Journal of Cancer Therapy
Vol.4 No.1A(2013), Article ID:26909,7 pages DOI:10.4236/jct.2013.41A010
Studying the Importance of VacA Gene of Helicobacter pylori in Identifying the Pathogenicity of Strains by Comparing It with the Disease Status of the Subjects
![]()
1Department of Clinical Laboratory Sciences, Microbiology Unit, Asmara College of Health Sciences, Asmara, Eritrea; 2Centre for Liver Research and Diagnostics, Deccan College of Medical Sciences, Hyderabad, India; 3Dr. Hari Singh Gour University, Sagar, India.
Email: *khajashakeel@gmail.com
Received September 20th, 2012; revised October 21st, 2012; accepted October 31st, 2012
Keywords: Helicobacter pylori; 16S rRNA; VacA; Rapid Urease Test; Culture
ABSTRACT
Introduction: H. pylori is a primary pathogen isolated by Warren and Marshall in 1983. They called it as Campylobacter Pylori and in 1989 Goodwin et al. renamed it as Helicobacter pylori. Helicobacter pylorus is one of the responsible factors for causing gastritis, Peptic ulcer disease and is strongly associated with gastric carcinoma and gastric Malt lymphoma. Materials and Methods: In this study 300 biopsies were collected at Deccan College of Medical Sciences and Allied Hospitals, Hyderabad, India. Of these, 101 patients had peptic ulcer, 95 patients had gastritis and 4 had gastric carcinoma. A total of four gastric biopsy specimens were collected. One was used for culturing H. pylori, one for histological lesions and the remaining two one each from the antrum & corpus was collected in phosphate buffered saline for the DNA analysis. Results: Helicobacter pylori were isolated from many of the biopsies and the identification of Helicobacter pylori was confirmed in 200 biopsies with colony characteristics, biochemical tests, and 16S rRNA amplification. The presence of VacA marker was detected by using appropriate primers. From the data obtained in our study, in a total of 169/200 isolates s1 region was found in 84.1% and s2 was found in 36/200 i.e. 18% of the isolates. In the middle region, m1a was found in 60/200 which was 30%, m1b was found in 60/200 i.e. 60% and m2 was found in 114/200 i.e. 57.1% of the isolates. Conclusion: Thus, VacA detection might be helpful for determination of which patients are at highest risk for severe clinical outcomes such as duodenal ulcer, gastric ulcer or gastric carcinoma and eventually, to define strategies for the treatment or prevention of H. pylori infection.
1. Introduction
Helicobacter pylorus is a gram-negative spiral bacterium. It is non-spore forming bacilli which is motile and usually posses multiple or single bipolar sheathed flagella. H. pylori is the human specific inhabitant. Its biochemical, genetic and physiological features distinguish it from the related genera Campylobacter and Arcobacter, emphasizing their unique contributions to the gastro intestinal micro biota [1]. It is a primary pathogen isolated by Warren and Marshall in 1983 and they called it as Campylobacter pylori. Later on in 1989 Goodwin et al. renamed it as Helicobacter pylori. Helicobacter pylorus is one of the responsible factors for causing gastritis and peptic ulcer disease and is strongly associated with gastric carcinoma and gastric Malt lymphoma [2].
H. pylori has co-evolved with humans to be transmitted from person to person and to persistently colonize the stomach. Their population structure is a model for the ecology of the indigenous microbiota [3]. Currently 26 formally named species have been designated and proposed names such as Helicobacter rappini [1]. There are many virulent factors of H. pylori that contribute in dissimilar ways to gastric mucosal damage. Among them are factors known to be required for the colonization and survival of H. pylori in the human stomach. These factors are urease and flagella that are expressed by all Heliocobacter species. H. pylori manufactures large amounts of urease that helps to break down urea into carbon dioxide and ammonium, which neutralizes the acid during colonization of the stomach.
Flagellar motility of Helicobacter species has been also shown to be essential for successful colonization of the host. Flagella allow the bacterium to swim across the viscous gastric mucus and reach the more neutral pH underneath the mucus.
Other major and important virulent genes of Helicobacter pylori is the immunodominant CagA protein encoded by the cytotoxin associated gene A (CagA), which has been one of the most intensely investigated putative virulence factors and many studies have reported the status of CagA in the pathogenicity of H. pylori strains.
