Journal of Cancer Therapy
Vol.07 No.07(2016), Article ID:68968,15 pages
10.4236/jct.2016.77056
Cytogenetic Profile in 7209 Indian Patients with de novo Acute Leukemia: A Single Centre Study from India
P. S. Kadam Amare1*, H. Jain1, S. Kabre1, Y. Deshpande1, P. Pawar1, S. Banavali1, H. Menon2, M. Sengar2, B. Arora2, N. Khattry2, G. Narula2, D. Sarang1, S. Kaskar1, B. Bagal2, H. Jain2, Uma Dangi2, P. G. Subramanian3, S. Gujral3
1Cancer Cytogenetics Department, Tata Memorial Hospital, Mumbai, India
2Department of Medical Oncology, Tata Memorial Hospital, Mumbai, India
3Hematopathology Laboratory, Department of Pathology, Tata Memorial Hospital, Mumbai, India

Copyright © 2016 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 17 June 2016; accepted 22 July 2016; published 25 July 2016
ABSTRACT
Background: Cytogenetics is one of the most important diagnostic parameters in the classification of acute leukemia. Recurrent chromosomal aberrations in acute leukemia have provided insights into the molecular mechanism of leukemogenesis. The variable frequencies of recurrent cytogenetic markers due to ethical/racial differences have been reported from Western and some Asian countries. Objective: We report cytogenetic data of largest cohort of 7209 adult and pediatric patients with de novo acute leukemia (AL) to determine the prevalence of various cytogenetic sub groups and compare with the Western and Asian population. Material & Methods: The AL patients included 2609 AML (adult: 2042, pediatric: 567), 3708 B-cell-precursor (BCP)-ALL (adult: 1300, pediatric: 2408) and 892 cases of T-ALL (adult: 480, pediatric: 412). Cytogenetic studies included conventional karyotyping and FISH using panel of probes. Results: The incidence of t(8;21) was high, comparable to other Asian countries. In comparison to our series and Western population, t(15;17) was more prevalent in Chinese population. Cytogenetic profiling of BCP-ALL revealed low prevalence of ETV6/RUNX1 in ours as well as other Asian population. The MLL aberrations in BCP-ALL and TLX1 & TLX3 aberrations in T-ALL occurred less frequently in our series as compared with Western population. Conclusion: The present study with a large cohort showed the heterogeneity of AL that involved various factors, such as age, gender and prevalence of distinct cytogenetic subgroups. Our data in comparison with other population based studies revealed differential distribution of some cytogenetic sub-groups indicating geographic heterogeneity due to differential environmental exposure which probably influenced underlying genetic susceptibility.
Keywords:
Cytogenetics, Acute Leukemia, Incidence, Asian Population, Geographic Heterogeneity

1. Introduction
Acute leukemia (AL) that includes genetically heterogeneous acute myeloid and acute lymphoblastic leukemias accounts for 25% of all leukemias in adults [1] . Cytogenetics is one of the most important diagnostic parameter in the classification of acute myeloid leukemia (AML) [2] [3] . In addition to its importance in diagnosis, prognostic significance of cytogenetic subtypes has also been identified by various cooperative groups such as SWOG, MRC and CALGB. This has further led to establishment of ELN and NCCN guidelines that include cytogenetic risk groups: 1) favorable which includes t(8;21), inv(16) and t(15;17) with overall survival (OS) rates of 55% - 65%; 2) intermediate with normal karyotype and; 3) adverse that frequently shows MDS-related abn(5), abn(7), inv(3), -17/abn(17p) and monosomal and/or complex karyotype with OS rates of 5% - 14% [2] [4] - [10] .
As in AML, recurrent chromosomal aberrations observed in acute lymphoblastic leukemia (ALL) have provided insights into the molecular mechanism of leukemogenesis [11] [12] . Around 80% of childhood and adult B-cell precursor-ALL (BCP-ALL) reveal recurring numerical and structural alterations such as translocations, deletions and copy number changes [13] - [18] .
Over the past three decades, cytogenetic alterations have been an integral part of diagnosis and prognostication of disease in ALL. High hyperdiploidy: 51 - 65 chromosomes, t(12;21): ETV6/RUNX1 and t(1;19): TCF3/ PBX1 are associated with favorable outcome, whereas t(9;22) and 11q23 (MLL) translocations, intrachromosomal amplification of chromosome 21 (iAMP 21) and hypodiploidy are associated with poor prognosis [13] [14] [17] [19] - [21] .
Recurring chromosomal abnormalities with variable prognosis have also been identified in 70% of T-ALL which includes hypodiploidy, 9p deletion, translocations involving T-cell receptor genes TCR-α and -β, transcription factor genes viz., TAL1, LYL1, TLX1/HOX 11 and TLX3/HOX11L2 [15] [22] - [24] . Fluorescence in situ hybridization (FISH) and conventional cytogenetics are the main strategies for cytogenetic workup of acute leukemia in disease management [13] [25] - [27] . Independent FISH analysis is used in several laboratories due to high sensitivity for detection of various cryptic structural and submicroscopic abnormalities, and also copy number changes in AL within short turnaround time [25] - [30] .
The variable frequencies of recurrent cytogenetic markers due to ethical/racial differences have been reported in some published studies from Western and some Asian countries [31] - [36] . In the current study, we present cytogenetic data of a largest cohort of 7209 patients with de novo acute leukemia from a single centre. The aim of the study is to analyze the cytogenetic pattern with prevalence of various recurrent cytogenetic markers and to compare it with reported literature.
2. Material and Methods
The study was conducted at the Department of Medical Oncology, Tata Memorial Hospital, Mumbai, India. It is the largest cancer diagnostic, treatment and research centre in India, with 40,000 - 50,000 new cancer cases being diagnosed every year. The present study includes 7209 untreated patients that were newly diagnosed with AL between January, 2008 and December, 2015. Of these, 4600 cases were of ALL and 2609 cases were of AML that included patients diagnosed with acute promyelocytic leukemia (APL). Among the 2609 AML cases, 567 were pediatric and 2042 were adult cases, whereas the 4600 cases of ALL consisted of 2408 cases of pediatric BCP-ALL, 1300 cases of adult BCP-ALL, 480 cases of adult T-ALL and 412 cases of pediatric T-ALL.
The initial diagnostic criteria included standard clinical and laboratory findings including immunophenotyping. On the other hand, cytogenetic workup by FISH and conventional cytogenetics is an integral part of routine classification, risk stratification with appropriate treatment decision and monitoring of the disease. Cytogenetic studies were performed using standard protocols preferably in bone marrow aspirate and in peripheral blood in around 10% of cases, and occasionally on bone marrow morphology smears. At least 15 - 20 GTG-banded metaphase cells were karyotyped and analysed as per ISCN 2009 and 2013 [37] [38] . In ALL, ploidy analysis was done by counting chromosome number in 30 - 40 metaphase cells. Ploidy was classified into various subgroups: Low and high hyperdiploidy, low and high hypodiploidy, near haploidy, diploidy and near triploidy/tetraploidy.
