Agricultural Sciences
Vol.08 No.10(2017), Article ID:80053,22 pages
10.4236/as.2017.810086
Horticultural Production of Ultra High Resveratrol Peanut
Godson O. Osuji*, Paul Johnson, Eustace Duffus, Sela Woldesenbet, Jeneanne M. Kirven
Plant Systems, College of Agriculture and Human Sciences, Prairie View A&M University, Prairie View, USA
Copyright © 2017 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY 4.0).
http://creativecommons.org/licenses/by/4.0/



Received: July 6, 2017; Accepted: October 28, 2017; Published: October 31, 2017
ABSTRACT
Background: Resveratrol naturally occurring antioxidant in peanut (Legume: Arachis hypogaea) has phytochemical human health dietary effects associated with reduced inflammatory cancer risks. Its levels in peanut are ultra-low and variable (0 to 26 µg∙g−1), which has made it difficult to market as a consistent high resveratrol produce. Objective: Understanding the regulation of resveratrol accumulation in peanut might lead to development of new techniques for optimizing and stabilizing its yield. Method: Peanuts were cultivated in horticultural field plots and treated with solutions of mineral salts (sulfate, potassium, phosphate, ammonium ion) that were optimized in stoichiometric (reactive) ratios. Peanut seed’s RNAs were subjected to Northern blot analysis for profiling the RNAs synthesized by glutamate dehydrogenase (GDH), and mRNAs encoding resveratrol synthase. The seed’s extracts were analyzed by GC-MS for determination of the resveratrol and fatty acid compositions. Result: Stoichiometric mixes of mineral ions induced the peanut GDH to synthesize some RNA that silenced the mRNAs encoding resveratrol synthase, phosphoglucomutase, isocitrate lyase, malate synthase, enolase, phosphoenolpyruvate carboxylase, malate dehydrogenase, and phosphoglycerate mutase in the control, KN-, and NPKS-treated but not in the NPPK-treated peanut. These resulted to decreased resveratrol content (6.0 µg∙g−1) in the control peanut but maximized it (1.15 mg∙g−1) in the NPPK-treated peanut. Therefore, resveratrol accumulation was optimized by coupling of glycolysis and citric- glyoxylic acid cycles to resveratrol biosynthesis. Fatty acid content of control (55.6 g∙kg−1) was higher than the NPKS-treated (48.5 g∙kg−1) and NPPK- treated peanut (44.9 g∙kg−1) meaning that malonyl-CoA intermediate in both fatty acid and stilbenoid pathways was diverted to support maximum resveratrol biosynthesis in the NPPK-treated peanut. Conclusion: The functional coupling of citric-glyoxylic acid cycles and glycolysis to optimize resveratrol biosynthesis may encourage development of horticultural technology specific for production of ultra-high resveratrol peanuts.
Keywords:
GC-MS, Fatty Acids, Glycolysis, Glutamate Dehydrogenase, mRNA Silencing, Mineral Salt Antagonism, Resveratrol Synthase

1. Introduction
Peanut (Arachis hypogaea) belongs to the botanical family Fabaceae (also known as Leguminosae, and commonly known as the bean or pea family). Like most other legumes, peanuts harbor symbiotic nitrogen-fixing bacteria in root nodules. Peanut is known for its food and dietary values, peanut fatty acid is the most superior vegetable oil. Peanut proteins are very rich in the essential amino acids; vitamins, minerals, and medicinal nutraceuticals are abundant, resveratrol is the most studied [1] [2] [3] [4] . Resveratrols are polyphenolic antioxidants detected in peanut, Japanese knotweed, grapes and berries. They are synthesized by resveratrol synthase. Resveratrols exist as several isoforms, with the trans- isoform being most abundant. Resveratrol has various phytochemical human health beneficial effects associated with long life extension, increased antioxidant activity, reduced inflammatory cancer risk, reduced cardiovascular disease [5] [6] ; its effect on metabolic syndrome [7] is the best characterized [8] [9] . Of increasing human health interest is pterostilbene, a methylated resveratrol compound present in blueberries [10] . Pterostilbene reduced the cholesterol levels in hamsters [11] , and ameliorated diabetes, neurological and vascular diseases [12] . Resveratrol 3-O-β-D-glucoside is credited with anti-HIV activity [13] [14] . The societal importance of resveratrol is highlighted in red wine which contains up to 5.0 mg/L resveratrol [15] . The human health beneficial link between red wine consumption and the resveratrol is an expanding area of conversation in the public domain [16] .