Along with this, the other most important gene is VacA which is the major toxin secreted by H. pylori [4]. The active cytotoxin is produced by approximately 50% of H. pylori clinical isolates, [5] and its presence is associated epidemiologically with tissue damage and peptic ulcers [6]. The vacuolating cytotoxin gene A (VacA), which encodes VacA protein, is present in all H. pylori strains, but the structure among VacA alleles, particularly in two regions is different [7].
One of these regions is the mid region of the gene, which may be type m1 (subtype a) or m2 (subtypes a or b). The other variable region of VacA gene is in the second half of the signal sequence, which may be type s1 (subtypes a, b and c) or s2. H. pylori and its encoded products are associated with or without vacuolating activity in vitro [7-10].
Like wise, H. pylori strains containing certain types of VacA alleles have been associated with an increased risk of peptic ulcer disease. Atherton and colleagues showed that strains with VacA mid-region type m1 are more strongly associated with gastric epithelial damage (epithelial degeneration, mucus depletion, and microscopic erosion) than mid-region type m2. Meanwhile, s1a strains are associated with increased mucosal neutrophil and lymphocyte infiltration in vivo. These results suggest that s1a/m1 strains have the most virulent allelic type activity, [7,10]. While VacA s2 strains are rarely associated with ulcer disease and are uncommon in many populations [10-14].
Over 90% of patients with duodenal ulcer disease harbored VacA s1a strains, while patients with Vacs2 strains were not more likely to have ulcer disease than uninfected patients [10]. Likewise, in a study of the United Kingdom population, all patients with ulcer disease had VacA s1 strains and no VacA s2 strains were associated with this disease [15]. Other studies from western Europe also indicate a strong correlation between the VacA s1 genotype and peptic ulceration [15- 17]. Recent studies show that most H. pylori strains in Japan have an s1a/m1 genotype. However, regardless of the VacA genotype, the infection leads to peptic ulcer or gastric cancer.
In addition to these typical differences between VacA alleles, there is also considerable variation among different H. pylori strains with regard to the production of vacuolating cytotoxin. Thus, the vacuolating cytotoxin phenotypes of an H. pylori strain is not only dependent on the amino acid sequence of VacA but may also be modulated by other strain-specific factors, such as the level of VacA transcription or the efficiency with which VacA is secreted. [18] The considerable variation in these determinants may be relevant to the occurrence of different clinical outcomes in H. pylori.
In this study, we employed a PCR-based method to define the VacA genotypes of 200 individual H. pylori isolates from antrum and corpus biopsy specimens obtained from 300 patients undergoing endoscopy. Then it was compared with histopathology slides to locate any pathological changes in the subjects who are positive for VacA genotype.
2. Materials and Methods
Three hundred subjects were enrolled in the studies, who were undergoing gastroscopy in the Department of Gastroenterology, Deccan College of Medical Sciences and Allied Hospitals, Hyderabad, India, for the evaluation of upper gastrointestinal symptoms. The Institutional Ethical Committee (IEC) of the hospital approved the study. Of these, 101 patients (mean age 47 years) had peptic ulcer including 61 duodenal ulcers, 25 gastric ulcers, and 15 complete ulcers; 95 patients (mean age 41 years) had gastritis including 48 supper gastritis, 17 atrophic gastritis and 30 erosive gastritis; and 4 patients (mean age 56 years) had gastric adenocarcinoma.
A total of four gastric biopsy specimens (two from the antrum and two from the corpus) were collected from each patient after taking informed consent from the subjects to take part in the study. One antral biopsy collected in brucella broth supplemented with 2% fetal calf serum (FCS) was used for culturing H. pylori, one corpus biopsy for histological lesions and the remaining two biopsies one each from the antrum & corpus were collected in phosphate buffered saline for the DNA analysis.
2.1. Inclusion and Exclusion Criteria
In this study those patients who were having epigastric pain, indigestion, radiation, Hypochlorhydria and vomiting sensation have been included. And those subjects who were taking any medicine like proton pump inhibitors or any antibiotic therapy one week ahead of endoscopy has been excluded from the study.
