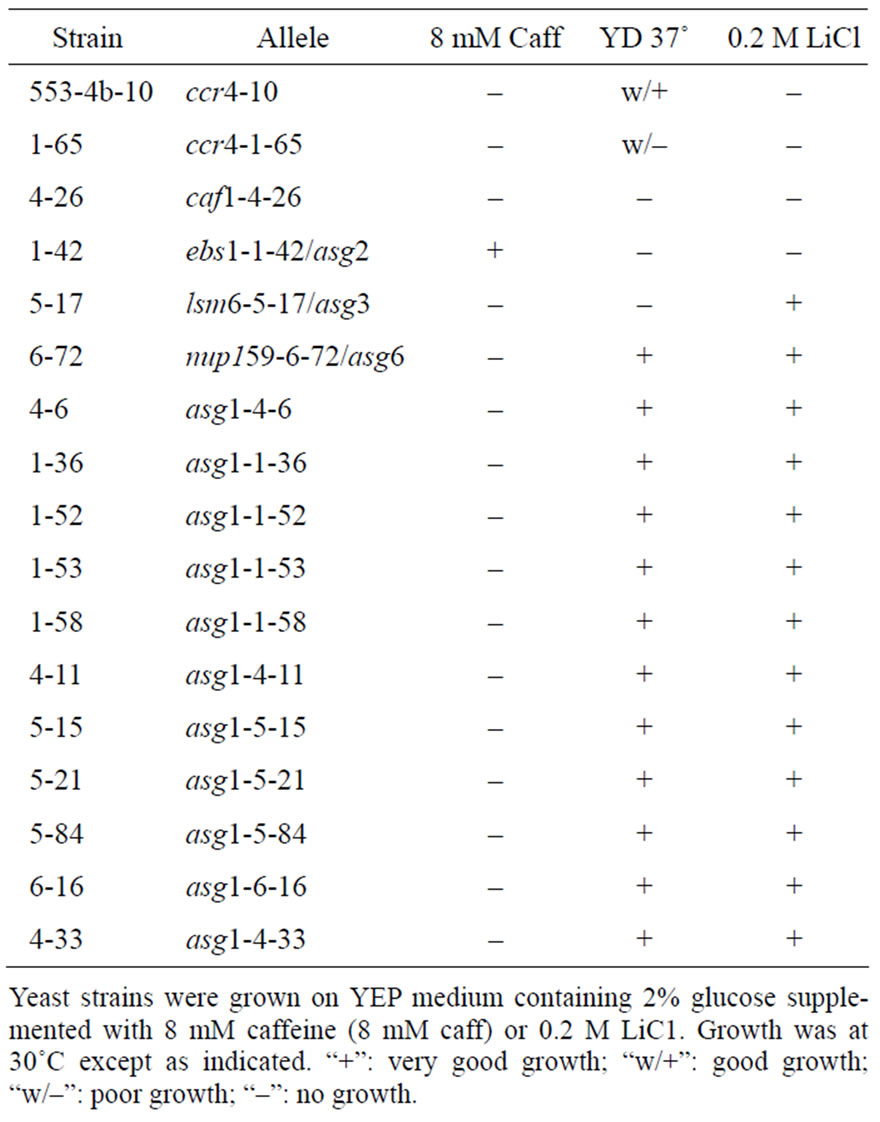
Table 2. Phenotypic characterization of spt10 revertants.
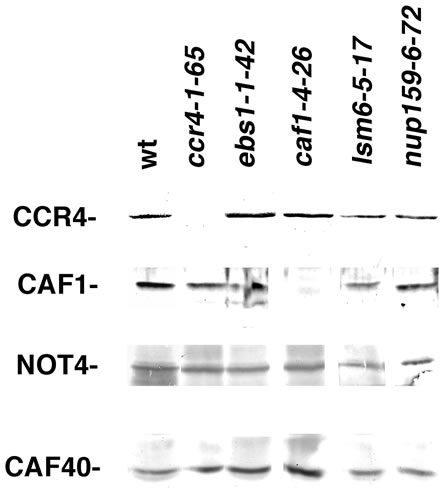
Figure 1. nup159, lsm6 and ebs1 alleles do not affect the expression of CCR4-NOT components. Western analysis was conducted on yeast extracts with antibodies directed against CCR4, CAF1, NOT4, and CAF40. Left lane- (wt) strain 553-4b; other lanes-strain 553-4b except containing the allele as indicated.
vertants 553-4b-4-26 and -6-72 partially failed to complement the caf1 mutation, although not completely, so that we were unable to conclusively identify these revertants as containing caf1 mutations. To clarify this result, the mutation in revertant 553-4b-4-26 was cloned by complementation using a genomic library and the complementing gene was shown to contain the complete CAF1 gene following DNA sequence analysis. Subsequent retransformation of the CAF1 containing genomic clone into revertant 553-4b-6-72 failed to complement its defects, demonstrating that 553-4b-4-26 is the only caf1 mutation produced by this screen. Crosses with strains containing either a dbf2 or not4 mutation showed that none of the remaining revertants carried mutations in either of these genes.
