Journal of Cancer Therapy
Vol.1 No.2(2010), Article ID:2050,7 pages DOI:10.4236/jct.2010.12017
Proteomic Profile Modification of Anaplastic Medulloblastoma after in-Vivo Radiotherapy: A Case Study
![]()
1Department of Genetics, Biology and Biochemistry, University of Turin, Turin, Italy; 2Department of Genetics, Biology and Biochemistry, University of Turin, Turin, Italy; 3Division of Pediatric Pathology, Ospedale Infantile Regina Margherita, Turin, Italy; *Corresponding author: Molecular Biotechnology Centre (MBC), Torino, Italy.
Email: cristina.zanini@unito.it
Received April 21st, 2010; revised May 17th, 2010; accepted May 24th, 2010.
Keywords: Proteomics, Anaplastic Medulloblastoma, Radiotherapy
ABSTRACT
Medulloblastoma (MDB) is an aggressive tumor of Central Nervous System (CNS). Radiotherapy after radical surgery has an important role in treatment of standard and high risk patients and is followed by intensive chemotherapy. To explore modifications of protein expression induced by in vivo radiotherapy proteomic analysis was performed on a case of Anaplastic MDB. 2D-gel electrophoresis and MALDI-TOF mass spectrometry detected qualitative differences of protein expression in Anaplastic MDB at diagnosis and in relapse after radiotherapy. Relevant proteomic data were confirmed by western blot and Real-Time PCR analysis, validating the presence of Sthatmin 1 (STMN1), Heat shock protein 60 (HSP60), HSP27 and Disulfide Isomerase (ER60) among the six proteins present in both samples. The most relevant modification induced by radiotherapy was a drastic reduction of the total number of proteins (60.6%) and the appearance of few new proteins. The modifications and the striking simplification of proteins expressed by the tumor after radiotherapy may allow to tailor subsequent chemotherapy on a rational basis. A proteomic guided chemotherapy may be of great benefit to patients.
1. Introduction
Medulloblastoma (MDB) is an aggressive tumor of central nervous system (CNS) located in the cerebellum and represents the more frequent and studied malignant tumor of CNS of paediatric age [1]. MDB less frequently affects adult patients [2,3]. With modern protocols survival is high (70-80%) but neuropsychological damages on developing brain may be severe. Radiotherapy is one of the relevant causes of therapy related brain damage. The aim of modern protocols is to grade tumor aggressiveness relying on solid prognostic factors. The intensity of treatment may be modulated by risk group (low- standard-high). Such an approach may limit the use of radiotherapy to less favorable groups of patients and protect infants and young children from radiotherapic damages [4].
Proteomic studies on MDB were mainly limited to cell lines DAOY and D283 which shared Ded protein, an antiapoptotic principle [5]. Modifications induced in vivo by radiotherapy have not been investigated with a proteomic approach. The presented case offered the unique opportunity to investigate an Anaplastic MDB at diagnosis and at relapse after sole radiotherapy.
2. Materials and Methods
2.1 Patient
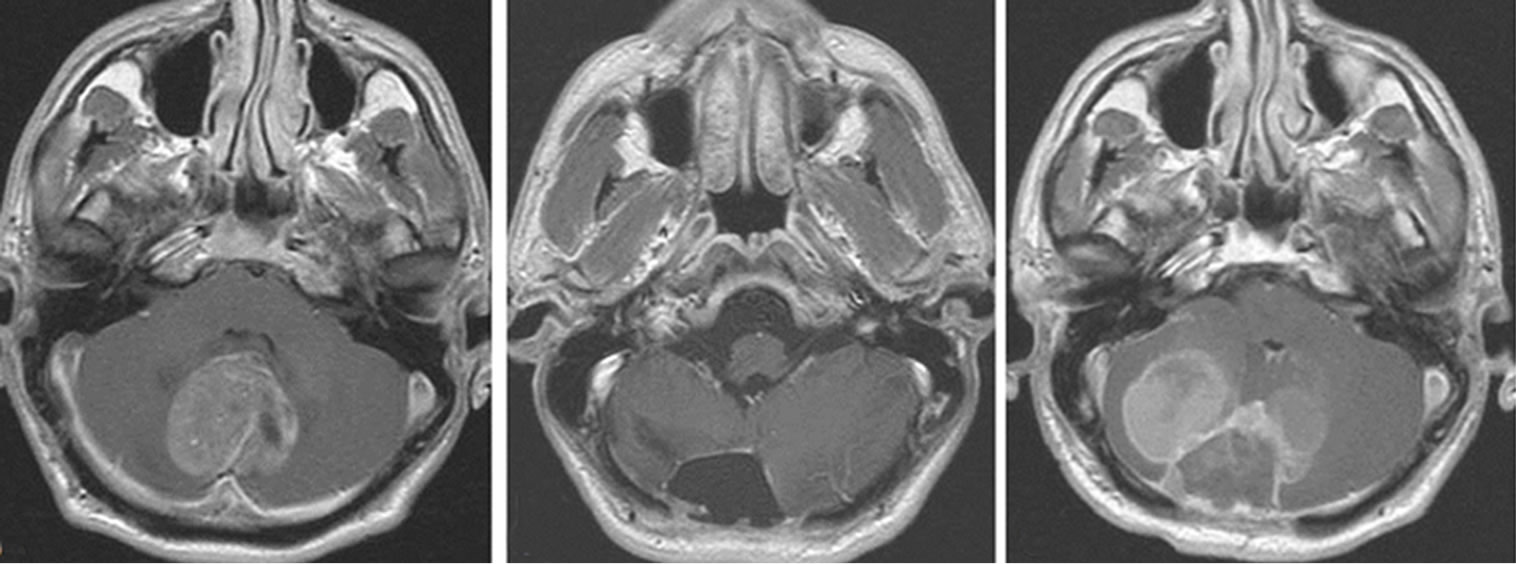
The patient, a 26 year old white male, presented with a large tumor of the cerebellar vermis (Figure 1(a)) which was totally resected (Figure 1(b)). Histological diagnosis was Anaplastic MDB, according to WHO 2007 classification [1]. The patient underwent radiotherapy, which delivered 54 Gray on the whole neuraxis with regression of clinical symptoms. Eight months later a local relapse developed (Figure 1(c)) and a second neurosurgery was performed.