2.2. Bacterial Strains and Growth Conditions
The collected biopsy specimens were transported to the laboratory, the specimens were best processed within 1 hr, if delayed the specimen was preserved at 4˚C for 4 - 24 hr. The biopsy was inoculated in to the Chocolate Brucella agar medium (Difco Laboratories, Detroit, USA) supplemented with 7% sheep blood and antibiotics Vancomycin (6 mg/mL), Amphoteracin-B (2 mg/mL) and Polymixin-B (2500 units/mL). The plates were incubated in a microaerophillic conditions at 37˚C for 3 - 5 days with 90% - 100% humidity. Then the plates were examined after 72 hrs for H. pylori (Figures 1-3). The isolated colonies of H. pylori were again sub cultured in a solid media. Rapid Urease method (RUT), a touch cytology method, was also done by inoculating few colonies into 250 µL urea broth comprising phenol red indicator. The presence of urease activity was observed by a rapid change in color from yellow to pink, which indicates the presence of H. pylori. The culture was considered to be positive for H. pylori by observing small, translucent, tiny colonies, which on staining gave Gram negative staining.
2.3. DNA Extraction and PCR Amplification
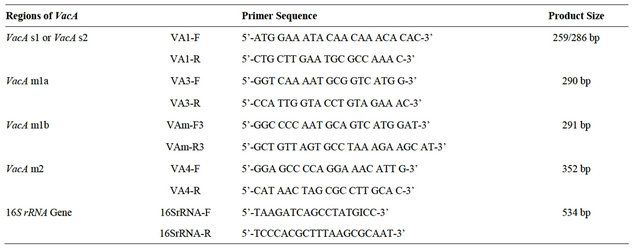
Genomic DNA isolation from 200 pure H. pylori cultures was done using a standard protocol described previously [13]. The isolated DNA was then amplified for the presence of target genes viz., 16S rRNA, vacA genes. Ethidium bromide was used to visualize the presence of amplified genes and photographic registries were performed using a Bio-Rad gel documentation system. Primers used in the study are shown in Table 1.

Figure 1. Rapid urease test for Helicobacter pylori identification. From left E. coli, H. pylori and Campylobacter jejuni. The pink colour of H. pylori indicating positive RUT.
Figure 2. Helicobacter pylori (primary isolation) on Brucella agar supplemented with 7% sheep blood.
Figure 3. Helicobacter pylori (pure culture) on Brucella agar supplemented with 7% sheep blood.
2.4. Genotyping of VacA by Polymerase Chain Reaction
To check the presence of VacA reported 4 sets of primers were used. Different regions were selected for structural screening of VacA [19]. They are second half of the single region (s-region) and mid-region (m-region). The different primer sets used were Va1, Va3, Vam3 and Va4 whose sequences are described in Table 1.
The VacA single sequence and mid region were typed by allelic type-specific PCR as described previously. In brief, each strain was typed as VacA single region type s1a, s1b, or s2 by performing three separate PCR assays. Product sizes were differentiated on a 2% agarose gel. The vacA mid-regions were typed as m1 or m2 by performing two separate PCR assays, each with two allelic type-specific primers. Conditions for thermal cycling were 35 cycles of 94˚C for 60 s, 52˚C for 60 s, and 72˚C for 60 s.
2.5. Histopathological Analysis
Histopathological analysis was mainly performed to detect the presence of H. pylori and to ascertain the presence of significant preneoplastic and neoplastic lesions. Two sections of 4 µm size were cut from each block: one section was stained with a modified Giemsa stain, and the other section with Haematoxylin & Eosin (H & E) to assess the presence of intestinal metaplasia and dysplasia. A single pathologist Zakia Abid (Z. A.) who was blinded to the patient’s clinical conditions evaluated all the histologic sections.The results of Histopathology is not included in the study.
3. Results
3.1. Test Specimens
300 biopsies were collected for the study. Of these, 101 patients had peptic ulcer including 61 duodenal ulcers, 25 gastric ulcers (Figures 4 and 5) and 15 complete ulcers; 85 patients had gastritis including 48 supper gastritis, 17
Table 1. Primer used in the study.
atrophic gastritis and 30 erosive gastritis; and 4 patients had gastric adenocarcinoma. All the 300 biopsies were screened for the presence of Helicobacter pylori but only 200 biopsy samples were found to be positive for Helicobacter pylori and confirmed with colony characteristics, Biochemical tests and 16S rRNA PCR amplification using species-specific primers for Helicobacter pylori.