In addition to conventional cytogenetics, FISH was performed in every specimen by using a panel of probes- LSI dual fusion RUNX1/RUNX1T1, PML/RARA, break apart CBF-B, EVI1, MLL, RARA, LSI 5q31, 5q33/5p15, 7q22, 7q36/SE7, TP53/CEP 17 in AML; dual fusion BCR/ABL1, ETV6/RUNX1 ES, TCF3/PBX1, break apart MLL, TCF3, CEP 4, 6, 10, 17 in BCP-ALL; and break apart probe TCR-A, TCR-B, TLX1, TLX3, MLL and LSI 9p/CEP 9 in T-ALL. MLL translocations were characterized using dual fusion MLL/MLLT3 for t(9;11), MLL/ MLLT2 for t(4;11), MLL/MLLT1 for t(11;19) and MLL/MLLT4 for t(6;11) (Abbott Molecular, Zytovision, Kreatech Diagnostic and Cytocell, UK). Minimum 100 - 200 interphase cells and 5 - 10 metaphase cells were analyzed in each specimen. The cut off threshold for trisomy and dual fusion was 2% and for break apart probe and LSI deletion probe was 5%. The cut off threshold for CEP and LSI on BMA smear was 5% and 10%, respectively.
Age-related prevalence and variable ploidy association of recurrent chromosomal alterations were analyzed by the Pearson’s chi-square test for determining the statistical significance (SPSS version 20).
3. Results
3.1. Incidence of Recurrent Cytogenetic Abnormalities in AML Patients
Among the 2042 adult AML patients, 1240 were males and 802 were females (M/F ratio of 1.5) in the age range of 16 - 86 years (median age 38 years). Among the 567 pediatric AML patients, 394 were males and 173 were females (M/F ratio of 2.2) in the age range of 5 months - 15 years (median age 8 years). Of these 567 pediatric AML cases, 42 were in the age range of 5 months - 1 year.
3.2. Adult AML
Among the 2042 AML patients, 9% (179 cases) showed the translocation, t(15;17), whereas 1.4% (3 cases) revealed RARA variant translocation, t(11;17)(q13;q21), t(11;17)(q23;q21) and t(v;17). The detection rate of PML/RARA by FISH was 98% (220/225) (Table 1).
Among 1906 patients with successful results, frequency of t(8;21), inv(16)/t(16;16) and MLL translocations was 15% (280 cases), 4.4% (85 cases) and 5% (103 cases), respectively. In the t(8;21) positive group, 98% patients were below 50 years. The complex variants of t(8;21) were detected in 11% (30/280) cases, whereas two
Table 1. Incidence of recurrent cytogenetic aberrations in present series, adult AML (n = 2042), pediatric AML (n = 567) and comparison with other populations.
Western population: 3, 6, 8, 39, 40, 42, 44, 45, 46, 48, 49, 50; Asian population: 31, 32, 35, 43, 47.
cases (0.1%) revealed (iAMP21). Overall frequency of t(8;21), inv(16) and t(15;17) was 62% - 70% in children and young adults (1 - 38 years). Of 1906 patients, 103 patients with MLL translocation comprised of 36 cases (2%) with t(9;11)(p22;q23), 13 cases (0.7%) with t(6;11)(q27;q23), 3 cases (0.16%) with t(11;19)(p13;q23), 2 cases (0.1%) with t(11;17)(q21;q23) and 45 cases (2%) with t(v;11). The MLL amplification was detected in two cases (0.1%) above 50 years. Trisomy 8 (8.4%: 16/1906) was another common abnormality (Table 1 and Figures 1(a)-(e)).
Figure 1. (a) LSI RUNX1/RUNX1T1 dual fusion probe on metaphase cell shows der(8) with RUNX1/RUNX1T1 (Yellow signal) and residual RUNX1T1 (Red signal) indicating variant of t(8;21); (b) LSI dual colour CBF-β break apart probe on metaphase and interphase shows 1R1G1Y signal pattern indicative of inv(16); (c) LSI dual fusion RUNX1/RUNX1T1 probe on metaphase and interphase cell shows RUNX1 amplification on der(21) (iAMP21); (d) LSI dual colour MLL break apart probe on metaphase and interphase cell shows MLL amplification; (e) LSI dual colour MLL break apart probe on metaphase and interphase cell shows der(9) with residual MLL (Red signal) indicating t(9;11); (f) LSI dual colour EVI1 break apart probe on metaphase cell shows 1R1G1Y signal pattern with residual EVI1 (Red signal) on another chromosome indicating EVI1 translocation; (g) Triple colour LSI 5q31,5q33/hTERT 5p15 probe on metaphase and interphase cell shows absence of Red & Green signal on der(5) indicating del(5)(q31q33); (h) Triple colour LSI 7q22, 7q36/SE7 on metaphase and interphase cell shows absence of Red & Green signal on der(7) indicating del(7)(q22); (i) Triple colour LSI 7q22, 7q36/SE7 on metaphase and interphase cell shows absence of Red signal on der(7) indicating del(7)(q36).
In the adverse cytogenetic group, frequency of EVI1 aberrations inv(3)/t(3;3) was 1.5% (14/919 cases). Among the 3.4% (47/1387 cases) of abn(5) positive patients and 6.3% (87/1387 cases) of abn(7) positive patients, 5q deletion (96%) and 7q deletion (58%) were most common, respectively. Both these MDS-related abnormalities were frequently detected (80% - 83%) in patients above 30 years. The abn(17) [two cases of -17 and 13 cases of del(17p)] occurred in 1.6% (15/919) cases, especially in those above 38 years (Table 1) (Figures 1(f)-(i)).
3.3. Pediatric AML
The incidence of t(8;21), t(15;17), MLL translocations and inv(16)/t(16;16) in pediatric cases was 26% (148/567), 7.2% (41/567), 8% (45/567) and 5% (18/342) respectively. Frequency of complex variant of t(8;21) was 3.5% (20/567). Of 567 cases, 45 patients with MLL translocations comprised 20 cases (3.5%) with t(9;11), 4 cases (0.7%) with t(6;11), 3 cases (0.5%) with t(11;19), 1 case (0.2%) with t(4;11) and 7 cases (1.2%) with variant MLL translocation. The incidence of abn(5) and abn(7) was 2% (6/295) and 8.8% (26/295), respectively. Among these, 5q deletion (100%) and 7q deletion (62%) were most common (Table 1).
3.4. Incidence of Recurrent Cytogenetic Abnormalities in BCP- ALL Patients
Among the 2408 patients with pediatric Pre-B ALL, 1592 were males and 816 were females (M/F ratio of 2) in the age range of 2 months - 15 years (median age 5 years) and 137 cases were ≥1 years. Among the 1300 adult patients, 930 were males and 370 were females (M/F ratio of 2.5) in the age range of 16 - 77 years (median age 28 years).
3.5. Pediatric BCP-ALL
Ploidy analysis was performed in 2016 patients (84%) due to failure of cultures and unavailability of BM aspirate. The ploidy analysis revealed diploidy in 923 cases (46%), high hyperdiploidy in 540 cases (27%), low hyperdiploidy in 339 cases (17%), low hypodiploidy in 45 cases (2.2%), high hypodiploidy 128 cases (6.3%), haploidy in 7 cases (0.3%) and near triploidy/tetraploidy in 33 cases (1.6%) (Table 2). Trisomy 4, 6, 10, 17 were detected either as a sole or in combination in 41% (407/986) cases (Table 2).