Peanut seeds and leaves have ultra-variable contents (0 to 26 µg∙g−1 fresh wt.) of resveratrol depending on the agronomic conditions of cultivation [17] [18] [19] . The molecular basis for the variability has not been understood, but the variability has prevented the marketing of high resveratrol peanut as a consistent added value dietary produce. Peanut is the most popular nut-snack in the world [20] . Therefore, several attempts are in progress to stabilize and enhance the resveratrol contents of peanut [21] . Treatment of peanut with microbes increased resveratrol although such treatment could leave the product unsafe and inedible [22] . UV light irradiation of peanut suspension cells and tissues, and abiotic stressing of wounded seeds have elicited resveratrol accumulation [23] . Genetic engineering of microbes and plants [24] , chemical synthesis [25] and biotechnology [26] [27] have been applied to produce cost effective resveratrol concentrations that are focused for the food industry and commerce. However, use of cultured tissue and recombinant strategies are yet to gain extensive acceptance among consumers of dietary natural products.
Resveratrol biosynthetic pathway starts from the conversion of phenylalanine to p-coumaroyl-Coenzyme A; the condensation of which with malonyl-Coenzyme A by resveratrol synthase produces resveratrol [28] . Metabolism of phenylalanine provides p-coumaroyl-CoA, whereas fatty acid biosynthesis provides the malonyl-CoA [29] . The derivation of phenylalanine from phosphoenolpyruvate suggests possible committed upstream involvement of glycolytic, citric, and glyoxylic acid cycles [30] in the optimization of resveratrol accumulation. Resveratrol accumulation is regulated by aluminum chloride [31] , and plant defense compounds [4] [18] [21] [32] , but regulation by fertilizer mineral ions was not studied. It is necessary to study mineral ion regulation synergism-antagonism [33] of resveratrol accumulation because peanuts are naturally cultivated in field plot stress conditions. Transcriptional regulation of resveratrol synthase activity has been confirmed in plant systems [18] [21] [32] ; the peanut resveratrol synthase being differentially controlled by a family of genes [33] . Identification of the physiological machines that regulate the enzyme would expand the understanding of the biosynthetic pathway; and might lead to improved horticultural approaches to maximize the accumulation of this valuable nutraceutical in other crops as well.
Our earlier studies had demonstrated the responses of peanut mRNAs encoding primary metabolic enzymes (phosphoglucomutase, phosphoglyceromutase, enolase, phosphoenolpyruvate carboxylase, malate dehydrogenase etc.) to those mineral salt combinations that induced the synthesis of some RNA by glutamate dehydrogenase (GDH) [30] [34] [35] [36] [37] . GDH is a target site of mineral ion action in plants [38] . Our earlier studies on GDH kinetics had observed the signal integration/discrimination [39] and biomass enhancement [34] functions of the enzyme. The principles and practice of the induction of GDH-synthesized RNAs have been applied to explain some hitherto inexplicable biological phenomena including metabolic detoxification of xenobiotics in plants [40] , regulation of fatty acid contents by mRNA encoding lipoxygenase [41] ; the regulation of cellulosic biomass and fatty acid accumulation by mRNAs encoding acetyl CoA carboxylase, nitrate reductase, phosphate translocator [35] . Most of the research on peanut had focused on genetic and plant breeding scientific logics. The method in plant biochemistry adopted hereunder was different being based on horticultural regulation of plant metabolism [35] . The aim was to identify target step(s) in intermediary carbon-nitrogen metabolism that regulate resveratrol accumulation. Peanuts enriched in resveratrol would further stabilize the incomes of limited resource farmers who cultivate the crop. In the light of transcript silencing by homologous RNAs synthesized by GDH [34] [42] , the theoretical logic is that induction of GDH isomerization and RNA synthesis by horticulturally optimized mineral ion treatments of peanut will coordinate the expression of resveratrol synthase mRNAs with the mRNAs encoding some enzymes in intermediary carbon-nitrogen metabolism, leading to production of several peanut metabolic variants that are substantially enriched with resveratrol. Results presented hereunder show that treatment of peanut with stoichiometric combinations of macro-mineral ions induced GDH to synthesize some RNAs that regulated the abundance of mRNAs encoding resveratrol synthase and several enzymes in intermediary carbon-nitrogen metabolism [43] - [48] thereby producing ultra-high resveratrol peanuts.