2.2 Proteomics
2D-electrophoresis using pH range of 3-10 (Bio-Rad) was performed as previously described [6] on snap frozen tissue of the primary tumor to detect a complete proteomic profile of the sample. Since the majority of proteins were included in the 4-7 pH range, to analyze the proteins involved in radiotherapy response, further analysis of primary tumor and its relapse was limited to such range. More than 450 detectable protein spot were found on primary tumor and 270 in the relapse gel with the aid of image analysis software PDQuest version 7.2 (Bio-Rad). As previously described [7], the spots excised from 2D-electrophoresis Colloidal Comassie stained gels were destained (in 50% ACN and 5 mM NH4CO3), dehydrated in 100% ACN and digested overnight with 5 µl trypsin solution (0,1 mg/ml trypsin and 5 mM NH4CO3). The mass spectrometer used in this work was a MALDI-TOF (MALDI micro MX, Waters, MA, USA) with a delayed extraction unit. Peptide spectra were obtained in reflection mode in the range 800-3000 Da. Database searching was performed using the 25 most intense measured peptide masses against the Swiss-Prot database (one missed cleavage with the trypsin enzyme selected, oxidation of methionine as potential variable modification, peptide tolerance of 100 ppm, taxa human) using the free search program MASCOT (http://www.matrixscience.com).
Only protein identifications with significant Mascot score (p < 0.05) were taken into consideration.
2.3 Western Blotting
The presences of Stathmin 1 (STMN1) was investigated by western blot. Protein samples (20 µg) were separated in a 10% polyacrylamide gel and transferred to nitrocellulose membrane. STM1 rabbit polyclonal antibody (Cell Signaling) was diluited using manufacturer's instruction and incubated overnight at 4˚C. After incubation with the appropriate secondary antibody, immunoreactive bands were detected with an enhanced chemiluminescence (EuroClone). The membranes were after stripped and reprobed for β-Tubulin (rabbit monoclonal, 9F3; Cell Signaling, 1:1000 diluition) as a protein loading control.
2.4 Real Time PCR
The presence of Heat Shock Proteins 27 (HSP27), HSP60, ER60 in both tumor samples was investigated by Real-Time PCR analysis in order to validate proteomic findings. Ribosomal protein (S14) was used as reference gene to normalize cDNA across samples. Primers were designed using Primer 3 with the Beacon Designer Software (PREMIER Biosoft International) for: 1) HSP27 forward GCTGACGGTCAAGACCAAGG-3’ and reverse 5’-ATGTAGCCATGCTCGTCCTG-3’; 2) HSP60 forward 5’-GCTGAAGATGTTGATGGAGA-3’ and reverse 5’-TTGTCACCAAACCCTGGAGC-3’; 3) ER60 forward 5’-TTCCTGTTGCTATCAGAACTG3’ and reverse 5’-ACCTCTCCAGAGCCTTCCCA-3’. Realtime-PCR was performed with the ABI PRISM 7900 Ht Applied BioSystem instrument.
3. Results and Discussion
The present case offered the unique opportunity of study the original tumor (Figure 1(a)) that underwent radical resection (1(b)) and relapse (1(c)) after radiotherapy. MRI control scan, two weeks after second surgery, detected dorso-lumbar leptomeningeal seeding. Chemotherapy was started with Cisplatinum and Vincristin. Despite chemotherapy the patient died few weeks later.