3.2. Biochemical Tests General Procedure
To identify bacteria, we must rely heavily on biochemical testing. The types of biochemical reactions each organism undergoes act as a “thumbprint” for its identification. A series of differential media were inoculated. After incubation, each medium was observed to see if specific end products of metabolism are present. This was done by adding indicators to the medium that react specifically with the end product being tested, producing some form of visible reaction, such as a color change. The results of these tests are then compared to known results for that organism to confirm its identification. The isolated bacteria were tested for the presence of Oxidase, Catalase and Urease enzymes.
Oxidase: Oxidase test was done for the 200 isolates and a positive reaction was indicated by the appearance of purple color in the smeared area within 10 sec. All the 200 isolated were positive for oxidase.
Catalase: Catalase test was done for the 200 isolates and the positive reaction was observed with effervescence and indicated the presence of catalase enzyme. All the 200 isolates were positive for catalase enzyme.
Urease: Urease test was done for the 200 isolates and the positive reaction was indicated by the color change from yellow to pink. All the 200 isolates were positive for urease.
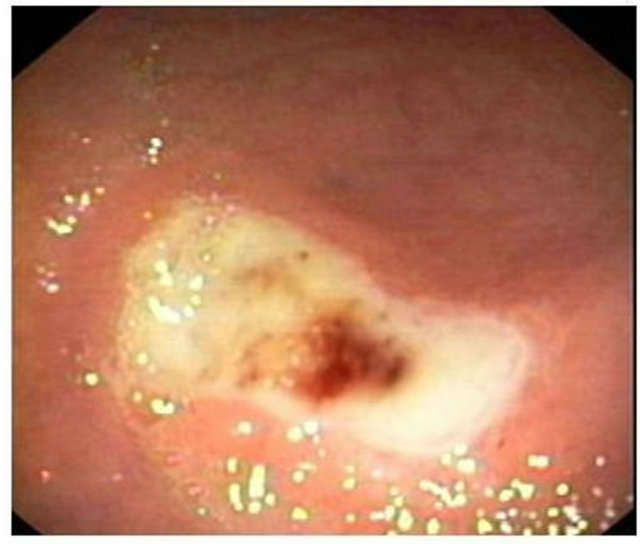
Figure 4. Partial view of gastric ulcer from Helicobacter pylori infected subjects.
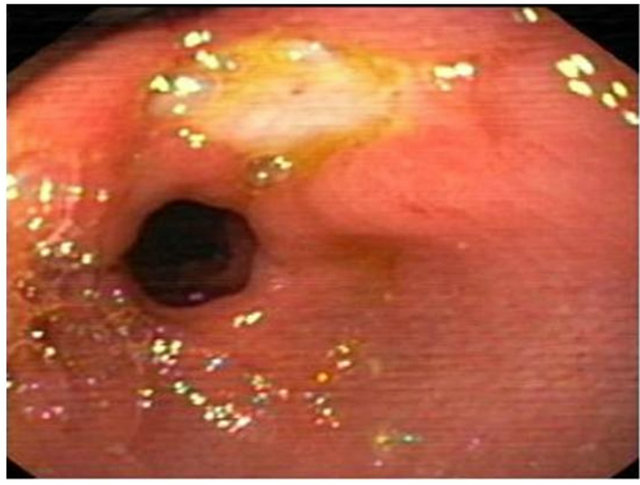
Figure 5. Partial view of doudenal ulcer from Helicobacter pylori infected subjects.
3.3. Helicobacter pylori DNA Isolation
Genomic DNA of H. pylori was isolated from the obtained strains and was detected by running the samples on 1% agarose gel and was observed under UV transilluminator. The shining bands appeared indicate the presence of genomic DNA.