2. Materials and Methods
Treatment of peanuts with stoichiometric mixes of mineral salt solutions: Peanut (Arachis hypogaea L. Cv. Virginia) seeds were planted in 120 × 120 × 30 cm (width x length x depth) horticultural boxes, each filled with mixture of two bags (18 kg) top soil with compost (Landscapers Pride, New Waverly, Texas, USA) and three bags (2.8 cu ft.) of professional growing mix (Sungro Horticulture, Bellevue, Washington, USA). Each box was set up on level ground in the field on weed-blocking plastic mat, in the University farm, Waller County, Texas, USA. About 25 - 30 seeds were planted per box in early June. There was replanting to make up for ungerminated seeds. The applied mineral salt compositions were based on stoichiometric combinations to mimick the binomial subunit polypeptide compositions of the GDH isoenzymes [36] and to interact with target molecules in molar ratios. Mineral salt compositions and solutions (Table 1) were prepared as described [34] . Weeds were manually removed. Commercial fertilizer (N:P:K: 13:13:13) from American Plant Food Corporation, Texas, USA was included in the study. Treatments were in triplicate boxes, and were watered every other day. Mineral solutions were applied three times, first at pre-flowering stage (2 weeks after seed germination), second at flowering, and finally at post- flowering. When the leaves turned yellow (peanut maturity), pods were harvested, allowed to dry on the greenhouse floor for about 2 weeks, weighed, shelled by hand, and the kernels (seeds) and shells weighed separately. Seeds were stored at −30˚C.
Purification and assay of GDH: GDH was purified by electrophoresis [37] from peanut seeds harvested from the control or mineral salts-treated boxes. RNA synthetic activity of GDH [36] was assayed in combined deamination and amination substrate solutions of 0.1 M Tris-HCl buffer (pH 8.0) containing the four NTPs (0.6 mM each), CaCl2 (3.5 mM), L-glu (3.23 µM), NAD+ (0.375 µM), NH4Cl (0.875 mM), α-ketoglutarate (10.0 mM), NADH (0.225 mM), 5 Units RNase inhibitor, 1 Unit DNase 1, and 5 µg of actinomycin D. Reaction was started by adding 0.2 mL of whole gel-eluted GDH charge isomers containing 3 - 9 µg protein per mL. Final volume of the reaction was brought to 0.4 mL with 0.1 M Tris-HCl buffer pH 8.0. Reactions were incubated at 16˚C overnight and stopped by phenol-chloroform (pH 5.5) extraction of the enzyme. RNA was precipitated with ethanol, and dissolved in minimum volume of molecular biology quality water. RNA yield and quality were determined by photometry and by agarose gel electrophoresis. Assays were carried out in duplicate to verify the reproducibility of the results. The GDH isoenzyme patterns for each of the triplicate
Table 1. Hierarchical responses of peanut yield to stoichiometric mixes of mineral ions to produce metabolic variants of peanut that are linked at the molecular level by GDH- synthesized RNA.
seed yields per treatment of peanut were determined. Replicate seed yields that gave similar/identical GDH patterns per experimental treatment were combined and used for metabolic, total RNA, and molecular analyses.
Metabolic analyses: Dry and milled (composited) seeds (100 g) per experimental treatment, sent to Intertek American Analytical Chemistry Laboratories, Champaign, IL USA were custom analyzed for resveratrol and for fatty acid compositions. Authentic standards applied in the HPLC (Shimadzu 6) for resveratrol were emodin at 19.8 ppm, pterostilbene at 191.6 ppm, trans-resveratrol at 181 ppm, transresveratrol-3-D-glucoside at 6 ppm, and cis-resveratrol at 20 ppm. Supelco mixed fatty acids methylene chloride 37 (FAME 37) standards for the GC 10 Agilent 6890_3 fatty acid fractionation were applied at 200 ppm. EZChrom Elite Version 3.3.2 SP2 software was used to acquire and to process the chromatograms and spectra.
Total RNA: Total RNA was extracted from peanut seeds harvested from the control or mineral salt-treated boxes using the acidic phenol/chloroform (pH 4.5) method [49] .
Complementary DNA synthesis, cloning, characterization, and probe selection: cDNAs were synthesized with 2 µg of each product RNA synthesized by the whole gel-eluted GDH charge isomers using random hexamer primer. Restriction fragment PCR amplification; adapter ligation; sequencing gel fractionation; and purification of cDNA fragments [31] were conducted according to the methods of Display Systems Biotech, Vista, CA, USA. Selected cDNA fragments were subcloned into pCR4-TOPO vector and transformed into TOP10 One Shot Chemically Competent Escherichia coli (Invitrogen, Carlsbad, CA), followed by overnight growth on selective plates. Up to ten positive transformant colonies were picked per plate and cultured overnight in LB medium containing 50 µg/mL of kanamycin. Plasmid DNA was purified with a plasmid kit (Novagen, Madison, WI). The insert cDNA was sequenced with T3 and T7 primers by Functional Biosciences, Inc. (Madison, WI, USA). To identify the GDH-synthesized RNAs that were homologous to mRNAs encoding resveratrol synthase and related enzymes in carbon-nitrogen intermediary metabolism, the cDNA sequences were used as queries to search the NCBI nucleotide-nucleotide (excluding ESTs) BLAST (blastn), and non-redundant protein translation (blastx) databases. Complementary DNAs that displayed the highest alignment scores with the mRNAs were selected as the probes.