Table 2. Incidence of recurrent cytogenetic aberrations in present series, adult BCP-ALL (n = 1300), pediatric BCP-ALL (n = 2408) and comparison with other populations.
Western population: 12, 13, 14, 16, 19, 21, 51, 52, 53, 59, 60, 61, 62, 63, 64; Asian population: 36, 55, 56.
Among 2408 patients, BCR/ABL1 and MLL translocations were detected in 154 cases (6.3%) and 74 cases (3.1%), respectively. MLL translocations were characterized as t(4;11) in 28 cases (1.2%), t(11;19) in 8 cases (0.3%) and MLL variants in 21 cases (0.9%). Among the 28 cases of t(4;11), 22 cases (79%) were in ≥10 years age group, of which 11 cases (39%) were infants (2 months - 1 year). Translocation (1;19) was observed in 7.2% cases (169/2362).
Translocation t(12;21): ETV6/RUNX1 was detected in 12.2% cases (291/2982), of which loss of ETV6 allele in 162 cases (55%) and trisomy 21 in 80 cases (27%) were the most common additional abnormalities. The ETV6 allelic loss and trisomy 21 were also detected in 5% cases (102/2101) and 37% cases (783/2101), respectively in case of ETV6/RUNX1 negative group, whereas (iAMP21) was found in 23 cases(1%) (Table 2 and Figures 2(a)-(e)).


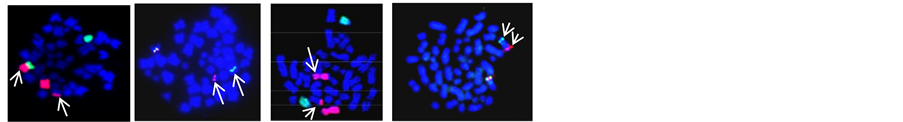
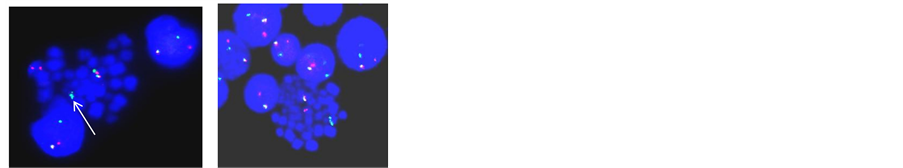
Figure 2. (a) LSI dual colour ETV6/RUNX1 ES translocation probe on metaphase and interphase cell shows ETV6/RUNX1 (Yellow signal) on der(21) with additional copy of RUNX1 and absence of ETV6 allele indicating ETV6/RUNX1 with trisomy 21 and loss of ETV6 allele; (b) LSI dual colour ETV6/RUNX1 ES translocation probe on metaphase and interphase cell shows RUNX1 amplification on der(21) (iAMP21); (c) LSI MLL/MLLT2 dual fusion probe on metaphase cell shows reciprocal MLL/MLLT2 (Yellow signal) on der(4) and der(11): t(4;11); (d) LSI TCF3/PBX1 translocation probe on metaphase and interphase cell shows TCF3/PBX1 (Yellow signal): t(1;19); (e) LSI dual colour TCF3 break apart probe on interphase and metaphase cell shows 1R1G1Y signal pattern in a specimen with no evidence of TCF3/PBX1 indicating variant t(17;19); (f) WCP 7 ( Red) and WCP 14 (Green) on metaphase cell shows t(?;7;14)(?;p12;q11.2); (g) LSI dual colour TCR-α break apart probe on metaphase cell shows residual TCR-α (Green signal) on der(11) indicating t(11;14)(p15;q11.2); (h) WCP 7 (Red) and WCP 14 (Green) on metaphase cell shows residual 7q36 region (Red signal) on 10q24 indicating t(7;10)(q34;q24); (i) LSI dual colour TCR-β break apart probe on metaphase cell shows residual TCR-β (Green signal) on 7p indicating t(7;7) (q34;q15)/inv(7)(p15q34); (j) LSI dual colour TLX1 break apart probe on metaphase and interphase cell shows green signal on der(14) indicating t(10;14)(q24;q11.2); (k) LSI dual colour TLX3 break apart probe on metaphase and interphase cell shows 1R1G1Y signal pattern indicating TLX3 translocation.
3.6. Age and Ploidy Distribution of Cytogenetic Subgroups in Pediatric Pre-B ALL
Hyperdiploidy (p = 0.0005), trisomy 4, 6, 10, 17 (p = 0.0005), MLL translocations (p = 0.025) and ETV6/RUNX1 (p = 0.0005) were associated with lower age group (1 - 10 years), whereas BCR/ABL1 (p = 0.0005) was associated with higher age group (11 - 15 years) (Table 3(a)). The ploidy distribution revealed association of BCR/ABL1, ETV6/RUNX1 and TCF3-PBX1 with diploidy (p = 0.0005) and of trisomy 4, 6, 10, 17 with hyperdiploidy (p = 0.0005) (Table 3(b)).
3.7. Incidence of Recurrent Cytogenetic Abnormalities in Adult BCP-ALL
Ploidy analysis was performed in 1015 of total 1300 cases (78%) due to poor mitotic index. Ploidy analysis revealed diploidy in 617 cases (61%), low hyperdiploidy in 147 cases (15%), high hyperdiploidy in 93 cases (9%), low hypodiploidy in 39 cases (4%), high hypodiploidy in 88 cases (9%), haploidy in 5 cases (0.5%) and near triploidy/tetraploidy in 26 cases (2.5%). The trisomy 4, 6, 10, 17 were detected either alone or in combination in 19% (83/432) cases (Table 2).
The incidence of BCR/ABL1 was observed in 27% (357/1300) cases, MLL translocation in 2.4% (31/1300) cases and TCF3/PBX1 in 4.4% (56/1268) cases. The variant t(17;19) was detected in 0.6% of the 166 cases
Table 3. (a) Cytogenetic markers and age distribution in Pediatric and adult BCP-ALL; (b) Cytogenetic markers and ploidy distribution in Pediatric and adult BCP-ALL.
(1/166). Thirty one patients with MLL translocation included 19 cases (1.5%) of t(4;11), two cases (0.15%) of t(11;19), one case (0.07%) each of t(9;11) and t(6;11) and eight cases (0.6%) of variant MLL translocations (Table 2).
ETV6/RUNX1 was observed in three cases (0.2%) in the age group 21 - 26 years, whereas (iAMP21) was observed in five (0.4%) young adults (16 - 20 years) apart from one, 56 years old.
3.8. Age and Ploidy Distribution of Cytogenetic Subgroups in Adult BCP-ALL
The incidence of t(9;22) and t(1;19) was higher in patients under 40 years (p = 0.0005 and p = 0.009, respectively). Ploidy distribution revealed association of BCR/ABL1 with diploidy (p = 0.007) and association of trisomy 4, 6, 10, 17 with hyperdiploidy (p = 0.0005) (Table 3).