To investigated radiotherapy response proteomic analysis of primary tumor and relapse was performed. Figures 1 (d) and (e) demonstrated that these two tissue samples (Anaplastic Medulloblastoma at diagnosis and at relapse) presented remarkable different proteomic patterns, with striking difference in the distribution and number of proteins or spot intensity. From gel comparison sixty-one proteins (Tables 1-3) appeared differentially expressed between primary tumor and its relapse after radiotherapy. Among the proteins identified, 37 (60.6%) proteins were only present in the original tumor and lost after radiotherapy (Table 1), and 18 (29.5%) were only found in the relapse tumor after radiotherapy (Table 2); 6 (9.8%) proteins were actually present in both samples (Table 3). Several protein biologically related to MDB growth and aggressiveness were present only in the primary tumor: these included MEIS1, which has DNA-binding property and a role in embryogenesis and in CNS development [8], Annexin VIII involved in regulation of process of apoptosis, some Heat Shock Proteins (HSP70 protein 5 and 8) and several enzymes or enzyme inhibitors (Peroxiredoxin-2, Alpha-1-antiproteinase, alfaenolase, Cyclin dependent kinase 9, GTPase activating protein, etc). An impressive modification of the protein repertoire was induced by radiotherapy. Only six proteins were expressed in both tumor samples (Table 3). From these STM1, HSP60, HSP27 and ER60 were further investigated. The presence in both samples of STMN1 was confirmed by western blot (Figure 2(a)), while that of HSP27, HSP60 and Disulfide Isomerase by Real-Time PCR. Figure 2(b) illustrated the ratio between messenger RNA levels in Anaplastic MDB