RNA analyses: Equal amounts (20 µg) of total RNA, and RNAs synthesized by GDH from the control and mineral salts-treated peanuts were loaded, briefly electrophoresed on 2% agarose gels, stained with ethidium bromide, and photographed to verify RNA quality and equality of loading. RNA was electro-transferred from the electrophoresed gel onto Brightstar-Plus nylon membrane (Applied Biosystems, Foster City, CA, USA) as described before [41] .
The cDNAs that were used as Northern probes were those homologous to mRNAs encoding resveratrol synthase. For the labeling of the cDNA probes, cDNA inserts were amplified by PCR from the corresponding plasmids (15 ng) using T3 and T7 promoter primers (2 µM each), [32P]-dATP (6000 Ci/mmol, 20 mCi/mL), dCTP/dGTP/TTP mix 50 mM, (2 µL), and Taq polymerase (1 U), in a final volume of 50 µL. Amplification was according to Display Systems Biotech (Vista, CA, USA)‘touch-down’ PCR procedure (denature: 94˚C, 1 min for the first 10 cycles: 94˚C, 30 sec; anneal: 60˚C, 30 sec for the first cycle, then reduced the temperature 0.5˚C each cycle until an annealing temperature of 55˚C was reached after 10 cycles; extension: 72˚C, 1 min. continued another 25 cycles with 94˚C, 30 sec; 55˚C, 30 sec; 72˚C, 1 min; final extension 72˚C, 5 min). Nylon membranes with immobilized RNA were prehybridized with ULTRAhyb buffer and hybridized with 32P-labeled cDNA inserts as probes overnight at 68˚C [41] . Solutions of labeled cDNA were first heated in boiling water bath for 10 min before adding to the prehybridized membrane. After hybridization, the membranes were washed (45 min, 68˚C) with NorthernMax (Applied Biosystems, Foster City, CA, USA) low stringency wash solution, then by NorthernMax high stringency wash solution (30 min, 68˚C). The membrane was autoradiographed by exposure to X-ray film within intensifying screens at −80˚C. Northern membranes were re-washed several times (30 min, 68˚C) with the high stringency solution until non-specifically bound probes were removed. Northern band intensities were digitalized using UN-SCAN-IT gel digitalizing software (Silk Scientific, Inc., Orem, Utah, USA) in order to estimate the mRNA relative abundances.
3. Results and Discussion
Treatment of peanut with mineral salt solutions: Mineral salt compositions [34] were formulated to mimic a wide array of reacting quantities, and the stoichiometric ratios of the subunit polypeptide compositions of GDH (Table 1). The mineral ion ratios made for interlocking internal repeats in the ionic compositions thus limiting stochastic variability in peanut plot treatments. The differences in yield from one treated peanut to the next in the hierarchy of yields were statistically insignificant between most of the peanuts especially the KN- and NPKS-treated peanuts. However, the GDH isoenzyme profiles were different from one mineral salt treated peanut to the other [35] thus demonstrating the individuality of each metabolic variant. The peat moss was common to all the experimental boxes, thereby minimizing environmental variability in soil physical and chemical characteristics. Mineral treatments were applied after seed germination, at flowering, and after flowering so that at all times the mineral concentration in the soil was nearly constant and in that way, the environmental effects of the mineral ions on GDH isomerization did not attenuate and/or fluctuate most of the time (synchronization). These horticultural management conditions assured the most efficient production metabolism of the peanuts as natural bioreactor through stoichiometric synchronization of the isomerization of GDH, synthesis of RNA by GDH, and silencing of mRNAs by homologous RNAs synthesized by GDH [35] . GDH isomerization and synthesis of RNA as the target sites of mineral ion action are due to the binomial distribution of its three subunits in the hexameric isoenzymes, on the basis of the twin nonallelic GDH1 and GDH2 gene structure, with the gene (GDH1) encoding the more acidic subunits (α, and a) being heterozygous and codominant, whereas the other gene (GDH2) encoding the less acidic subunit (β) is homozygous [50] .