3.4. Confirmation of VacA Subtypes Using Specific Primers
The obtained DNA was confirmed using 16S rRNA primers in the PCR and by 1.5% agarose gel running along with the ladder. The amplification of the DNA indicated the presence of H. pylori genes and the bands appeared were found to be of 534 base pairs when compared with the ladder, which can be seen from the gel image Lane M represents 100 bp molecular markers. Lane represents 1 - 6 16S rRNA amplification of H. pylori DNA isolated from the culture. Lane 7 represents positive control of band at 534 bp Figure 6. The presence of VacA marker was detected by using appropriate primers (Table 1) in the PCR and the products obtained were detected in 1.5% agarose gel and comparing the bands obtained with the ladder used. This can be seen from Figures 7-11. The distribution of alleles was studied from the obtained results. (Table 2) Study of the combination of these alleles (Table 3) can be correlated with the disease status. In H. pylori the VacA alleles are classified in to two regions, namely signal and middle regions indicated by s (signal) and m (middle) respectively. First region (s) contains two types of alleles they are s1, s2 and m1a, m1b, and m2 in mid region. Individually the patient with duodenal ulcers (DU) has prevalence of s1 region (73.77%) than s2 region with (16.3%), mla (11.4%), mlb (32.7%) and m2 (60.6%). In gastric ulcer cases high prevalence of s1 (100%) is observed, s2 being (17.5%). m1a region 1 2 3 4 5 m 6 7 8 9
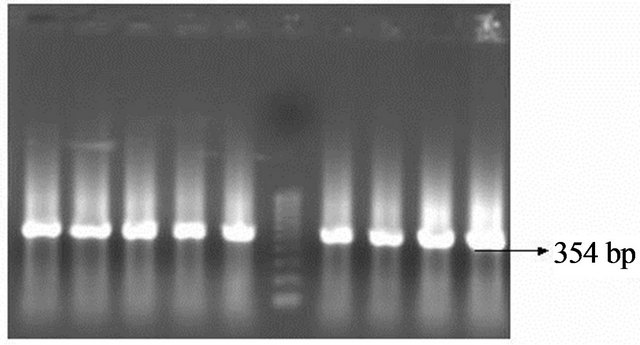
Figure 6. 16S rRNA amplification of Helicobacter pylori DNA. Lanes 1-9 indicate Helicobacter pylori DNA and Lane m indicates molecular wt marker of 100 bp.
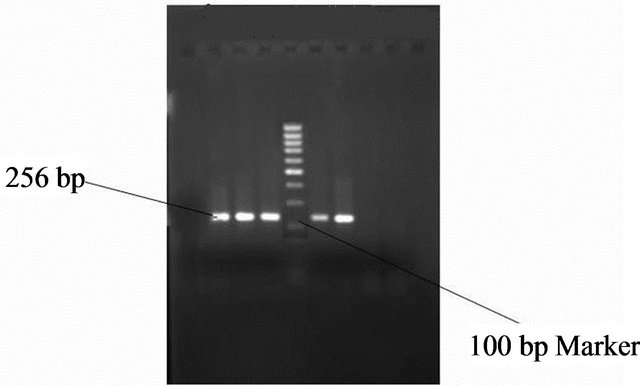
Figure 7. Gel photo showing VacA s1 region of Helicobacter pylori VacA gene.
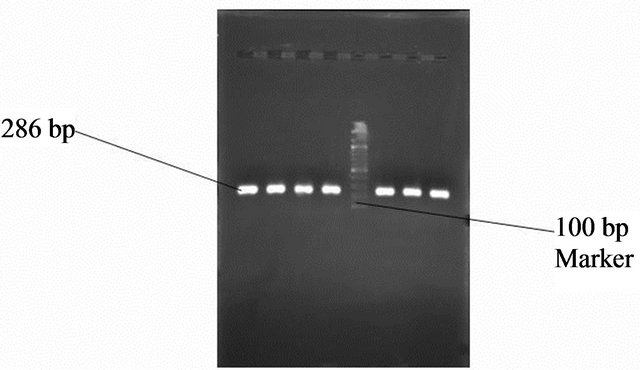
Figure 8. Gel photo showing VacA s2 region of Helicobacter pylori VacA gene.
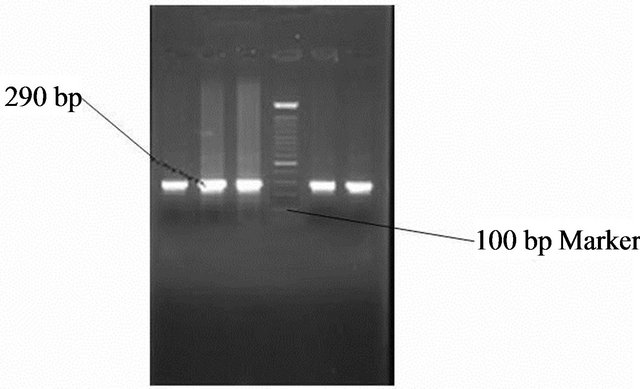
Figure 9. Gel photo showing Vac m1a region of Helicobacter pylori VacA gene.