and levels in MDB after radiotherapy. In agreement to proteomic data, HSP27, HSP60 and ER60 proteins were expressed in both tumor with the similar levels. These conserved proteins, which possibly represented an essential protein asset linking the original tumour to the post radiotherapy relapse, were considered of biological relevance. STM1, a cytosolic phosphoprotein involved in cell proliferation and mobility, has been reported as a marker of aggressiveness in MDB [9]. HSPs originally described as chaperonines also play a role in tumor development and progression in several tumors [10] with possible modulated influence depending on histotype. ER60 showed a role in protein folding in Endoplasmic Reticulum and, with its precursor ER57, interacts with important signalling targets and III
 (a)
(a)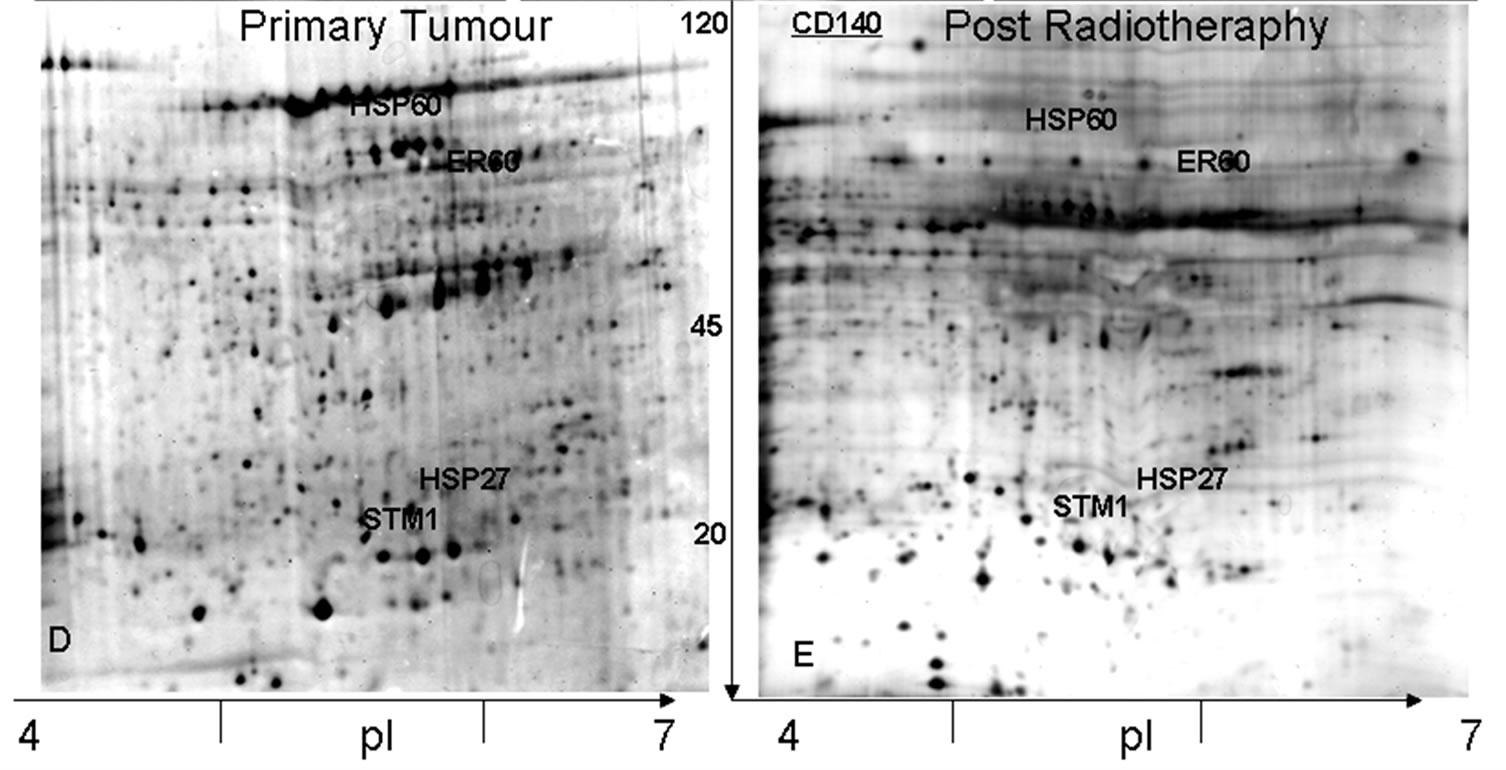 (b)
(b)
Figure 1. NMR images of the patient. (a) at diagnosis T1- weighted NMR demonstrating a large vermian mass; (b) after radical surgery no residual tumor is detectable; (c) at relapse tumor infiltrates cerebellar hemispheres. Comassie blue stained 2D-electrophoresis patterns of Anaplastic MDB. 2D-electrophoresis was performed on immobilized pH 4-7 strips, followed by SDS-PAGE on 10%, 7cmx8cm gels. (d) 2D-electrophoresis pattern of primary tumor shows a large number of proteins. (e) At relapse tumor 2Delectrophoresis demonstrates an impressive reduction of the protein profile. Proteins validate by other techniques are marked by name. Total spots excised from D and E are reported in details in Table 1
beta Tubulin [11].