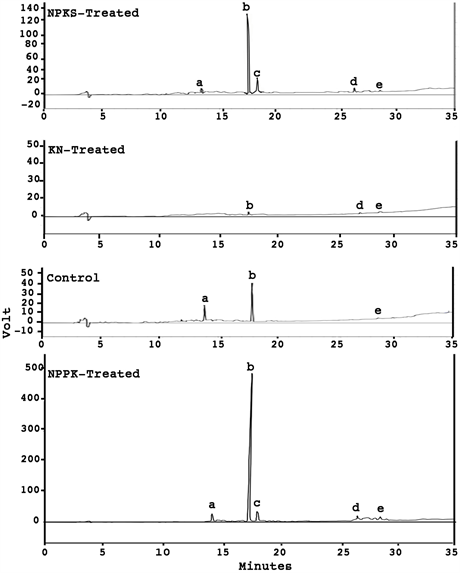
Mineral ion-dependent optimization of resveratrol accumulation: The differential chromatographic profiles (Figure 1) of resveratrols vividly showed that cis-resveratrol, trans-resveratrol, and trans-resveratrol-3-D-glucoside were lowest in accumulation in the KN-treated (9.0 µg∙g−1) and control untreated (6.0
Figure 1. Sketch of liquid Chromatographic profiles for resveratrol compounds extracted from KN-treated, control untreated, NPPK-treated, and NPKS-treated metabolic variants of Virginia peanuts seeds. Emodin served as the internal standard. Peak (a) is trans- resveratrol-3-D-glucoside; peak (b) is trans-resveratrol; peak (c) is cis-resveratrol; peak (d) is pterostilbene; and peak (e) is emodin.
µg∙g−1) peanuts; followed by that of the NPKS-treated (327.0 µg∙g−1); and were highest in the NPPK-treated (1.15 mg∙g−1) peanut. Trans-resveratrol was the major constituent, comprising 98.5% of the resveratrols in the NPPK-treated, 96.5% of the resveratrols in the NPKS-treated, 100% of the resveratrols in the KN-treated, and 41.0% of the resveratrols in the untreated peanut. The NPPK- treated peanut’s resveratrol content was the highest in field-cultivated peanut ever reported [17] [18] [19] . KN-treated peanut had no detectable trans-resveratrol- 3-D-glucoside and cis-resveratrol; control untreated peanut had no detectable cis-resveratrol but had trace amounts of trans-resveratrol-3-D-glucoside (3.5 µg∙g−1); NPKS-treated had trace amounts of both trans-resveratrol-3-D-glucoside (6.1 µg∙g−1) and cis-resveratrol (5.6 µg∙g−1); and the NPPK-treated had high levels of both trans-resveratrol-3-D-glucoside (6.6 µg∙g−1) and cis-resveratrol (10.3 µg∙g−1). The differential metabolic profiles of resveratrol (Figure 1) may be indicative of a complex regulation of intermediary carbon-nitrogen metabolism by the mineral ions. Peanut treated with NPPK possessed ultra-high resveratrol accumulation similar to grapes, Japanese know-weed, and berries [2] [5] [10] .
Integration of resveratrol biosynthesis, glycolysis, and glyoxylic-citric acid cycles: The ultra-high resveratrol peanut is due to integration of biochemical pathways at the mRNA level (Figure 2). Two cDNA probes specific to the mRNAs encoding resveratrol synthase were synthesized. The first probe (plasmid 5b, Table 2) shared seven-fold sequence repeats with peanut mRNA encoding resveratrol synthase; four of the repeats were minus/minus, whilst three repeats were plus/minus. This is a unique chemistry of GDH-synthesized RNA that accounts for its efficiency in binding tenaciously in vivo to target mRNA during silencing reaction, and in vitro during Northern hybridization reaction. Very high stringency washes were employed to sufficiently clean the Northern membranes (Figure 2(b)) of loosely-bound probes. The second cDNA probe (plasmid 2X, Table 2) shared sequence homologies with the mRNAs encoding resveratrol synthase and several regulatory enzymes in the citric-glyoxylic acid cycles and glycolysis. The cDNA probe 2X shared plus/plus sequence homology with the mRNA encoding resveratrol synthase. In addition, there is a network of sequence homologies between the probe and the mRNAs encoding the enzymes
Figure 2. Northern blot analyses of peanut resveratrol synthase mRNAs. Twenty micrograms total RNA was electrophoresed through 2% agarose gel, transferred to nylon membrane, then hybridized with 32Plabeled cDNA probe 5b, (Figure 2(b)), or probe 2X (Figure 2(d)); washed with high stringency solutions; then autoradiographed. Ethidium bromide-stained rRNA bands (Figure 2(a), Figure 2(c)) as indicators of equal loading. The arrows indicated the diminished resveratrol synthase transcript bands in the control, NPKS-and KN-treated peanuts.