3.9. Incidence of Recurrent Cytogenetic Abnormalities and Age and Ploidy Distribution in T-ALL
Among the 480 adult patients with T-ALL, 391 were males and 89 were females (M/F ratio of 4.3) in the age range of 16 - 78 years (median age 24 years). Among the 412 pediatric T-ALL cases, 329 were males and 83 were females (M/F ratio of 4) in the age range of 1 - 15 years (median age 10 years).
3.10. Adult T-ALL
Ploidy analysis was performed in 296 cases due to low mitotic index. The ploidy distribution was as follows: Diploidy in 182 cases (61%), low hyperdiploidy in 36 cases (12%), high hyperdiploidy in five cases (1.7%), low hypodiploidy in 13 cases (4.4%), high hypodiploidy in 45 cases (15%) and triploidy/tetraploidy in 15 cases (5%) (Table 4).
Heterozygous/homozygous 9p deletion occurred in 32% (154/480) cases, whereas MLL translocations occurred in 1.3% (6/460) cases, of which t(11;19) and MLL variant translocations occurred in 0.2% (1/460)and 1.1% cases(5/460), respectively.
TCR-α and TCR-β occurred in 18% cases (85/460), whereas TLX1 and TLX3 translocations occurred in 3.5% (16/460) and 7.6% (35/460) cases, respectively (Table 4 and Figures 2(f)-(k)).
There was no association of ploidy or variable age groups with cytogenetic subgroups, except for association of 9p deletion with young adults ≤40 years (p = 0.0005) and with diploidy (p = 0.03) (Table 5(a) and Table 5(b)).
Table 4. Incidence of recurrent cytogenetic aberrations in present series, adult T-ALL (n = 480), pediatric T-ALL (n = 405) and comparison with other populations.
Table 5. (a) Cytogenetic markers and age distribution in Pediatric and Adult T-ALL; (b) Cytogenetic markers and ploidy distribution in Pediatric and Adult T-ALL.
3.11. Pediatric T-ALL
The ploidy analysis of 244 cases revealed diploidy in 161 cases (66%), low hyperdiploidy in 24 cases (10%), high hyperdiploidy in three cases (1.2%), low hypodiploidy in six cases (2.5%), high hypodiploidy in 39 cases (16%) and triploidy/tetraploidy in 11 cases (4.5%).
The 9p deletion occurred in 44% (179/405) cases, whereas MLL translocations occurred in 5% (18/386) cases, of which t(9;11) occurred in four cases (1%), t(11;19) and t(6;11) in one case each (0.25%) and variant MLL translocations in 12 cases (3.1%). The incidences of TCR-α (14q11) and TCR-β (7q34) translocations was 21% (79/377 cases), TLX1 (10q24) was 2.4% (9/377 cases) and TLX3 (5q35) translocation was 11% (40/377 cases) (Table 4). The 9p deletion was associated with lower age group ≤10 years (p = 0.018) (Table 5(a) and Table 5(b)).
4. Discussion
The comprehensive analysis of heterogeneity of acute leukemia of the large cohort in the present study has demonstrated the involvement of various factors that involves age, incidence pattern, gender and genetic factors. Several studies from various locations have shown the geographic heterogeneity in acute leukemias [6] [20] [27] [30] - [32] [34] [36] [39] [40] . The cytogenetic pattern of acute leukemia in Indian population however is not clear due to lack of large scale studies. Hence, the main approach of the present study was to retrospectively analyze a large cohort to focus upon the epidemiology of various cytogenetic subtypes which may reveal the involvement of various etiological factors, thereby improving our understanding of the biology of the disease and thus help in better management of the disease.
Our combined approach of conventional cytogenetics and FISH significantly improved the detection rate of submicroscopic cryptic rearrangements within a short turnaround time. We could detect PML/RARA with RARA variants in 98% of APL cases. Table 1 summarizes the prevalence of recurrent chromosomal abnormalities in pediatrics and adult AML in present study and compares the incidence with different populations from USA, European and Asian countries.
In the present study, the median age of adult AML was lower (38 years) in comparison to Western population (40 - 45 years) and other Asian population [6] [8] [9] [31] - [34] [41] . In adult AML, the incidence of t(8;21) was higher (15%) than that of Western population (6% - 8%) and comparable to that in Chinese (8% - 15%) and Japanese (16%) populations [3] [6] [8] [31] [32] [35] [42] . Similarly, in pediatric AML, the frequency of t(8;21) was higher (26%) than that of Western population and comparable to that in Japanese population (25%) [8] [32] [39] [42] - [46] . However, the frequency of t(15;17) was comparable to that of Western population and was lower than that in Chinese population, both in adult and pediatric groups (14-18% and 16-18% respectively).
The higher incidence of t(15;17) in Chinese population could be due to high prevalence of APL in their population [3] [6] [8] [31] [39] [44] [46] [47] . Similar to literature reports, our data on age distribution of favorable subtypes t(8;21), t(15;17) and inv(16) in adult and pediatric AML also indicated that incidence of favorable markers are most common in children and young adults compared to older adults [3] [6] [8] [39] [44] [46] . Although overall MLL frequency in both adult and children were comparable with Western populations, our study showed lower (3%) incidence of t(9;11) in pediatric AML, similar to that in Chinese population [8] [31] [39] [44] - [48] . The translocation (4;11) occurred less frequently in pediatric AML probably due to lower number of infantile cases below one year in our series. The aberrations of chromosome 5, 7 and 8 were comparable with other studies however, these aberrations occurred with higher frequency compared to that in both adult and pediatric Chinese population [6] [31] [39] [40] [47] . The RUNX1 and MLL amplification frequencies (both 1%) of adult AML in our study have been reported as rare secondary events with dismal outcome [49] [50] .
In BCP-ALL, the age and gender distribution was comparable to reports from other geographic areas [14] [15] [17] - [19] [41] . BCP-ALL has various cytogenetic subtypes which have significant impact on risk stratification and hence remain strong independent indicators of disease outcome [13] [14] [16] [17] [19] - [21] . The incidence of high hyperdiploidy in children (27%) and adult ALL (9%) are similar to that of Western population [12] [16] [51] . The overall hyperdiploidy rate observed in our study was probably due to slightly higher incidence of low hyperdiploidy in our patients. As reported in literature, hyperdiploidy in childhood ALL was strongly associated with gain of chromosomes 4, 10, 17 and 21 (Table 3(b)) [12] [51] . The hypodiploidy incidence was comparatively higher in both pediatric and adult populations due to elevated frequency of high hypodiploidy. Near triploidy-tetraploidy, which is rare in childhood ALL was observed both in childhood and adult ALL with incidence of 1% - 2% [13] [52] . Near haploidy with chromosome numbers 23 - 29 is a rare hypodiploid group in pre-B-ALL occurring in 0.3% - 0.5% in ALL group in our study falling within the universal frequency of <1% [13] [53] .
The ETV6-RUNX1 is another common favorable subset of pediatric BCP- ALL that occurs with an average frequency of 25%, but showed lower incidence of 12% in our study. This finding is consistent with our previous study [27] and another cohort of 928 patients [Nahar A et al. in communication]. Data from various studies from Asian countries also indicated low prevalence (13% - 19%) of ETV6/RUNX1 [54] - [56] . Studies have reported incidence rates of <10% from other parts of India, however, these were smaller studies with sample size not large enough to focus on true prevalence [57] [58] . The peak incidence of ETV6/RUNX1 was in lower age group of 1 - 10 years as reported in our previous studies as well as in studies from other Asian countries [27] [52] [55] [59] . Trisomy 21 and ETV6 allelic loss were most frequent additional abnormalities in ETV6/RUNX1 positive group, however these abnormalities were common in overall BCP-ALL patients [27] [59] .