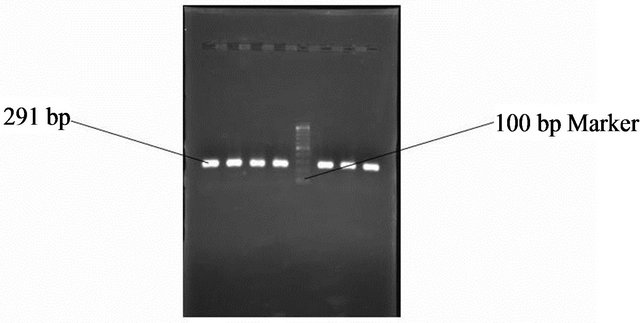
Figure 10. Gel photo showing Vac m1b region of Helicobacter pylori VacA gene.
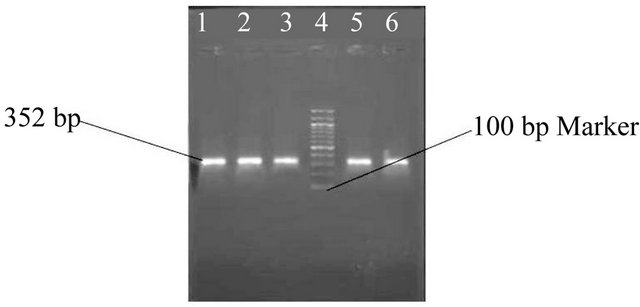
Figure 11. Gel photo showing VacA m2 region of Helicobacter pylori VacA gene.
with (20%), m1b is present in (25%), m2 with (60%). In nonulcer dyspepsia has high prevalence of s1 region (84.2%) s2 (20%) m1a with (47.3%) m1b (31.5%) and m2 (52.6%). In gastric carcinoma s1 region (100%) s2, m1a, m1b, regions are absent m2 is (100%) Table 2.
Table 2. Distribution of s and, m alleles in clinical isolates.
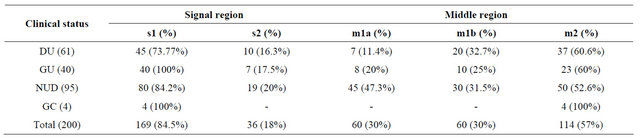
Table 3. Combination of s and m alleles in correlation with clinical status.
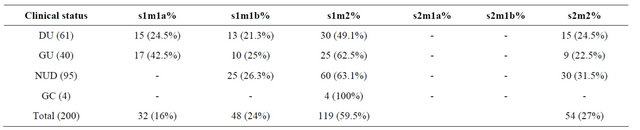
In this study in a total of 200 isolates, s1m2 is a more prevalent (59.5%) in our isolates followed by s2m2 (27%) s1m1b (24%) and s1m1a (16%) in duodenal ulcer cases s1m2 has higher prevalence with (49.1%) than s1m1b with (21.3%) s2m2 with (24.5%) and s1m1a with (24.5%) s2m1a s2m1b are totally absent. In gastric ulcer cases s1m1a has a prevalence of (42.5%), s1m1b has (25%), s1m2 has (62.5%) and s2m2 has (22.5%), while S2m1a, s2m1b are absent. In non-ulcer dyspepsia cases s2m2 has a prevalence of (31.5%), s1m1b (26.3%) and s1m2 has a prevalence of (63.1%) respectively, while s1m1a, s2m1a, s2m1b are absent. In gastric carcinoma cases s1m2 has a prevalence of (100%) while s1m1a, s1m1b, s2m1a, s2m1b and s2m2 are totally absent (Table 3).