Table 2. cDNA sequences of the GDH-synthesized RNAs homologous to peanut mRNAs encoding resveratrol synthase.
that catalyze the non-exergonic reactions of glycolysis, citric-glyoxylic acid cycles. Multiple sequence repeats are the exclusive structural characteristics of the GDH-synthesized RNA because of the binomial arrangement of the subunit polypeptides inside the GDH hexamers [42] .
Probe 2X (Table 2) shared six-fold plus/plus sequence homology with the mRNA encoding phosphoenol pyruvate carboxylase (PEPCase) in glycolysis; three-fold plus/plus sequence homology with the mRNA encoding enolase in glycolysis; two-fold plus/plus sequence homology with the mRNA encoding phosphoglyceromutase (PGlyM) in glycolysis; five-fold plus/plus and five-fold plus/minus sequence homologies with the mRNA encoding malate dehydrogenase (MD) in glyoxylic acid cycle; plus/plus sequence homology with the mRNA encoding malate synthase (MS) in glyoxylic acid cycle; two-fold plus/plus and four-fold plus/minus sequence homologies with the mRNA encoding isocitrate lyase (ICL) in glyoxylic acid cycle; five-fold sequence homologies with the mRNA encoding phosphoglucomutase (PGM) in glycolysis, two of which were minus/minus, one was plus/minus, another was minus/plus, and the fifth was plus/plus sequence repeats. Therefore a single RNA sequence (probe 2X) synthesized by GDH silenced the mRNAs encoding resveratrol synthase together with those encoding PEPCase, enolase, PGlyM, MD, MS, ICL, and PGM in KN-, NPKS- and control peanuts but not in NPPK-treated peanut thereby resulting to maximum accumulation of resveratrol in that peanut metabolic variant. The resveratrol metabolic network integrating glycolysis, gluconeogenesis, and citric acid cycle at the mRNA level give further confirmation to the biochemical role of PGM, PGlyM, enolase, PEPCase, malate synthase, MDH, and ICL in controlling carbon fluxes through intermediary carbon-nitrogen metabolism [40] . This meant that within the cellular metabolism, the seven mRNAs were not knocked out under NPPK-applied mineral ion regimen. GDH-synthesized RNA 2X (Table 2) is the molecule that integrated/discriminated the carbon-nitrogen biochemical pathways in the peanut. The molecular integration/discrimination of glycolysis, glyoxylic-citric acid cycles through the optimal abundance of the mRNAs (2.7 - 1.2 kb range) encoding resveratrol synthase, PGM, PGlyM, enolase, PEPCase, malate synthase, MDH, and ICL accounted for the apparent muddled transcript smear in the Northern blot of NPPK-treated peanut (Figure 2(d)).
The Northern Blots: Two bands covering the 1.2 - 1.4 kb range (Figure 2(b)) were obtained with the first Northern probe (plasmid 5b, Table 2) in agreement with the pair of mRNAs that encode peanut resveratrol synthase [3] [28] [33] . Digital estimation of the Northern bands (Figure 2(b)) showed that NPPK- treated peanut had the highest abundance of the bands; followed by the NPKS- treated in which the lowest molecular weight band was considerably diminished; to the untreated control where both bands were considerably diminished compared with that of NPPK-treated; and to the KN-treated where both bands were almost undetectable. This is in agreement with the inverse order of the seed yields (Table 1) with the KN-treated peanut being the highest yield and the NPPK-treated being the lowest yield. The metabolic profiles (Figure 1) showed that where cis-resveratrol and trans-resveratrol maximally accumulated (NPPK- treated peanut), all the resveratrol synthase transcripts were not silenced; but where cis-resveratrol minimally accumulated, most of the transcripts (Figure 2(b)) were silenced (KN-treated). Therefore, the higher molecular weight mRNAs of resveratrol synthase regulate the enzyme’s activity. Peanut resveratrol synthase is a dimer [51] . GDH-synthesized RNA probes have made it possible perhaps for the first time, to align the resveratrol concentrations and the resveratrol synthase mRNA abundance [18] [33] thereby confirming the transcriptional control of resveratrol synthase activity. The mechanism is that the mineral salt treatments induced the peanut GDH to isomerize and to synthesize a unique set of RNA which then silence the corresponding