The (iAMP21), a distinct high risk cytogenetic subtype was identified with 1% incidence in pediatric BCP- ALL patients predominantly in older children of 3 - 14 years age group which is in accordance with other reported studies [21] [60] .
In our study, the t(9;22) incidence in childhood ALL was 6%, whereas it showed variable frequencies of 2% - 6% in Western and other Asian populations [14] [36] [59] . The t(9;22), which is the most common chromosomal abnormality in adult ALL occurred in 27% of the adult patients, falling within the universal range of 25% - 30%. It was more prevalent in older children as well as adult ALL under 40 years. Literature also indicate rising incidences of t(9;22) in older patients [61] - [63] . Geographic heterogeneity of variable frequencies of BCR/ABL1 has been reported [14] [62] [63] .
The translocation (1;19) is another cytogenetic subgroup of BCP-ALL that occurs more frequently in children than adults. The incidence in both pediatric and adult patients in our study was closer to other reports [61] - [64] except in Chinese population which showed comparatively lower frequency [36] .
Translocations involving the MLL (11q23) gene occur in up to 5% of childhood and adult BCP-ALL [19] [61] - [63] . The lower incidence of 2% - 3% observed in the present study as well as in studies from other Asian countries indicate that MLL rearrangement incidence vary in different populations [36] . Data from previous studies from our lab supports the finding that MLL rearrangement incidence is low in Indian population [30] . The t(4;11) that is known for its adverse outcome is the common MLL translocation in childhood ALL with higher frequencies (60% - 80%) in infants [61] - [63] . The lower incidence of 38% of t(4;11) in children in the present study is due to the low number of infants in our study. The incidence of t(4;11) was also found to be lower in childhood AML in the present study indicating under representation of infantile leukemia in India. This could be due to poor tendency of referring infantile leukemia cases to tertiary centres. Pattern of age distribution in MLL and t(1;19) positive groups as observed in other studies, suggested that these translocations tend to occur in lower age group [19] [30] [52] [61] - [64] . The ploidy distribution revealed association of favorable t(12;21)/ ETV6-RUNX1, t(1;19) as well as unfavorable t(9;22), MLL translocation subtypes with diploidy.
T-ALL commonly affects young males than females. This was evident from the data obtained in the present study which revealed 3 - 4 times higher prevalence of T-ALL in young males [13] [15] [65] . Hypodiploidy is common in T-ALL as compared with Pre-B-ALL. It was comparatively higher in the present study in both pediatric and adult cases [13] [61] . On the contrary, MLL translocation frequency was lower in adult patients in our study than that of western population [66] . As reported previously, 9p deletion and translocation involving T-cell receptors TCR-α (14q11.2) and TCR-β (7q34), transcription factor genes TLX1 (HOX11) (10q24) and TLX3 (HOX11L2) (5q35) occurred in more than 50% of cases [22] - [24] [67] [68] . The incidence of TCR-α and TCR-β translocations in pediatric and adult T-ALL was similar to previous reports [65] [69] . Among the most common partner genes, the TAL1, LYL1, LMO1, LMO2, TLX1 and TLX3 of TCR-α and TCR-β translocations, TLX1 is associated with better outcome, whereas TLX3 has been found to have increased risk of treatment failure [22] [23] .
In comparison to previous reports, the incidence of TLX1 and TLX3 translocations in the present study was lower in both pediatric and adult patients [22] [65] [69] . The study also observed that 9p deletion was common in young age group and associated with diploidy as has been reported previously [15] [22] - [24] [46] [69] [70] .
5. Conclusion
The present study with a large cohort showed the heterogeneity of AL that involved various factors, such as age, gender and prevalence of distinct cytogenetic subgroups. Cytogenetic data when compared with data from other geographic areas revealed geographic heterogeneity due to differential environmental exposure which probably influenced the underlying genetic susceptibility.
Cite this paper
P. S. Kadam Amare,H. Jain,S. Kabre,Y. Deshpande,P. Pawar,S. Banavali,H. Menon,M. Sengar,B. Arora,N. Khattry,G. Narula,D. Sarang,S. Kaskar,B. Bagal,H. Jain,Uma Dangi,P. G. Subramanian,S. Gujral, (2016) Cytogenetic Profile in 7209 Indian Patients with <i>de novo</i> Acute Leukemia: A Single Centre Study from India. Journal of Cancer Therapy,07,530-544. doi: 10.4236/jct.2016.77056
References
- 1. Ries Lag, H.D., Krapcho, M., Mariotto, A., Miller, B.A., et al. (2006) SEER Cancer Statistics Review, 1975-2003. Bethesda MD: National Cancer Institute, Based on November 2005 SEER Data Submission Posted to the SEER Website 2006.
- 2. Union for International Cancer Control (2014) Acute Myelogenous Leukemia and Acute Promyelocytic Leukemia. Review of Cancer Medicines on WHO List of Essential Medicines.
- 3. Swerdlo, S.H., Campo, F., Harris, N.L., Jaffe, E.S., et al. (2008) Tumours of Hematopoietic and Lymphoid Tissues. Lyon France, IARC Press, 2008. World Health Organization Classification of Tumours, Vol 2.
- 4. Byrd, J.C., Mrózek, K., Dodge, R.K., Carroll, A.J., et al. (2002) Pretreatment Cytogenetic Abnormalities Are Predictive of Induction Success, Cumulative Incidence of Relapse, and Overall Survival in Adult Patients with de novo Acute Myeloid Leukemia: Results from Cancer and Leukemia Group B (CALGB 8461). Blood, 100, 4325-4336.
- 5. Dohner, H., Estey, E.H., Amadori, S., Appelbaum, F.R., et al. (2010) Diagnosis and Management of Acute Myeloid Leukemia in Adults: Recommendations from an International Expert Panel, on Behalf of the European Leukemia Net. Blood, 115, 453-474.
- 6. Grimwade, D., Hills, R., Moorman, A.V., Walker, H., et al. (2010) Refinement of Cytogenetic Classification in AML: Determination of Prognostic Significance of Rare Recurring Chromosomal Abnormalities amongst 5635 Younger Adults Treated in the United Kingdom Medical Research Council Trials. Blood, 116, 354-365.
- 7. Slovak, M.L., Kopecky, K.J., Cassileth, P.A., Harrington, D.H., et al. (2000) Karyotypic Analysis Predicts Outcome of Pre-remission and Post-Remission Therapy in Adult Acute Myeloid Leukemia: A Southwest Oncology Group/Eastern Cooperative Oncology Group Study. Blood, 96, 4075-4083.
- 8. Creutzig, U., van den Heuvel-Eibrink, M.M., Gibson, B., Dworzak, M.N., et al. (2012) AML Committee of the International BFM Study Group. Diagnosis and Management of Acute Myeloid Leukemia in Children and Adolescents: Recommendations from an International Expert Panel. Blood, 120, 3187-3205.