Additional complementation analysis grouped the remaining 14 revertants that displayed pleiotropic effects into four complementation groups (Table 2). Most of the complementation groups were represented by a single revertant, whereas complementation group designated asg1 contained 10 members.
ADH II enzyme activity assays were conducted on the 16 revertants from each of these complementation groups, the parent strain 553-4b, and 553-4b-10, which contains the previously characterized ccr4-10 allele (Table 3). The ADH II assay values that were obtained confirm that growth on ally1 alcohol was due to reduced expression of ADH2. As expected, values for many of the revertants were not as low as the ccr4-containing strain 553-4b-10 since many of the revertants identified were not Antimycin A sensitive (data not shown). On glucose, most rever
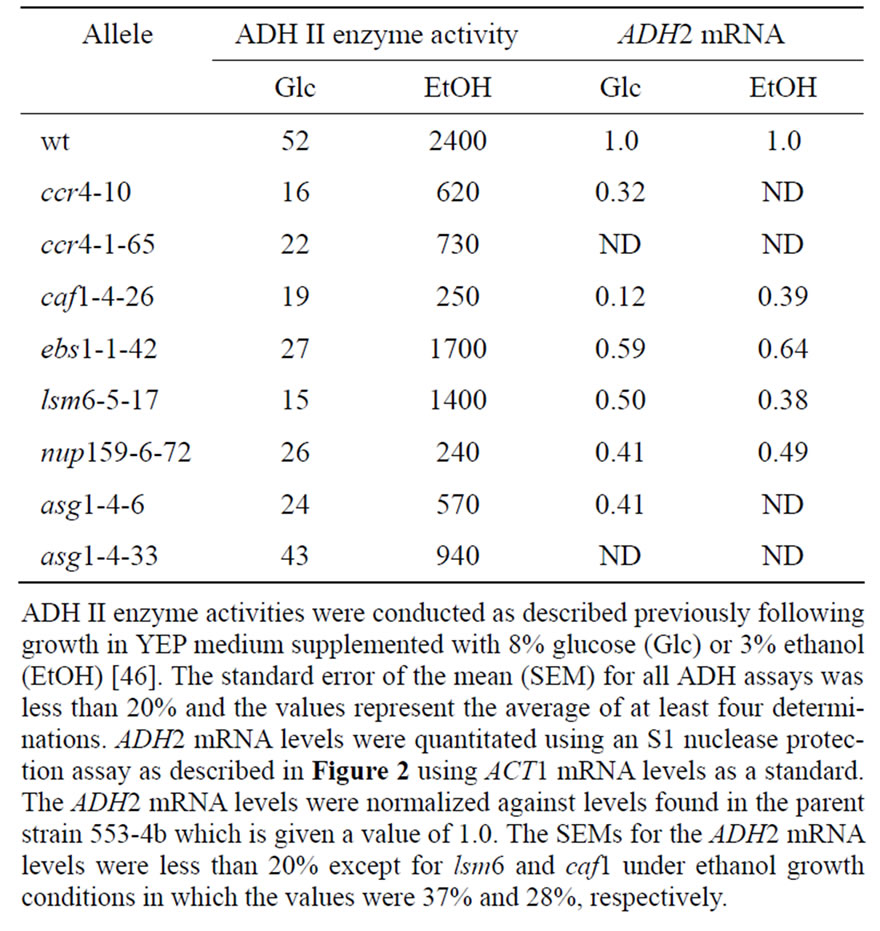
Table 3. ADH2 expression in spt10 revertants.
tants displayed at least a two-fold reduction of ADH II activity compared to the parental strain (Table 3). Assay values for these revertants grown in ethanol medium showed a wide range of values. Most of the activity levels in these revertants fell into an intermediate range around 1000 mU/mg of ADH II. For the ten alleles of asg1 the range of ADH II activities under ethanol conditions was from 440 to 1200 mU/mg. These results indicate that the spt10 revertants, like previously identified ccr4 and caf1 alleles, suppressed both spt10-induced ADH2 expression and the ability of ADH2 to derepress under non-fermentative growth conditions.
3.3. Cloning of ASG2, 3, and 6
In order to clone the respective genes for the remaining unidentified complementation groups, a representative from each group was transformed with either or both low and high copy yeast genome libraries. Depending on the phenotype of the representative mutant, selection for complementing plasmids was conducted on different media. Repeated attempts to complement asg1 failed, however, and complementing clones were only obtained for asg2, asg3 and asg6.