4. Discussion
From this study it was observed that s1m2 is more prevalent (59.5%) than s2m2 (27%), s1m1b (24%) and s1m1a (16%). Thus analyses of H. pylori isolates from diverse geographic locations permitted a comprehensive description of the s and m regions of VacA. Type s1 VacA strains are often associated with disease than type s2 strains [20]. Here in our study we observed the same that (84.5%) of our isolates are s1 VacA type and are associated with severe forms of the disease like peptic ulcer (DU + GU), NUD (chronic gastritis) and gastric carcinoma (GC). In the middle region m1 strains are associated with increased gastritis epithelial damage than m2 [10] but in contrary to the above in our isolates we have got 57% of m2 allele than m1 with (30%) m1a + m1b. In correlation with the different combination of VacA we got s1m2 combination at a maximum of (59.5%) followed by s1m1b (24%) and s2m2 (27%), but in reports s1m1 strains produce a higher toxin activity in vitro than s1m2 strains and significantly associate with peptic ulcer disease [21]. Here in our study, we found that s1m2 strains are associated with severe forms of the diseases and the most striking feature of the study was s2m2 type (27%) was equally prevalent with s1m1b (24%) type and pathogenecity is not at all related with any kind of diseases in other parts of world. This study demonstrates that the prevalence of s1 and m2 is high in our isolated strains. Among different types of VacA alleles s1m1a type was minimum, the s1m2 strain has maximum prevalence in duodenal ulcer cases and it is also responsible for the gastric carcinoma, gastric ulcer and nonulcer dyspepsia cases. But the strain s2m1a, s2m1b were totally absent in Hyderabad strains.
How ever studies of H. pylori virulence factors have been difficult to interpret because observations reported for one geographic region or ethnic group have not always been confirmed in different places. It is considered that differences in the frequency of particular H. pylori genotypes could play a role in the incidence of H. pylori associated diseases. Thus, VacA detection might be helpful for determination of which patients are at highest risk for severe clinical outcomes such as duodenal ulcer or gastric carcinoma and eventually, to define strategies for the treatment or prevention of H. pylori infection.
5. Acknowledgements
We would like to thank our colleagues at our department for their fruitful discussion during the research and their technical assistance during manuscript preparation.
REFERENCES
- J. Versalovic and J. G. Fox, “Taxonomy and Phylogeny of Helicobacter,” In: M. Achtman and S. Suerbaum, Eds., Helicobacter pylori: Molecular and Cellular Biology, Horizon Scientific Press, Wymondham, pp. 15-28.
- N. Figura, “Identification of H. pylori Strain or Factors Important in the Development of Duodenal Ulcer Disease,” Helicobacter, Vol. 2, Suppl. 1, 1997, pp. S3- 12.
- M. J. Blaser and J. C Atherton, “Helicobacter pylori Persistence: Biology and Disease,” Journal of Clinical Investigation, Vol. 113, No. 3, 2004, pp. 321-333.
- R. D. Leunk, P. T. Johnson, B. C. David, W. G. Kraft, D. R. Morgan, “Cytotoxic Activity in Broth-Culture Filtrates of Campylobacter pylori,” Journal of Medical Microbiology, Vol. 26, No. 2, 1988, pp. 93-99. doi:10.1099/00222615-26-2-93
- N. Figura, P. Guglielmetti, A. Rossolini, A. Barberi, G. Cusi, R. A. Musmanno, et al., “Cytotoxin Production by Campylobacter pylori Strains Isolated from Patients with Peptic Ulcers and from Patients with chronic Gastritis Only,” Journal of Clinical Microbiology, Vol. 27, No. 1, 1989, pp. 225-226.
- T. L. Cover and M. J. Blaser, “Purification and Characterization of the Vacuolating Toxin from Helicobacter pylori,” The Journal of Biological Chemistry, Vol. 267, No. 15, 1992, pp. 10570-10575.
- J. C. Atherton, P. Cao, R. M. Peek Jr., M. K. Tummuru, M. J. Blaser and T. L. Cover, “Mosaicism in Vacuolating Cytotoxin Alleles of Helicobacter pylori. Association of Specific VacA Types with Cytotoxin Production and Peptic Ulceration,” The Journal of Biological Chemistry, Vol. 270, No. 30, 1995, pp. 17771-17777.
- L. J. Van Doorn, C. Figueiredo, R. Sanna, S. Pena, P. Midolo, E. K. Ng, et al., “Expanding Allelic Diversity of Helicobacter pylori VacA,” Journal of Clinical Microbiology, Vol. 36, No. 9, 1998, pp. 2597-2603.
- R. Morales-Espinosa, G. Castillo-Rojas, G. Gonzalez-Valencia, S. P. de Leon, A. Cravioto, J. C. Atherton, et al., “Colonization of Mexican Patients by Multiple Helicobacter pylori Strains with Different VacA and CagA Genotypes,” Journal of Clinical Microbiology, Vol. 37, No. 9, 1999, pp. 3001-3004.