- 9. Medeiros, B.C., Othus, M., Fang, M., Roulston, D., et al. (2010) Prognostic Impact of Monosomal Karyotype in Young Adult and Elderly Acute Myeloid Leukemia: The Southwest Oncology Group (SWOG) Experience. Blood, 116, 2224-2228.
- 10. Mrozek, K., Marcucci, G., Nicolet, D., Maharry, K.S., et al. (2012) Prognostic Significance of the European Leukemia Net Standardized System for Reporting Cytogenetic and Molecular Alterations in Adults with Acute Myeloid Leukemia. Journal of Clinical Oncology, 30, 4515-4523.
- 11. Pui, C.H., Carroll, W.L., Meshinchi, S. and Arceci, R.J. (2011) Biology, Risk Stratification, and Therapy of Pediatric Acute Leukemias: An Update. Journal of Clinical Oncology, 29, 551-565.
- 12. Schultz, K.R., Jeanette Pullen, D., Sather, H.N., Shuster, J.J., et al. (2007). Risk- and Response-Based Classification of Childhood B-Precursor Acute Lymphoblastic Leukemia: A Combined Analysis of Prognostic Markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG). Blood, 109, 926-935.
- 13. Harrison, C.J., Moorman, A.V., Broadfield, Z.J., Cheung, K.L., et al. (2004) Three Distinct Subgroups of Hypodiploidy in Acute Lymphoblastic Leukemia. British Journal of Haematology, 125, 552-559.
- 14. Moorman, A.V., Chiltron, L., Wilkinson, J., Ensor, H.M., et al. (2010) A Population-Based Cytogenetic Study of Adults with Acute Lymphoblastic Leukemia. Blood, 115, 206-214.
- 15. (2015) PDQ Cancer Information Summaries. Childhood Acute Lymphoblastic Leukemia Treatment (PDQ®), Health Professional Version, PDQ Pediatric Treatment Editorial Board. Published Online: December 10.
http://www.ncbi.nlm.nih.govpubmedhealth/PMH0032725/ - 16. Paulsson, K., Forestier, E., Andersen, M.K., Autio, K., et al. (2013) High Modal Number and Triple Trisomies Are Highly Correlated Favorable Factors in Childhood B-Cell Precursor High Hyperdiploid Acute Lymphoblastic Leukemia Treated According to the NOPHO ALL 1992/2000 Protocols. Haematologica, 98, 1424-1432.
http://dx.doi.org/10.3324/haematol.2013.085852 - 17. Schultz K.R., Bowman, W.P., Aledo, A., Slayton, W.B., et al. (2009) Improved Early Event-Free Survival with Imatinib in Philadelphia Chromosome-Positive Acute Lymphoblastic Leukemia: A Children’s Oncology Group Study. Journal of Clinical Oncology, 27, 5175-5181.
http://dx.doi.org/10.1200/JCO.2008.21.2514 - 18. Zhang, Y. and Le Beau M.M. (2015) Cytogenetics and Molecular Genetics in Acute Lymphoblastic Leukemia.
http://www.uptodate.com - 19. Pui, C.-H., Chessells, J.M., Camitta, B., Baruchel, A., et al. (2003) Clinical Heterogeneity in Childhood Acute Lymphoblastic Leukemia with 11q23 Rearrangements. Leukemia, 17, 700-706.
http://dx.doi.org/10.1038/sj.leu.2402883 - 20. Pui, C.-H. and Campana, D. (2008) Childhood Leukemia. In: John, E., et al., Eds., Abeloff’s Clinical Oncology, Elsevier Inc., Livingstone, 2139-2169.
- 21. Harrison, C.J., Moorman, A.V., Schwab, C., Carroll, A.J., et al. (2014) An International Study of Intrachromosomal Amplification of Chromosome 21 (iAMP21): Cytogenetic Characterization and Outcome. Leukemia, 28, 1015-1021.
http://dx.doi.org/10.1038/leu.2013.317 - 22. Baak, U., Gokbuget, N., Orawa, H., Schwartz, S., et al. (2008) Thymic Adult T-Cell Acute Lymphoblastic Leukemia Stratified in Standard- and High-Risk Group by Aberrant HOX11L2 Expression: Experience of the German Multicenter ALL Study Group. Leukemia, 22, 1154-1160.
http://dx.doi.org/10.1038/leu.2008.52 - 23. Bergeron, J., Clappier, E., Radford, I., Buzyn, A., et al. (2007) Prognostic and Oncogenic Relevance of TLX1/HOX11 Expression Level in T-ALLs. Blood, 110, 2324-2330.
http://dx.doi.org/10.1182/blood-2007-04-079988 - 24. Sulong, S., Moorman, A.V., Irving, J., Strefford, J.C., et al. (2009) A Comprehensive Analysis of the CDKN2A Gene in Childhood Acute Lymphoblastic Leukaemia Reveals Genomic Deletion, Copy Number Neutral Loss of Heterozygosity and Association with Specific Cytogenetic Subgroups. Blood, 113, 100-107.
http://dx.doi.org/10.1182/blood-2008-07-166801 - 25. Amare Kadam, P.S., Raje, G., Pais, A. and Banavali, S. (2008) Co-Existence of TEL/AML1 and MLL Aberrations in B-Cell Precursor (BCP) ALL Discloses a Small Subclass of BCP-ALL. Cancer Genetics and Cytogenetics, 182, 27-32.
http://dx.doi.org/10.1016/j.cancergencyto.2007.12.012 - 26. Nordkamp, L.O., Mellink, C. and van der Schoot, Ellen. (2009) Karyotyping, FISH, and PCR in Acute Lymphoblastic Leukemia Competing or Complementary Diagnostics? Journal of Pediatric Hematology/Oncology, 31, 930-935.
http://dx.doi.org/10.1097/MPH.0b013e3181bc9c85 - 27. Pais, A., Amare Kadam, P., Raje, G., Banavali, S., et al. (2008) RUNX1 Aberrations in ETV6/RUNX1 Positive and ETV6/RUNX1 Negative Patients: Its Hemato-Pathological and Prognostic Significance in a Large Cohort (619 Cases) of ALL. Journal of Pediatric Hematology/Oncology, 25, 582-597.
http://dx.doi.org/10.1080/08880010802237450 - 28. Harrison, C.J., Moorman, A.V., Barber, K.E., Broadfield, Z.J., et al. (2005) Interphase Molecular Cytogenetic Screening for Chromosomal Abnormalities of Prognostic Significance in Childhood Acute Lymphoblastic Leukaemia: A UK Cancer Cytogenetics Group Study. British Journal of Haematology, 129, 520-530.
http://dx.doi.org/10.1111/j.1365-2141.2005.05497.x - 29. Amare Kadam, P., Baisane, C., Nair, R., Menon, H., et al. (2011) Characterization of Cryptic Rearrangements, Deletion, Complex Variants of PML, RARA in Acute Promyelocytic Leukemia. Indian Journal of Human Genetics, 17, 54-58.