For strain 553-4b-6-72 (asg6), complementing plasmids were selected on 8 mM caffeine plates. A single plasmid (6-72-1), derived from a YCp50-LEU2 genomic library, was identified and was shown to contain an 8.4 Kbp fragment of Chromosome IX. Three genes were identified on this plasmid that could potentially be responsible for complementing 553-4b-6-72: YIL113w, POR2 and NUP159. Given the function of NUP159 as involved in mRNA transport out of the nucleus and that a defect in such a gene could result in lowered gene expression, a nup159 strain (LGY101) was obtained carrying the rat7-1 mutation (RAT7 is the same as NUP159). Crossing strain LGY101 to strain 553-4b-6-72, resulted in diploids that failed to grow on 8 mM caffeine plates, indicating that the asg6-6-72 mutation could not complement the rat7-1 mutation. asg6-6-72 was therefore designated as an allele of NUP159. The NUP159 protein has recently been shown to play an important role in mRNA transport by aiding DBP5 remodeling of the mRNP complex [25], but otherwise has no direct role in mRNA expression.
Revertant 553-4b-5-17 (asg3) when transformed with the yeast genomic library YEp13 yielded one transformant that was caffeine resistant and that was also unable to grow on 10 mM ally1 alcohol media. The insert contained in plasmid 5-17-1 was determined to be a 6.5 Kbp fragment of chromosome IV containing three genes, ATP17, LSM6 and RGA2. RGA2 has been described as a GTPase activating protein, LSM6 is involved in RNA splicing processing and mRNA decapping [26,27] and ATP17 is a subunit of ATP synthase. A BamHI-SalI fragment of plasmid 5-17-1 that contained ATP17 and LSM6 was subcloned into pRS315, a single copy plasmid containing a LEU2 gene. This plasmid was able to complement the caffeine sensitive phenotype of 5-17, showing that one of these two genes, but not RGA2, was responsible for this complementation. Also, following transformation of a YCp50-LEU2, single copy yeast genomic library, into 553-4b-5-17, a single caffeine resistant colony was obtained that contained a plasmid with both ATP17 and LSM6 genes (data not shown). An lsm6- containing strain, AEMY19, was crossed to 5-17, and the resulting diploid strain failed to grow in the presence of 8 mM caffeine. This failure to complement indicates that the asg3-5-17 mutation is an allele of lsm6.
Strain 553-4b-1-42 (asg2) after transformation with a YEp13 high copy yeast genomic library yielded a single plasmid that could complement the ability to grow at 37˚C and the allyl alcohol phenotype. The insert was found to be a 4.3 Kbp region of chromosome IV containing a single gene, EBS1. EBS1 was initially reported to be involved in telomere maintenance based on its sequence similarity to EST1 [28] but more recently it has been shown to be the ortholog of the higher eukaryotic nonsense-mediated decay protein SMG7 [29]. Its involvement in nonsense mediated decay may be only one means that its is involved in mRNA expression, as it has also been demonstrated that EBS1 binds eIF4E, the mRNA cap binding protein required for translational initiation. In fact, EBS1 is a negative regulator of translation [30]. EBS1 also binds DCP1 [31], a component of the mRNA decapping complex [20]. Its exact role in decapping is not understood, as it does not clearly affect mRNA degradation rates (R. Parker, pers. comm), and more recent studies indicate that it does not affect MFA2 decapping [29]. Removal of amino acids 456-783 from EBS1, that overlap a putative RNA recognition motif, resulted in an EBS1-expressing plasmid that was unable to complement the 37˚C growth defect of the 553-4b-1-42, demonstrating that the EBS1 gene complemented 553- 4b-1-42 and that this deleted region is required for EBS1 function. We also showed that deleting ebs1 gave the same phenotype as the ebs1-1-42 allele in that it was capable of suppressing an spt10 deletion in its effects on either ADH2 or HIS4 (data not shown).