- J. C. Atherton, R. M. Peek, K. T. Tham, T. L. Cover and M. J. Blaser, “Clinical and Pathological Importance of Heterogeneity in VacA, the Vacuolating Cytotoxin Gene of Helicobacter pylori,” Gastroenterology, Vol. 112, No. 1, 1997, pp. 92-99. doi:10.1016/S0016-5085(97)70223-3
- S. R. Han, H. J. Schrieber, S. Bhadki, M. Loos and M. J. Maeuere, “VacA Genotypes and Genetic Diversity in Clinical Isolates of Helicobacter pylori,” Clinical and Diagnostic Laboratory Immunology, Vol. 5, No. 2, 1998, pp. 139-145.
- Y. Ito, T. Azuma, S. Ito, H. Miyaji, M. Hirai, Y. Yamazaki, et al., “Analysis and Typing of the VacA Gene From CagA-Positive Strains of Helicobacter pylori Isolated in Japan,” Journal of Clinical Microbiology, Vol. 35, No. 7, 1997, pp. 1710-1714.
- J. C. Stephens, A. M. Folwell, R. A. Swann and B. J. Rathbone, “Helicobacter pylori CagA Status, VacA Genotypes and Ulcer Disease [Abstract],” Gut, Vol. 39, Suppl. 1, 1996, p. A2.
- M. Kidd, A. J. Lastovica, J. C. Atherton and J. A. Louw, “Heterogeneity in the Helicobacter pylori VacA and Cag A Genes: Association with Gastroduodenal Disease in South Africa?” Gut, Vol. 45, No. 4, 1999, pp. 499-502. doi:10.1136/gut.45.4.499
- D. Basso, F. Navaglia, L. Brigato, M. G. Piva, A. Toma, E. Greco, et al., “Analysis of Helicobacter pylori VacA and CagA Genotypes and Serum Antibody Profile in Benign and Malignant Gastroduodenal Diseases,” Gut, Vol. 43, No. 2, 1998, pp. 182-186. doi:10.1136/gut.43.2.182
- L. J. Van Doorn, C. Figueiredo, R. Rossau, G. Jannes, M. Van Asbroek, J. C. Sousa, et al., “Typing of Helicobacter pylori VacA Gene and Detection CagA Gene by PCR and Reverse Hybridization,” Journal of Clinical Microbiology, Vol. 36, No. 5, 1998, pp. 1271-1276.
- S. Strobel, S. Bereswill, P. Balig, P. Allgaier, H. G. Sonntag and M. Kist, “Identification and Analysis of a New VacA Genotype Variant of Helicobacter pylori in Different Patient Groups in Germany,” Journal of Clinical Microbiology, Vol. 36, No. 5, 1998, pp. 1285-1289.
- G. Perales, J. Sanchez, A. Mohar, R. Lara-Lemus, A. Hernandez, R. Herreragoepfert, et al., “Single-Step PCR Amplification and Enzyme Restriction Analysis of the Entire Helicobacter pylori Cytotoxin VacA Gene for Genetic Variability Studies,” FEMS Microbiology Letters, Vol. 178, No. 1, 1999, pp. 55-62. doi:10.1111/j.1574-6968.1999.tb13759.x
- A. K. Mukhopadhyay, D. Kersulyte, J.-Y. Jeong, S. Datta, Y. Ito, A. Chowdhury, et al., “Distinctiveness of Genotypes of Helicobacter pylori in Calcutta, India,” Journal of Bacteriology, Vol. 113, No. 3, 2000, pp. 3219-3227. doi:10.1128/JB.182.11.3219-3227.2000
- J. C. Atherton, P. Cao, R. M. Peek, M. K. R. Tummuru, M. J. Blaser and T. L. Cover, “Mosaicism in Vacuolating Cytotoxin Alleles of Helicobacter pylori: Association of Specific VacA Types with Cytotoxin Production and Peptic Ulceration,” The Journal of Biological Chemistry, Vol. 270, No. 30, 1995, pp. 17771-17777.
- L. J. Van Doorn, C. Figueiredo, F. Mégraud, S. Pena, P. Midolo, D. M. Queiroz, et al., “Geographic Distribution of VacA Allelic Types of Helicobacter pylori,” Gastroenterology, Vol. 116, No. 4, 1999, pp. 823-830. doi:10.1016/S0016-5085(99)70065-X
NOTES
*Corresponding author.