Radiotherapy has a key role in the treatment of MDB in children as well as in adult patients [12,13] and almost all protocols for high risk patients include radiotherapy treatment as a second step, after radical surgery, followed by chemotherapy. Tumoral cell death induced by radiotherapy is obtained by DNA damages but relevant differences in radiosensitivity were observed in tumors of different histotype and the factors conferring resistance to radiotherapy are not completely understood [14]. MDB is a radiotherapy sensitive neoplasia but mechanism that modulate cell resistance in vivo are not well known. A proteomic approach has been utilized on epithelial cancers to correlate protein expression to response to radiotherapy [15,16]. These studies on colonic or breast cancer did not analyze protein expression modifications on RT treated tumors in vivo. Recently, utilizing a murine model of MDB and MDB cell lines, tumor cell modifications after the exposure to a course of total body radiation were investigated [17]. This study re
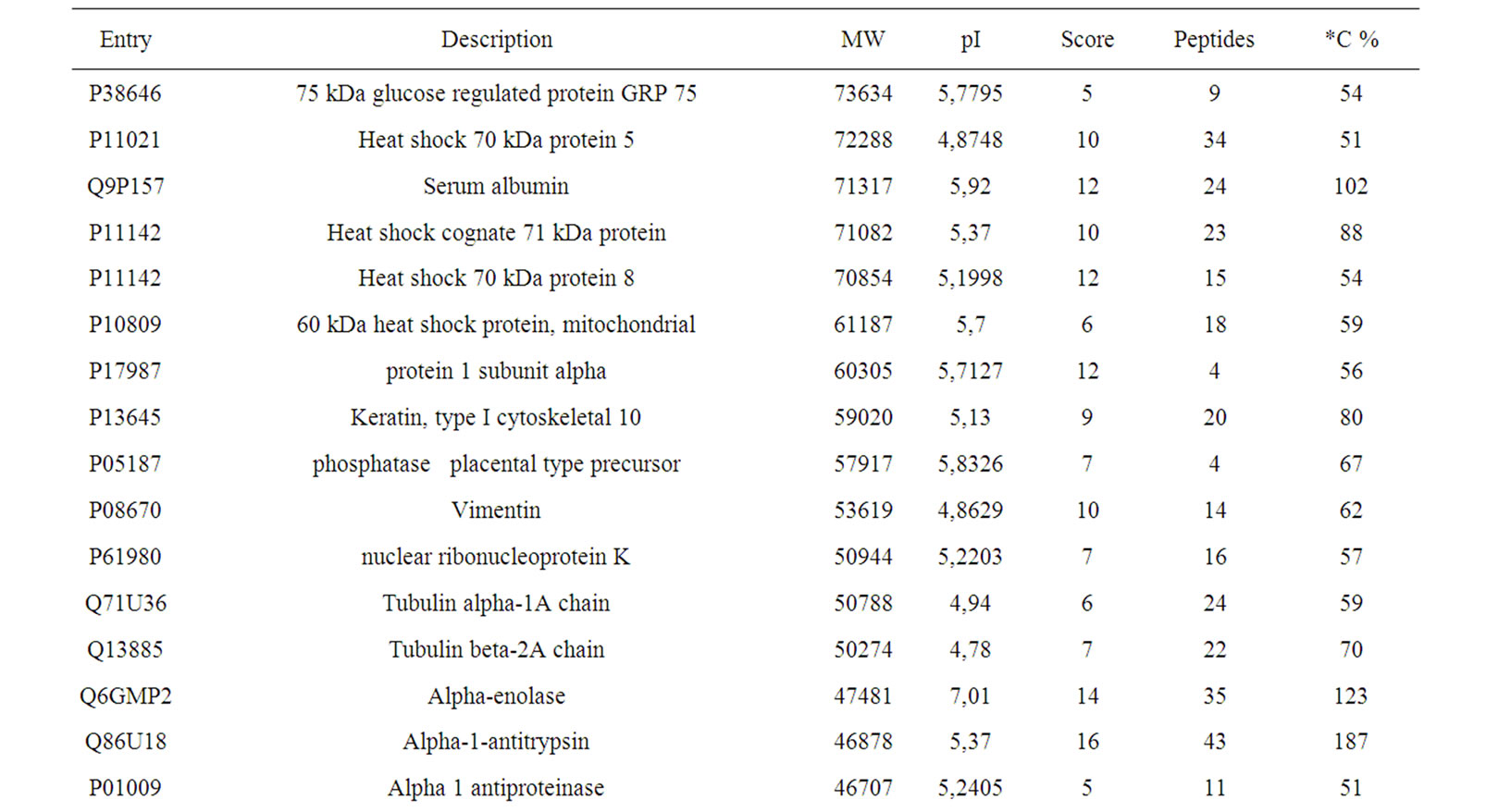


Table 1. Proteins identified by MALDI-TOF only on the sample of the primary tumor
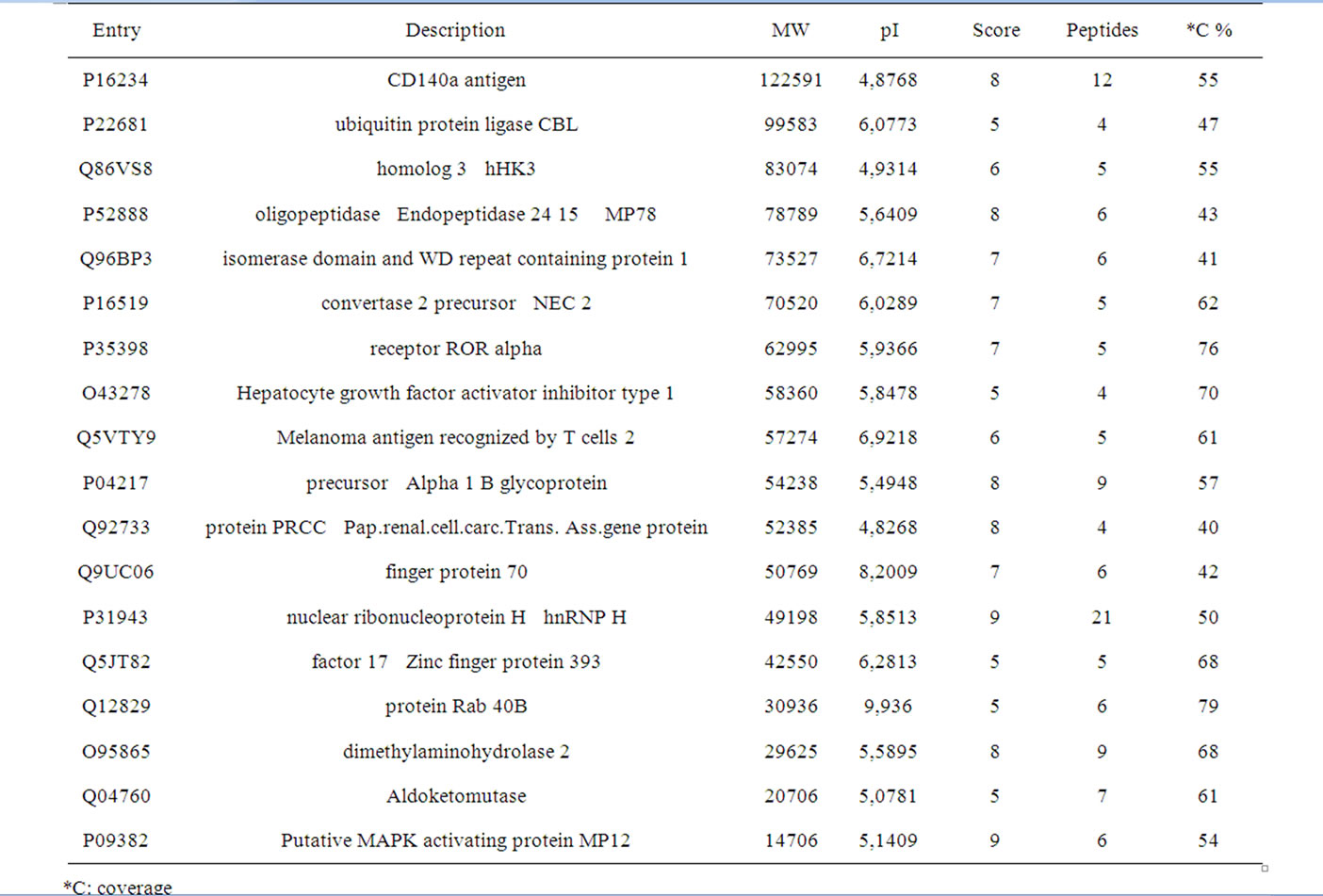
Table 2. Proteins identified by MALDI-TOF only on the post-radiotherapy sample
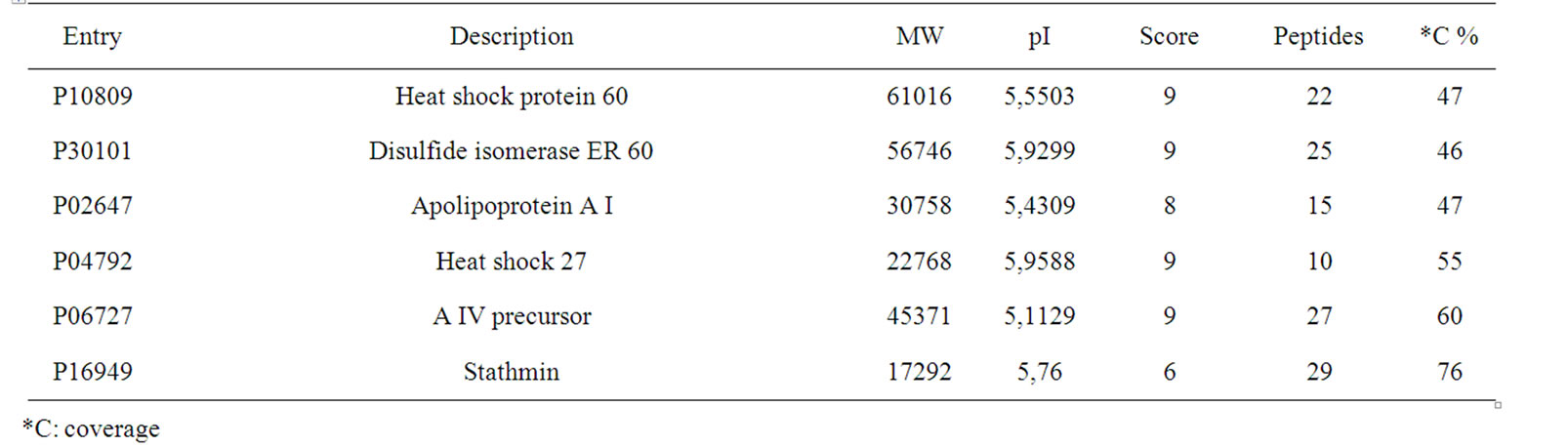
Table 3. Proteins identified by MALDI-TOF in both tumor samples
vealed important intracellular signalling modifications in tumor cells with activation of PI3K pathway. Experimental data may not be directly compared to the in vivo effects of radiotherapy on human patients due to higher complexity of histology in human MDB [18,19] and the sophisticated approach of patients radiotherapy treatment. Modifications of protein expression induced in vivo by radiotherapy alone on human MDB are practically unknown. The case