3.4. spt10 Suppressors Affect ADH2 Expression at the Transcriptional Level
The identification of three additional suppressors of spt10 as factors involved in mRNA degradation (LSM6 and EBS1), translation (EBS1), and transport (NUP159) coincide with the role of the other spt10 suppressors, CCR4, CAF1 and NOT4, that have known roles in posttranscriptional control of mRNA metabolism, including that of mRNA degradation, translation, and elongation [17-20,32]. We subsequently investigated whether the effects of these new spt10 suppressors were the result of reducing ADH2 mRNA expression. ADH2 mRNA levels were quantitated by S1 analysis and are summarized in Table 3 (see also Figure 2, where the zero time points are representative assays conducted under ethanol growth conditions). Each of the lsm6-17, ebs1-1-42, nup159-6-72 and asg1-4-6 alleles reduced spt10-enhanced ADH2 mRNA expression by around two-fold or more under glucose growth conditions and by a similar level following ethanol growth conditions. It should be mentioned that the spt10 allele does not affect ADH II enzyme levels following ethanol growth conditions [1,3]. Therefore, the effects of these suppressor mutations on ADH II activity following non-fermentative growth represent direct effects on ADH2 expression. These results indicate that these alleles reduce ADH II enzymatic levels by primarily reducing the steady state levels of ADH2 mRNA levels. Therefore, as previously found for CCR4 and CAF1 [3,11] (Table 3), these new spt10 suppressors are required for activating ADH2 expression. It should be noted, however, that under ethanol growth conditions the actual steady state reduction in ADH2 mRNA levels by lsm6 and nup159 did not appear to directly correlate with the degree of reduction in the levels of ADH II enzyme activity (Table 3). The cause for this is unclear but could result from the unknown nature of the mutation or possibly even from pleiotropic effects on the translatability or expression of the mRNA.
3.5. lsm6, ebs1, and nup159 Alleles Do Not Affect Expression of CCR4-NOT Complex Components
One likely cause for the suppression of spt10 and of ADH2 gene expression by the lsm6, ebs1 and nup159 alleles is that they reduce the levels of key CCR4-NOT complex components. The levels of CCR4-NOT complex proteins were subsequently determined by Western analysis in strains carrying these mutant alleles. As shown in Figure 1, lsm6, ebs1 and nup159 had no effect on the relative abundance of CCR4, CAF1, and other complex components. We also found that the integrity of the CCR4-NOT complex, as evidenced by co-immunoprecipitation, to be unaffected by the lsm6, ebs1, and nup159 alleles (data not shown). These data indicate that lsm6, ebs1, and nu159 affect ADH2 expression by a means other than through indirect effects on the CCR4-NOT complex.
3.6. lsm6, ebs1, nup159 Decrease or Have No Effect on ADH2 mRNA Half-Lives
Both LSM6 and EBS1 have known or suggested roles in

Figure 2. nup159, lsm6 and ebs1 alleles increase or have no effect on ADH2 mRNA stability. S1 nuclease protection assays were performed on 40 mg of total RNA using an oligonucleotide specific to the 5’ region of ADH2 and to ACT1 [17]. ADH2 mRNA stability was determined as previously described [17] following shifting of yeast from ethanol-containing medium to glucose-containing medium at time zero. The ebs1, nup159 and lsm6 alleles are the same as described in Table 2.
controlling the rate of mRNA degradation, with lsm6 slowing mRNA decapping and EBS1 interacting with the decapping complex or controlling at least nonsense mediated decay [26,27,29]. It was theoretically possible, however, that the lsm6 and ebs1 alleles identified in this spt10 screen were mutations that enhanced the rate of ADH2 mRNA degradation, thereby suppressing spt10 effects on the initiation of ADH2 synthesis and reducing ADH2 expression under non-fermentative growth conditions. To test this hypothesis we examined the rate of ADH2 mRNA degradation in each of these mutated strains. ADH2 mRNA half-lives were determined following pre-growth of yeast under ethanol (non-fermentative conditions) and shifting them to glucose growth conditions upon which ADH2 syntheses is rapidly shut off. Although in an spt10 background there is a low level of ADH2 expression under glucose growth conditions, this level is much less than found under non-fermentative conditions (Table 3 and Figure 2) allowing mRNA halflives to be readily calculated.
The ADH2 mRNA half-life was found to be about 12 min in agreement with previous results [17,33]. Both lsm6-5-17 and nup159-6-72 slowed ADH2 mRNA degradation (Figure 2 and summarized in Table 4) while the ebs1-1-42 allele had no effect on ADH2 mRNA stability. These results indicate that lsm6, nup159, ccr4 [17,33] and caf1 (data not shown) all increase the stability of ADH2 mRNA. Their effects on reducing the steady state levels of ADH2 mRNA must hence even be greater than the effects that are summarized in Table 3. Clearly, the reduction in ADH2 mRNA levels caused by these alleles is not occurring through direct effects on enhancing ADH2 mRNA degradation. These results, in contrast, suggest that interfering with post-transcriptional processes can reduce the level of mRNA synthesis.