http://dx.doi.org/10.4103/0971-6866.86174 - 30. Pais, A., Amare Kadam, P.S., Raje, G., Sawant, M., et al. (2005) Identification of Various MLL Gene Aberrations That Lead to MLL Mutations in Patients with Acute Lymphoblastic Leukemia (ALL) and Infants with Acute Leukemia. Leukemia Research, 29, 517-526.
http://dx.doi.org/10.1016/j.leukres.2004.11.016 - 31. Cheng, Y., Wang, Y., Wang, H., Chen, Z., et al. (2009) Cytogenetic Profile of De Novo Acute Myeloid Leukemia: A Study Based on 1432 Patients in a Single Institution of China. Leukemia, 23, 1801-1806.
http://dx.doi.org/10.1038/leu.2009.107 - 32. Kurosawa, S., Miyawaki, S., Yamaguchi, T.,Kanamori, H., et al. (2013) Prognosis of Patients with Core Binding Factor Acute Myeloid Leukemia after First Relapse. Haematologica, 98, 1525-1531.
http://dx.doi.org/10.3324/haematol.2012.078030 - 33. Nakase, K., Bradstock, K., Sartor, M., Gottlieb, D., et al. (2000) Geographic Heterogeneity of Cellular Characteristics of Acute Myeloid Leukemia: A Comparative Study of Australian and Japanese Adult Cases. Leukemia, 14, 163-168.
http://dx.doi.org/10.1038/sj.leu.2401638 - 34. Lazarevic, V., Horstedt, A.S., Johansson, B., Antunovic, P., et al. (2014) Incidence and Prognostic Significance of Karyotypic Subgroups in Older Patients with Acute Myeloid Leukemia: The Swedish Population-Based Experience. Blood Cancer Journal, 4, e188.
http://dx.doi.org/10.1038/bcj.2014.10 - 35. Wakui, M., Kuriyama, K., Miyazaki, Y., Hata, T., et al. (2008) Diagnosis of Acute Myeloid Leukemia According to the WHO Classification in the Japan Adult Study Group AML-97 Protocol. International Journal of Hematology, 87, 144-151.
http://dx.doi.org/10.1007/s12185-008-0025-3 - 36. Li, X., Li, J., Hu, Y., Xie, W., et al. (2012) A Comprehensive Cytogenetic Classification of 1466 Chinese Patients with De Novo Acute Lymphoblastic Leukemia. Leukemia Research, 36, 720-726.
http://dx.doi.org/10.1016/j.leukres.2011.12.016 - 37. ISCN (2009) An International System for Human Cytogenetic Nomenclature. In: Shaffer, L.G., Slovak, M.L., Karger, S.and Campbell, L.J., Eds., S. Karger, Basel.
- 38. ISCN (2013) An International System for Human Cytogenetic Nomenclature. In: Shaffer, L.G., McGowan-Jordan, J. and Schmid, M., Eds., S Karger, Basel, Switzerland.
- 39. Harrison, C.J., Hills, R.K., Moorman, A.V., Grimwade, D., et al. (2010) Cytogenetics of Childhood Acute Myeloid Leukemia: United Kingdom Medical Research Council Treatment Trials AML 10 and 12. Journal of Clinical Oncologyogy, 28, 2674-2681.
http://dx.doi.org/10.1200/JCO.2009.24.8997 - 40. Hasle, H., Alonzo, T.A., Auvrignon, A., Behar, C., et al. (2007) Monosomy 7 and Deletion 7q in Children and Adolescents with Acute Myeloid Leukemia: An International Retrospective Study. Blood, 109, 4641-4647.
http://dx.doi.org/10.1182/blood-2006-10-051342 - 41. Dores, G.M., Devesa, S.S., Curtis, R.E., Linet, M.S. and Morton, L.M. (2012) Acute Leukemia Incidence and Patient Survival among Children and Adults in the United States, 2001-2007. Blood, 119, 34-43.
http://dx.doi.org/10.1182/blood-2011-04-347872 - 42. Krauter, J., Wagner, K., Schafer, I., Marschalek, R., (2009) Prognostic Factors in Adult Patients up to 60 Years old with Acute Myeloid Leukemia and Translocations of Chromosome Band 11q23: Individual Patient Data-Based Meta-Analysis of the German Acute Myeloid Leukemia Intergroup. Journal of Clinical Oncology, 27, 3000-3006.
http://dx.doi.org/10.1200/JCO.2008.16.7981 - 43. Horibe, K., Tsukimoto, I. and Ohno, R. (2001) Clinicopathologic Characteristics of Leukemia in Japanese Children and Young Adults. Leukemia, 15, 1256-1261.
http://dx.doi.org/10.1038/sj.leu.2402194 - 44. Manola, K.N. (2009) Cytogenetics of Pediatric Acute Myeloid Leukemia. European Journal of Haematology, 83, 391-405.
http://dx.doi.org/10.1111/j.1600-0609.2009.01308.x - 45. Ravindranath, Y., Chang, M., Steuber, C.P., Becton, D., et al. (2005) Pediatric Oncology Group (POG) Studies of Acute Myeloid Leukemia (AML): A Review of Four Consecutive Childhood AML Trials Conducted between 1981 and 2000. Leukemia, 19, 2101-2116.
http://dx.doi.org/10.1038/sj.leu.2403927 - 46. Mrozek, K., Heerema, N.A. and Bloomfield, C.D. (2004) Cytogenetics in Acute Leukemia. Blood Reviews, 18, 115-136.
http://dx.doi.org/10.1016/S0268-960X(03)00040-7 - 47. So, C.-C., Wan, T.S., Chow, J.L., Hui, K-C., et al. (2011) A Single-Center Cytogenetic Study of 629 Chinese Patients with De Novo Acute Myeloid Leukemia Evidence of Major Ethnic Differences and a High Prevalence of Acute Promyelocytic Leukemia in Chinese Patients. Cancer Genetics, 204, 430-438.
http://dx.doi.org/10.1016/j.cancergen.2011.06.003 - 48. Schoch, C., Schnittger, S., Klaus, M., Kern, W., Hiddemann, W. and Haferlach, T. (2003) AML with 11q23/MLL Abnormalities as Defined by the WHO Classification: Incidence, Partner Chromosomes, FAB Subtype, Age Distribution, and Prognostic Impact in an Unselected Series of 1897 Cytogenetically Analyzed AML Cases. Blood, 102, 2395-2402.
http://dx.doi.org/10.1182/blood-2003-02-0434 - 49. Angelova, S., Spassov, B., Nikolova, V., Christov, I., et al. (2015) Is the Amplification of C-MYC, MLL and RUNX1 Genes in AML and MDS Patients with Trisomy 8, 11 and 21 a Factor for a Clonal Evolution in Their Karyotype? Genet, 49, 25-32.
http://dx.doi.org/10.3103/s0095452715030032 - 50. Haferlach, C., Grossmann V., Zenger M., et al. (2011) Gene Amplifications Are Rare Events in AML and MDS and Are Associated with Complex Karyotype, TP53 Deletions and Very Poor Survival [Abstract]. ASH. 2011, Abstract 2524.
- 51. Heerema, N.A., Sather, H.N., Sensel, M.G., Zhang, T., et al. (2000) Prognostic Impact of Trisomies 10, 17 and 5 among Children with Acute Lymphoblastic Leukemia and High Hyperdiploidy (> 50 Chromosomes). Journal of Clinical Oncology, 18, 1876-1887.