homologous mRNAs. The silencing of the mRNA may be complete or partial depending on the structure and abundance of the respective GDH-synthesized RNA [35] . In the KN-treated peanut (Figure 2(b)) the GDH induced by KN mineral salt mix synthesized a lot of the RNA complement of probe 5b (Figure 3). Therefore, the resveratrol synthase mRNAs were silenced. In the cases of NPKS-treated and the untreated control peanuts, the induced GDHs synthesized a limited quantity of the RNA complement of probe 5b. Therefore, the resveratrol synthase mRNAs were incompletely silenced in the NPKS-treated and the untreated control peanuts. In the case of the NPPK-treated peanut, the induced GDH synthesized almost an undetectable amount of the RNA complement of probe 5b. Therefore, the resveratrol synthase mRNAs were not silenced. This was the strategic systems biology approach for investigating the regulation of resveratrol accumulation by resveratrol synthase transcripts. Agronomic mineral ion signals embodied in the
Figure 3. Distribution of sequences homologous to mRNAs encoding resveratrol synthase in the RNA synthesized by the GDH of some peanut metabolic variants. RNAs synthesized by the charge isomers of whole-gel purified GDH of metabolic variants (KN-, control, NPKS-, NPPK-treated peanuts) were electrophoresed through 2% agarose gel. The electrophoresed gels were trans-blotted onto nylon membranes followed by membrane hybridization with 32P-labeled cDNA (probe 5b) of GDH-synthesized RNA homologous to resveratrol synthase. The membranes were washed with high stringency solutions and autoradiographed.
NPPK mix prevented the peanut GDH from synthesizing the RNA complement of probe 5b. The more the complementary RNA is synthesized by GDH, the more is the silencing of the homologous mRNA.
Results (Figure 2(d)) for Northern probe 2X (Table 2) were dissimilar from the first blot (Figure 2(b)) in that bands were obtained for the NPPK-treated peanut, but the other peanuts gave no bands. Northern bands produced by the NPPK-treated peanut stretched from 1.2 - 2.7 kb range. The 2.7 kb band represented the mRNA encoding PEPCase [52] [53] ; the 2.3 kb band represented the mRNA encoding malate dehydrogenase [54] [55] ; the 2.1 kB band represented the mRNA encoding PGM [35] ; the 2.0 kb band represented the mRNA encoding ICL [56] [57] [58] ; the 1.9 kb band represented the mRNA encoding PGlyM [59] [60] ; the 1.80 - 1.83 kb bands represented the mRNAs encoding enolase and malate synthase [35] ; and the 1.2 - 1.4 kb bands represented the resveratrol synthase family of mRNAs [33] . The absence of Northern bands under the KN- treated, NPKS-treated, and the untreated control peanuts in Figure 2(d) in sharp contrast with the results in Figure 2(b) suggested functional cross-talks (signal integration/discrimination) between the mRNAs encoding resveratrol synthase and those encoding MD, PGM, ICL, PGlyM, MS, enolase, and PEPCase as explained previously [30] [35] [36] [39] . The ribosomal RNA bands presented in Figure 2 demonstrated equality of RNA loading per gel well.
The resveratrol biosynthetic pathway: The mRNAs encoding MD, PGM, PGlyM, MS, enolase, and PEPCase, in peanut were readily detected by their specific cDNA probes [30] . Therefore, chemical inability of cDNA probe 2X (Table 2) to detect the mRNAs encoding resveratrol synthase, MD, PGM, PGlyM, MS, enolase, PEPCase in the KN-treated, NPKS-treated, and control untreated peanuts (Figure 2(d)) suggested there could be thresh-hold minimum abundance requirement factor for the mRNAs. The low levels below the minimum thresh- hold abundance of the mRNAs encoding resveratrol synthase were responsible for the low yields of resveratrol in the untreated control, KN-treated, and NPKS-treated peanuts. Conversely, the maximum levels above the thresh-hold abundance for the resveratrol synthase mRNAs (Figure 2(b)) was responsible for the ultra-high resveratrol content of the NPPK-treated peanut (Figure 1). The usual biochemical comparison limited to the control untreated and the NPPK-treated peanuts would have failed to unravel the comprehensiveness of the trends integrated by the agronomic system comprising control, KN-, NPKS-, and NPPK-treated peanuts.