4. DISCUSSION
CCR4 has been considered an activator of transcription since it first was identified as a suppressor of spt10 effects at ADH2 and as being required for ADH2 derepression following non-fermentative growth [1,3]. The mechanism by which it and other factors related to it, such as CAF1 or NOT4, are required for maintaining ADH2 mRNA levels has remained, however, obscure [5, 6,11,21]. The identification of the CCR4-NOT complex as involved in mRNA deadenylation, with CCR4 and CAF1 as deadenylases [17-20,32,34], indicate that ccr4 and caf1 deletions actually stabilize ADH2 mRNA and hence raise its mRNA levels, not lower them [17,33]. In this report we identify three new factors, NUP159, LSM6 and EBS1, whose defects, like those of caf1 and ccr4, suppress spt10-enhanced ADH2 expression and decrease the ability of ADH2 to derepress. The known or putative roles of NUP159, LSM6, and EBS1 in the post-transcriptional regulation of mRNA suggests, therefore, a functional link to CCR4 and CAF1 and a common mechanism by which they all affect ADH2 expression.
The lsm6-5-17, nup159-6-72 and ebs1-1-42 alleles effect on ADH2 mRNA levels were not the result of increasing the rate of ADH2 mRNA degradation. In fact, lsm6 and nup159 stabilized ADH2 mRNA, indicating that their effects on the synthesis of the mRNA was at least two-fold greater than that observed from monitoring ADH2 steady state mRNA levels. Similar conclusions
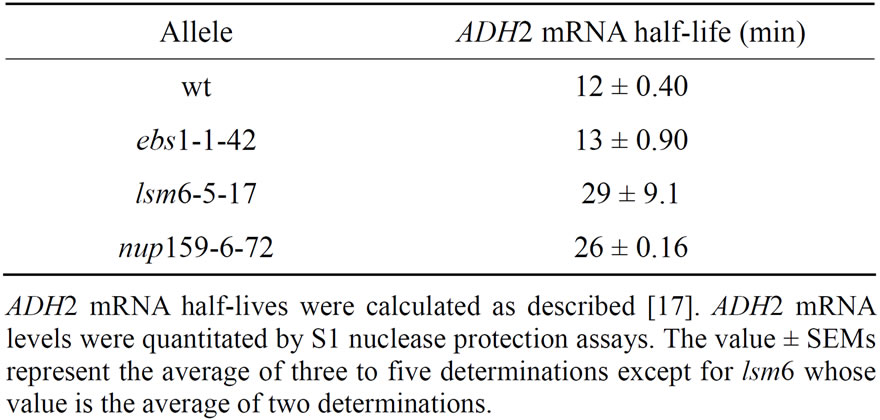
Table 4. Effect of spt10 revertants on ADH2 mRNA half-life.
can be made for ccr4 and caf1 defects that caused increased stabilization of ADH2 mRNA and yet, concurrently, reduced overall ADH2 mRNA levels [3,17,33] (Table 3). We also showed that the lsm6, nup159 and ebs1 alleles did not alter the abundance of CCR4-NOT complex components, indicating that their effects were not indirectly through lowering CCR4 and CAF1 abundance in the cell. In addition, the role of EBS1 in negatively regulating translation [30] cannot directly explain how it would function as an activator of ADH2.
spt10 results in a low level of ADH2 expression under glucose growth conditions by an unknown mechanism that could involve affecting ADH2 chromatin structure [2, 35,36]. The observation that lsm6, nup159, ebs1, ccr4 and caf1 can each suppress spt10 effects at ADH2 could suggest that these factors function downstream of spt10 on chromatin structure. However, our previous analyses indicated that CCR4 acted independently of SPT10 to affect ADH2 expression [36]. Also, the fact that lsm6, nup159, ebs1, caf1 and ccr4 reduce ADH2 expression under ethanol growth conditions when spt10 has no effect on ADH2 transcription further supports an effect of these alleles on ADH2 independently of spt10.
One possible model for how CCR4 and CAF1 function in activation is that they influence action of the RNA polymerase II holoenzyme through their contacts to SRB-9, -10, and -11 mediator components [21]. Alternatively, CCR4 and CAF1 could serve as coactivators by somehow mediating the action of activators such as ADR1 at ADH2. However, this latter model seems unlikely in that ADR1 is not required for spt10-enhanced ADH2 expression [1,11].
Based on the results presented in this paper, we suggest a different model for how CCR4, CAF1, LSM6, NUP159 and EBS1 act as activators at the ADH2 locus. The primary roles for these factors appear to be the mRNA degradative process, in mRNA transport from the nucleus, or in mRNA elongation. Studies have increasingly linked transcription initiation processes with posttranscriptional processing and transport of mRNA [37]. It is, therefore, conceivable that interfering with mRNA degradation and transport could result in slower processing and movement of mRNA through the cell. These effects could then result indirectly in reducing transcriptional initiation of particularly sensitive genes. A variant of this model would be that proteins involved in posttranscriptional mRNA processing are loaded onto the mRNA during its synthesis. Defects in these processing components would interfere with this loading process that again indirectly influences the corresponding initiation of transcription from certain promoters. In this regard CCR4-NOT complex is known to play a critical role in mRNA elongation [15,16,38]. Its function as an activator of transcriptional elongation would therefore be a very direct way for components of this complex to function as activators of ADH2 expression. For example, if transcription through the ADH2 gene were particularly prone to transcriptional arrests that the CCR4-NOT complex would be especially able to bypass by restimulating transcription [16], then a positive role for CCR4-NOT proteins would be substantiated. But no evidence for ADH2 susceptibility to transcriptional arrest is known and the EBS1, NUP159 and LSM6 factors do not have clear roles in elongation.