presented was someway unique because the relapse occurred after radiotherapy alone without any contribution of chemotherapy. The radiotherapy induced differential protein expression showed a somewhat new scenario with three relevant aspects: 1) a reduced asset of proteins in MDB after radiotherapy, 2) the appearance of new proteins and 3) few conserved proteins. A better understanding of radiotherapy induced changes on MDB cells may offer the opportunity of a more rational selections of drug regimens or suggest innovative approaches
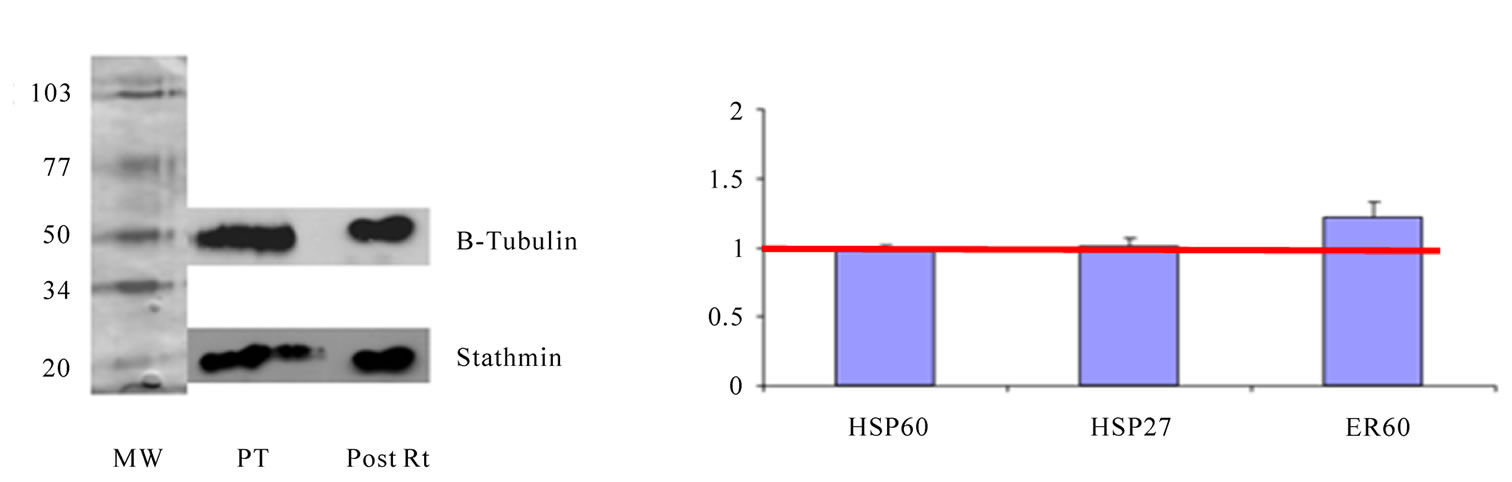
Figure 2. (a) Western blot analysis of primary tumor and post radiotherapy (Rt) stained by anti-STM1 and anti-β-Tubulin; (b) Real-Time PCR analysis of proteins expressed in both tumor in primary tumor and post radiotherapy. Results are present as fold increase in post radiotherapy vs primary tumor control. Data presented were normalized to S14 mRNA expression and are mean ± SD of three experiments
taking advantage of the reduced protein expression after radiotherapy [14].
Data presented confirmed the importance of Sthatmin as a marker of aggressiveness in Anaplastic MDB as this protein was one of the few proteins expressed in both of the tumor samples.
In fact the radiotherapy induced changes of protein expression, as evidenced by proteomic analysis, may represent an escape strategy of the tumor to survive to radiotherapy and a reversal to a more undifferentiated, stem cell-like status. On the other hand, the new protein expression patterns displayed by radiotherapy treated cells may modify cell sensibility to specific cytotoxic drugs or suggest innovative treatment strategies as the findings on the mouse model shows [17]. The expression of CD140a (PDGFRa) may be of particular interest as one of the possible target of anti-receptor small molecules of recent development [20].