- 52. Raimondi, S., Zhou, Y., Sheila, A., Rubnitz, J.E., Pui, C.-H. and Behm, F.G. (2006) Near-Triploidy and Near-Tetraploidy in Childhood Acute Lymphoblastic Leukemia: Association with B-Lineage Blast Cells Carrying ETV6/RUNX1 Fusion, T-Lineage Immunophenotype and Favorable Outcome. Cancer Genetics and Cytogenetics, 169, 50-57.
http://dx.doi.org/10.1016/j.cancergencyto.2006.04.006 - 53. Safavi, S., Forestier, E., Golovleva, I., Barbany, G., et al. (2012) Loss of Chromosomes Is the Primary Event in Near-Haploid and Low-Hypodiploid Acute Lymphoblastic Leukemia. Leukemia, 27, 248-250.
http://dx.doi.org/10.1038/leu.2012.227 - 54. Woo, H.Y., Kim, D.W., Park, H., Seong, K.W., et al. (2005) Molecular Cytogenetic Analysis of Gene Rearrangements in Childhood Acute Lymphoblastic Leukemia. Journal of Korean Medical Science, 20, 36-41.
http://dx.doi.org/10.3346/jkms.2005.20.1.36 - 55. Liang, D.C., Shih, L.Y., Yang, C.P., Hung, I.J., et al. (2010) Frequencies of ETV6-RUNX1 Fusion and Hyperdiploidy in Pediatric Acute Lymphoblastic Leukemia Are Lower in Far East than West. Pediatric Blood & Cancer, 55, 430-433.
http://dx.doi.org/10.1002/pbc.22628 - 56. Nakao, M., Yokota, S., Horike, S., Kaneko, H., et al. (1996) Detection and Quantification of TEL/AML1 Fusion Transcripts by Polymerase Chain Reaction in Childhood Acute Lymphoblastic Leukemia. Leukemia, 10, 1463-1470.
- 57. Siddaiahgar, S., Awagbad, S. and Latha, M. (2015) Clinical, Immunophenotype and Cytogenetic Profile in Children at Tertiary Health Care Centre in India. Muller Journal of Medical Sciences and Research, 6, 112-118.
http://dx.doi.org/10.4103/0975-9727.160676 - 58. Sazawal S., Bhatia K., Gutierras M.I., Saxena R., et al. (2004) Paucity of TEL-AML1 Translocation by Multiplex RT-PCR in B-Lineage Acute Lymphoblastic Leukemia (ALL) in Indian Patients. American Journal of Hematology, 76, 80-82.
http://dx.doi.org/10.1002/ajh.20000 - 59. Rubnitz, J.E., Wichlan, D., Devidas, M., Shuster, J., et al. (2008) Prospective Analysis of TEL Gene Rearrangements in Childhood Acute Lymphoblastic Leukemia: A Children’s Oncology Group Study. Journal of Clinical Oncology, 26, 2186-2191.
http://dx.doi.org/10.1200/JCO.2007.14.3552 - 60. Attarbaschi, A., Mann, G., Panzer-Grumayer, R., Rottgers, S., et al. (2008) Minimal Residual Disease Values Discriminate between Low and High Relapse Risk in Children with B-Cell Precursor Acute Lymphoblastic Leukemia and an Intrachromosomal Amplification of Chromosome 21: The Austrian and German Acute Lymphoblastic Leukemia Berlin-Frankfurt-Munster (ALL-BFM) Trials. Journal of Clinical Oncology, 26, 3046-3050.
http://dx.doi.org/10.1200/JCO.2008.16.1117 - 61. Moorman, A.V., Harrison, C.J., Buck, G.A., Richards, S.M., et al. (2007) Karyotype Is an Independent Prognostic Factor in Adult Acute Lymphoblastic Leukemia (ALL): Analysis of Cytogenetic Data from Patients Treated on the Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 Trial. Blood, 109, 3189-3197.
http://dx.doi.org/10.1182/blood-2006-10-051912 - 62. Mancini, M., Scappaticci, D., Cimino, G., Nanni, M., et al. (2005) A Comprehensive Genetic Classification of Adult Acute Lymphoblastic Leukemia (ALL): Analysis of the GIMEMA 0496 Protocol. Blood, 105, 3434-3441.
http://dx.doi.org/10.1182/blood-2004-07-2922 - 63. The Group Francais de Cytogenetique Hematologique (1996) Cytogenetic Abnormalities in Adult Acute Lymphoblastic Leukemia: Correlations with Hematologique Findings Outcome. A Collaborative Study of the Group Francais de Cytogenetique Hematologique. Blood, 87, 3135-3142.
- 64. Foa, R., Vitale, A., Mancini, M., Cuneo, A., et al. (2003) E2A-PBX1 Fusion in Adult ALL: Biological and Clinical Features. British Journal of Haematology, 120, 484-487.
http://dx.doi.org/10.1046/j.1365-2141.2003.04113.x - 65. Raimondi, S. (2007) T-Lineage Acute Lymphoblastic Leukemia (T-ALL). Atlas of Genetics and Cytogenetics in Oncology and Haematology, 11, 328-339.
- 66. Ferrando, A.A., Armstrong, S.A., Neuberg, D.S., Sallan, S.E., et al. (2003) Gene Expression Signatures in MLL-Rearranged T-Lineage and B-Precursor Acute Leukemias: Dominance of HOX Dysregulation. Blood, 102, 262-268.
http://dx.doi.org/10.1182/blood-2002-10-3221 - 67. Kim, M., Yim, S.H., Cho, N.S., Kang, S.H., et al. (2009) Homozygous Deletion of CDKN2A (p16, p14) and CDKN2B (p15) Genes Is a Poor Prognostic Factor in Adult but Not in Childhood B-Lineage Acute Lymphoblastic Leukemia: A Comparative Deletion and Hypermethylation Study. Cancer Genetics and Cytogenetics, 195, 59-65.
http://dx.doi.org/10.1016/j.cancergencyto.2009.06.013 - 68. Kawamata, N., Ogawa, S., Zimmerman, M., Kato, M., et al. (2008) Molecular Allelokaryotyping of Pediatric Acute Lymphoblastic Leukemia by High Resolution Single Nucleotide Polymorphism Oligonucleotide Genomic Microarray. Blood, 111, 776-784.
http://dx.doi.org/10.1182/blood-2007-05-088310 - 69. Berger, R., Dastugue, N., Busson, M., Akker, J., et al. (2003) t(5;14)/HOX11L2-Positive T-Cell Acute Lymphoblastic Leukemia. A Collaborative Study of the Groupe Francais de Cytogénétique Hématologique (GFCH). Leukemia, 17, 1851-1857.
http://dx.doi.org/10.1038/sj.leu.2403061 - 70. van Grotel, M., Meijerink, J.P.P., van Wering, E.R., Langerak, A.W., et al. (2008) Prognostic Significance of Molecular-Cytogenetic Abnormalities in Pediatric T-ALL Is Not Explained by Immunophenotypic Differences. Leukemia, 22, 124-131.
http://dx.doi.org/10.1038/sj.leu.2404957
NOTES
*Corresponding author.