Finally, it has been recently shown that the nuclear pore factors may have diverse and direct effects on chromatin structure and hence gene expression [39]. Given these interactions between mRNA transport and transcription, the NUP159 protein and the CCR4-NOT complex, which has been shown to interact with the mRNA export pathway [40], may exert their respective effects on ADH2 chromatin structure and expression by virtue of their effects on mRNA export. Since the SPT10 protein is specifically involved in affecting chromatin structure [2,35,36], this could be one means by which defects in NUP159 and CCR4-NOT components suppress spt10 effects.
In contrast to the above models, given that CCR4, CAF1, LSM6, NUP159 and EBS1 defects might be expected to have broad affects on the mRNA degradation and/or transport of many genes [39-41], it remains possible that the mRNA for certain key transcriptional initiation factors might be specifically sensitive to corresponding defects in mRNA degradation or transport. For instance, the mRNA for holoenzyme or core initiation components might be specifically affected by such defects, resulting again, indirectly, in reduced ADH2 gene expression. However, whole genome microarray analysis with ccr4, caf1 and not4 deletions did not identify specific core transcriptional factors under the regulation of any of the CCR4-NOT factors [10].
In that ccr4, caf1, lsm6 and nup159 defects all reduce ADH2 steady state mRNA levels and at the same time reduce the rate of ADH2 mRNA degradation indicates an additional difficulty with interpreting the actual mechanisms by which factors control gene initiation. A number of factors thought to control initiation of transcription (HPR1, RTF1, PAF1, SPT5) have been shown to be involved in post-initiation processes [33,38,42-45]. Whether these original effects on transcription initiation were merely the consequences of indirect effects derived from their post-initiation sites of action has not been determined. At least for the case of CCR4-NOT factors understanding how their effects on ADH2 mRNA degradation rates can exert opposite effects on ADH2 synthesis may shed new light on the total integration of mRNA metabolism.
5. ACKNOWLEDGEMENTS
We would like to thank Dr. C. Cole and J. Beggs for yeast strains, B. Lauze for manuscript preparation, and members of the Denis laboratory for aid in conducting ADH assays and Western analysis. This research is supported by NIH grants GM41215 and GM87864 and Hatch project H291. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution no. 2156.
REFERENCES
- Denis, C.L. (1984) Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics, 108, 833-844.
- Eriksson, P.R., Mendiratta, G., McLaughlin, N.B., Wolfsberg, T.G., Mariño-Ramírez, L., Pompa, T.A., Jainerin, M., Landsman, D., Shen, C.H. and Clark, D.J. (2005) Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Molecular and Cellular Biology, 25, 9127-9137. doi:10.1128/MCB.25.20.9127-9137.2005
- Denis, C.L. and Malvar, T. (1990) The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics, 124, 283-291.
- Chang, M., French-Cornay, D., Fan, H.Y., Klein, H., Denis, C.L. and Jaehning, J.A. (1999) A complex containing RNA polymerase II, Paflp, Cdc73p, Hprlp, and Ccr4p plays a role in protein kinase C signaling. Molecular and Cellular Biology, 19, 1056-1067.
- Liu, H.Y., Toyn, J.H., Chiang, Y.C., Draper, M.P., Johnston, L.H. and Denis, C.L. (1997) DBF2, a cell-cycle regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO Journal, 16, 5289-5298. doi:10.1093/emboj/16.17.5289
- Liu, H.Y., Badarinarayana, V., Audino, D.C., Rappsilber, J., Mann, M. and Denis, C.L. (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO Journal, 17, 1097-1106. doi:10.1093/emboj/17.4.1096
- McKenzie, E.A., Kent, N.A., Dowell, S.J., Moreno, F., Bird, L.E. and Mellor, J. (1993) The centromere and promoter factor, 1, CPF1, of Sacchaomyces cerevisiae modulates gene activity through a family of factors including SPT21, RPD1, (SIN3), RPD3 and CCR4. Molecular and General Genetics, 240, 374-386.
- Schild, D. (1995) Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics, 140, 115-127.