4. Conclusions
Proteomic analysis of post “in vivo” radiotherapy of Anaplastic Medulloblastoma disclosed relavant modifications of protein profile. The reduction of proteins present in the relapse after radiotherapy alone was impressive: almost 40% of the total protein repertoire was lost and few “new” proteins were detected. The expression of CD 140 as a new radiotherapy induced protein could be of particular interest as a possible target of anti-receptor therapeutic approach. A proteomic guided chemotherapy based upon protein depauperation induced by radiotherapy could take advantage of the limited protein expression and target these proteins, precluding to the tumor any survival alternative.
A detailed study on a large number of cases may disclose new opportunities for combined therapy based on solid scientific data of radiotherapy effects and not only on long lasting empirical experience.
5. Ackowledgments
This study was partially supported by the “ONCOLOGY SPECIAL PROJECT” Compagnia di San Paolo/FIRMS and Regione Piemonte, Turin, Italy and by UGI-Unione Genitori Italiani contro il tumore dei bambini-ugi@ugitorino.it.
REFERENCES
- F. Giangaspero, C. G. Eberhart, H. Haapasalo, T. Pietsch, O. D. Wiestler and D. W. Ellison, “WHO Classification of Tumors of Central Nervous System,” In: D. N. Louis, H. Ohgaki, O. D. Wiestler and W. K. Cavenee, Eds., IARC Press, Lyon, 2007, pp. 132-140.
- C. Ang, D. Hauerstock, M. C. Guiot, et al., “Characteristics and Outcome of Medulloblastoma in Adults,” Blood Cancer, Vol. 51, 2008, pp. 603-607.