- Chen, J., Rappsilber, J., Chiang, Y.C., Russell, P., Mann, M. and Denis, C.L. (2001) Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. Journal of Molecular Biology, 314, 683-694. doi:10.1006/jmbi.2001.5162
- Cui, Y., Ramnarain, D.B., Chiang, Y.C., Ding, L.H., McMahon, J.S. and Denis, C.L. (2008) Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Molecular Genetics and Genomics, 279, 323-337. doi:10.1007/s00438-007-0314-1
- Draper, M.P., Salvadore, C. and Denis, C.L. (1995) Identification of a mouse protein whose homolog in yeast is a component of the CCR4 transcriptional regulatory complex. Molecular and Cellular Biology, 15, 3487-3495.
- Collart, M.A. and Struhl, K. (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters [published erratum appears in EMBO J. 1993 12, 2990]. EMBO Journal, 12, 177-186.
- Collart, M.A. and Struhl, K. (1994) NOT1 (CDC39), NOT2 (CDC36), NOT3 and NOT4 encode a globalnegative regulator of transcription that differentially affects TATA-element utilization. Genes & Development, 8, 525-537. doi:10.1101/gad.8.5.525
- Badarinarayana, V., Chiang, Y.C. and Denis, C. L. (2000) Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics, 155, 1045-1054.
- Denis, C.L., Chiang, Y.C., Cui, Y. and Chen, J. (2001) Genetic evidence supports a role for the yeast CCR4- NOT complex in transcriptional elongation. Genetics, 158, 627-634.
- Kruk, J.A., Dutta, A., Fu, J., Gilmour, D.S. and Reese, J.C. (2011) The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes & Development, 25, 581-593. doi:10.1101/gad.2020911
- Chen, J., Chiang, Y.C. and Denis, C.L. (2002) CCR4, a 3’-5’ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO Journal, 21, 1414-1426. doi:10.1093/emboj/21.6.1414
- Daugeron, M.C., Mauxion, F. and Seraphin, B. (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Research, 29, 2448- 2455. doi:10.1093/nar/29.12.2448
- Tucker, M., Staples, R.R., Valencia-Sanchez, M.A., Muhlrad, D. and Parker, R. (2002) CCR4p is the catalytic sub-unit of Ccr4p/Pop2p/Notp mRNA deadenylasecomplex in Saccharomyces cerevisiae. EMBO Journal, 21, 1427-1436. doi:10.1093/emboj/21.6.1427
- Tucker, M., Valencia-Sanchez, M.A., Staples, R., Chen, J., Denis, C.L. and Parker, R. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell, 104, 377-386. doi:10.1016/S0092-8674(01)00225-2
- Liu, H.Y., Chiang, Y.C., Pan, J., Salvadore, C., Chen, J., Audino, D.C., Badarinarayana, V., Palaniswamy, V., Anderson, B. and Denis, C.L. (2001) Characterization of CAF4 and CAF16 reveal a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. Journal of Biological Chemistry, 276, 7541-7548. doi:10.1074/jbc.M009112200
- Casamassimi, A. and Napoli, C. (2007) Mediator complexes and eukaryotic transcriptional regulation: an overview. Biochimica, 89, 1439-1446. doi:10.1016/j.biochi.2007.08.002
- Bai, Y., Salvadore, C., Chiang, Y.C., Collart, M., Liu H.Y. and Denis, C.L. (1999) The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4,and NOT5. Molecular and Cellular Biology, 19, 6642-6651.
- Cui, Y. and Denis, C.L. (2003) In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS and SPT5 enhance upstream poly(A) site utilization. Molecular and Cellular Biology, 23, 7887-7901. doi:10.1128/MCB.23.21.7887-7901.2003
- Noble, K.N., Tran, E.J., Alcazar-Roman, A.R., Hodge, C.A., Cole, C.N. and Wente, S.R. (2011) The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Genes & Development, 25, 1065-1077. doi:10.1101/gad.2040611
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M. and Seraphin, B. (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO Journal, 19, 1661-1671. doi:10.1093/emboj/19.7.1661
- Tharun, S. and R. Parker (2001) Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Molecular Cell, 8, 1075-1083. doi:10.1016/S1097-2765(01)00395-1
- Zhou, J., Hidaka, K. and Futcher, B. (2000) The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Molecular and Cellular Biology, 20, 1947-1955. doi:10.1128/MCB.20.6.1947-1955.2000
- Luke, B., Azzalin, C.M., Hug, N., Deplazes, A., Peter, M. and Lingner, J. (2007) Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Research, 35, 7688-7697. doi:10.1093/nar/gkm912
- Ford, A.S., Guan, Q., Neeno-Eckwall, E. and Culbertson, M.R. (2006) Ebs1p, a negative regulatory of gene expression controlled by the Upf proteins in the yeast Saccharomyces cerevisiae. Eukaryotic Cell, 5, 301-312. doi:10.1128/EC.5.2.301-312.2006
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshorn, D., Narayan, V., Srinivasan, M., Pochart, P., Gureshi-Emili, A., Li, Y., Goodwin, B., Conover, D., Kalbfleishch, T., Vijayadamodar, G., Yang, M., Johnston, M., Field, S. and Rothberg, J.M., (2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature, 403, 623-627. doi:10.1038/35001009
- Ohn, T., Chiang, Y.C., Lee, D.J., Yao, G., Zhang, C. and Denis, C.L. (2007) CAF1 plays an important role in mRNA deadenylation separate from its contact to CCR4. Nucleic Acids Research, 35, 3002-3015. doi:10.1093/nar/gkm196
- Cui, Y., Chiang, Y.C., Viswanathan, P., Lee, D.J. and Denis, C.L. (2012) SPT5 physically interacts with CCR4 and affects mRNA degradation but does not control mRNA deadenylation. American Journal of Molecular Biology, in Press.
- Lee, D., Ohn, T., Chiang, Y.C., Liu, Y, Quigley, G., Yao, G. and Denis, C.L. (2010) PUF3 acceleration of deadenylation in vivo can operate independently of CCR4 activity, possibly involving effects on the PAB1-mRNP structure. Journal of Molecular Biology, 399, 562-575. doi:10.1016/j.jmb.2010.04.034
- Natsoulis, G., Winston, F. and Boeke, J.D. (1994). The SPT10 and SPT21 genes of Saccharomyces cerevisiae. Genetics, 136, 93-105.
- Shen, C.H., Leblanc, B.P., Neal, C., Akhavan, R. and Clark, D.J. (2002). Targeted histone acetylation at the yeast CUP1 promoter requires the transcriptional activator, the TATA boxes and the putative histone acetylase encoded by SPT10. Molecular and Cellular Biology, 22, 6406-6416. doi:10.1128/MCB.22.18.6406-6416.2002
- Orphanides, G. and Reinberg, D. (2002) A unified theory of gene expression. Cell, 108, 439-451. doi:10.1016/S0092-8674(02)00655-4
- Gaillard, H., Tous, C., Botet, J., Gonzalez-Aguilera, C., Quintero, M.J., Viladevall, L., Garcia-Rubio, M.L., Rodriguez-Gil, A., Marin, A., Anno, J., revuelta, J.L., Chavez, S. and Aguilera, A. (2009) Genome-wide analysis of factors affecting transcription elongation and DNA repair: a new role for PAF and Ccr4-Not in transcription-coupled repair. PLoS Genetics, 5, e1000364. doi:10.1371/journal.pgen.1000364
- Capelson, M., Doucet, C. and Hetzer, M.W. (2010) Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harbor Symposia on Quantitative Biology, 75, 585-598. doi:10.1101/sqb.2010.75.059
- Derr, S.C., Azzouz, N., Fuchs, S.M., Collart, M.A., Strahl, B.D., Corbett, A.H. and Laribee, R.N. (2011) The Ccr4- not complex interacts with the mRNA export machinery. PLoS One, 6, e18302. doi:10.1371/journal.pone.0018302
- Atchison, J.D. and Rout, M.P. (2012) The yeast nuclear pore complex and transport through it. Genetics, 190, 855-883. doi:10.1534/genetics.111.127803
- Hartzog, G.A., Wada, T., Handa, H. and Winston, F. (1998). Evidence that Spt4, Spt5 and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes & Development, 12, 357-369. doi:10.1101/gad.12.3.357
- Squazzo, S.L., Costa, P.J., Lindstrom, D.L., Kumer, K.E., Simic, R., Jennings, J., Link, A.J., Arndt, K.M. and Hartzog, G.A. (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO Journal, 21, 1764-1774.
- Strasser, K., S. Masuda, Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A., Aguilera, A., Struhl, K., Reed, R. and Hurt, E. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature, 417, 304-307. doi:10.1038/nature746
- Wada, T., Takagi, T., Yamaguchi, Y., Ferdous, A., Imai, T., Hirose, S., Sugimoto, S., Yano, K., Hartzog, G.A., Winston, F., Buratowski, S. and Handa, H. (1998). DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes & Development, 12, 343-356. doi:10.1101/gad.12.3.343
- Cook, W.J., Mosley, S., Audino, D.C., Rovelli, A., Mullaney, D., Stewart, G. and Denis, C.L.(1994) Mutations in the zinc-finger region of the yeast regulatory protein ADR1 affect both DNA binding and transcriptional activation. Journal of Biological Chemistry, 269, 9374-9379.
NOTES
*This research was supported by NIH and the USDA.
#Corresponding author.