- L. Riffaud, S. Saikali, E. Leray, et al., “Survival and Prognostic Factors in a Series of Adults with Medulloblastomas,” Journal of Neurosurgery, Vol. 111, 2009, pp. 478-487.
- A. Gajjar, M. Chintagumpala, D. Ashley, et al., “Riskadapted Craniospinal Radiotherapy Followed by HighDose Chemotherapy and Stem-Cell Rescue in Children with Newly Diagnosed Medulloblastoma (St Jude Medulloblastoma-96): Long-Term Results from a Prospective, Multicentre Trial,” Lancet Oncology, Vol. 7, 2006, pp. 813-820.
- A. Peyrl, K. Krapfenbauer, I. Slavc, et al., “Protein Profiles of Medulloblastoma Cell Lines DAOY and D283: Identification of Tumor-Related Proteins and Principles,” Proteomics, Vol. 3, 2003, pp. 1781-1800.
- G. Barbero, F. Carta, G. Giribaldi, et al., “Protein/RNA Coextraction and Small Twodimensional Polyacrylamide Gel Electrophoresis for Proteomic/Gene Expression Analysis of Renal Cancer Biopsies,” Analytical Biochemistry, Vol. 349, 2006, pp. 62-71.
- F. Carta, P. P. Demuro, C. Zanini, et al., “Analysis of Candidate Genes through a Proteomicsbased Approach in Primary Cell Lines from Malignant Melanomas and their Metastases,” Melanoma Research, Vol. 15, 2005, pp. 235-244.
- M. Mojsin, M. Stevanovic, “PBX1 and MEIS 1 Upregulate SOX3gene Expression by Direct Iteraction with a Consensus Binding Site within the Basal Promoter Region,” Biochemical Journal, Vol. 425, 2009, pp. 107- 116.
- M. F. Kuo, H. S. Wang, Q. T. Kuo, et al., “High Expression of Stathmin Protein Predicts a Fulminant Course in Medulloblastoma,” Journal of Neurosurgery: Pediatrics, Vol. 4, 2009, pp. 74-80.
- C. Zanini, G. Giribaldi, G. Mandili, et al., “Inhibition of Heat Shock Proteins (HSP) Expression by Quercetin and Differential Doxorubicin Sensitization in Neuroblastoma and Ewing’s Sarcoma Cell Lines,” Journal of Neurochemistry, Vol. 103, 2007, pp. 1344-1354.
- Y. J. Qi, Q. Y. He, Y. F. Ma, et al., “Proteomic Identification of Malignant Transformation-Related Proteins in Esophageal Squamous Cell Carcinoma,” Journal of Cellular Biochemistry, Vol. 104, 2008, pp. 625-635.
- U. Selek, F. Zorlu, P. Hurmuz, et al., “Craniospinal Radiotherapy in Adult Medulloblastoma,” Strahlentherapie und Onkologie, Vol. 183, 2007, pp. 236-240.
- F. Spreafico, M. Massimino, L. Gandola, et al., “Survival of Adults Treated for Medulloblastoma Using Paediatric protocols,” European Journal of Cancer, Vol. 41, 2005, pp. 1304-1310.
- B. B. Ma, R. G. Bristow, J. Kim, L. L. Siu, “Combined modality Treatment of Solid Tumors Using Radiotherapy and Molecular Targeted Agents,” Journal of Clinical Oncology, Vol. 21, 2003, pp. 2760-2776.
- A. S. Allal, T. Kahne, A. K. Reverdin, et al., “Radioresistance-Related Proteins in Rectal Cancer,” Proteomics, Vol. 4, 2004, pp. 2261-2269.
- L. Smith, O. Qutob, M. B. Watson, et al., “Proteomic Identification of Putative Biomarkers of Radiotherapy Resistence: A Possible Role of the 26S Proteasome?” Neoplasia, Vol. 11, 2009, pp. 1194-1207.
- D. Hambardzumyan, O. J. Becher, M. K. Rosenblum, et al., “PI3K Pathway Regulates Survival of Cancer Stem Cells Residing in the Perivascular Niche Following Radiation in Medulloblastoma in Vivo,” Genes, Vol. 22, 2008, pp. 436-448.
- C. S. McManamy, J. Pears, C. L. Weston, et al., “Nodule Formation and Desmoplasia in MedulloblastomasDefining the Nodular/Desmoplastic Variant and its Biological Behavior,” Brain Pathology, Vol. 17, 2007, pp. 151-164.
- F. J. Rodriguez, C. Eberhart, B. P. O’Neill, et al., “Histopathologic Grading of Adult Medulloblastomas,” Cancer, Vol. 9, 2007, pp. 2557-2565.
- W. N. Hait, “Targeted Cancer Therapeutics,” Cancer Research, Vol. 69, 2009, pp. 1263-1267.