The other factor was the absence of the mRNAs encoding PGM and the related carbon-nitrogen metabolic enzymes in the KN-treated, NPKS-treated, and untreated control peanuts (Figure 2(d)). Phosphoglucomutase (PGM, EC 2.7.5.1) generates glucose-1-phosphate (Glc-1-P) from glucose-6-phosphate (Glc-6-P). The enzyme is localized both in the plastids and cytosol [43] . The plastidic PGM is essential for starch synthesis to store photosynthates in leaves during the day, and in the degradation of starch [44] . The cytosolic PGM is involved in sucrose catabolism to provide intermediates for glycolysis, citric acid cycle, fatty acid, amino acid, glyoxylic acid cycle etc metabolic pathways [45] . Therefore, PGM plays an important role in the distribution of Glc-6-P to glycolytic, starch, citric acid, amino acid, and fatty acid pathways [46] [47] . Resveratrol derives from the condensation of malonyl-Coenzyme A and p-coumaroyl-CoA, [28] [29] , which are intermediate products of glycolytic reaction that are dependent on Glc-6-P availability. In NPPK-treated peanut where the mRNA encoding PGM was not silenced (Figure 2(d)), there was abundant above thresh-hold concentration of Glu-6-P to power the supply of acetyl-CoA, malonyl-CoA, and p-coumaroyl- CoA for the synthesis of ultra-high quantities of resveratrol.
Malonyl-CoA is the point of bifurcation of stilbenoid from the fatty acid biosynthetic pathways [28] . Therefore, in further verification of the resveratrol pathway, the fatty acid yields (Table 3) were found to align in decreasing order from KN-treated (56.96 g∙kg−1), untreated control (55.60 g∙kg−1), NPKS-treated (48.45 g∙kg−1), and to NPPK-treated (44.88 g∙kg−1) peanuts and in reverse order to the resveratrol contents (Figure 1) and the relative abundance of resveratrol synthase mRNAs (Figure 2) because high resveratrol synthase activity (NPPK- treated peanut) implies the depletion of malonyl CoA and diversion to resveratrol synthesis instead of fatty acid synthesis. On the other hand, low resveratrol synthase activities (KN-treated, untreated control, and NPKS-treated peanuts) implies the accumulation of malonyl CoA and its diversion to fatty acid biosynthesis instead of resveratrol biosynthesis.
The integration of resveratrol biosynthesis, glycolysis, and glyoxylic-citric acid cycles by RNA probe 2X (Table 2) to become a single physiological machine complex defined the huge biochemical dimensions of the resveratrol biosynthetic pathway (Figure 4). Glc-6-P, phosphoenol pyruvate, glycerate-3-phosphate, malate, isocitrate, as well as acetyl CoA, malonyl CoA, and p-coumaroyl CoA are the metabolites that feed into the resveratrol pathway. Whenever probe 2X is synthesized in the peanut (KN-, control, NPKS-treated) and some or all the in-bound metabolites are unavailable, the resveratrol biosynthetic pathway functions minimally leading to low production of resveratrol by peanut (Figure 1). But when probe 2X is not synthesized (NPPK-treated peanut), all the in-bound metabolites are available; and the resveratrol biosynthetic pathway (Figure 4) is maximally functional, leading to production of ultrahigh resveratrol peanut. Therefore, GDH-synthesized RNA probe 2X encodes the signal for the physiological machine complex that maximally expands the resveratrol biosynthetic pathway (Figure 4) and the accumulation of ultrahigh resveratrol contentsin peanut.
When the GDH-synthesized RNA did not knock down the mRNAs encoding the enzymes of biomass and fatty acid metabolism, peanut yield was doubled [34] [35] [36] [37] . The NPPK treatment gave the lowest ever yield [35] of peanut (Table 1). Therefore, the drastic knock down of the biomass yield metabolism accounted in part for the ultra-high resveratrol contents of the NPPK- treated peanut.
The horticultural system comprising control, KN-, NPKS-, and NPPK-treated peanuts captured the identity and structure of probe 2X with physiological function to couple resveratrol biosynthesis to glycolysis, citric-glyoxylic acid cycles. Probe 2X was synthesized in the control, KN-, and NPKS-treated peanuts, but
Table 3. Gas Chromatography Fatty Acid Profiles of Peanut.
Notes: ₱g/kg.
not in the NPPK-treated peanut with the highest accumulation of resveratrol. The practical importance of 2X is that its suppression might guide horticultural technology development to achieve increased accumulation of resveratrol in sustainably-cultivated peanuts. The ultra-high resveratrol peanut seeds could be market-ready as in-shell snack nut to restaurant chain customers [20] thereby providing a convenient alternative to chemically synthesized resveratrol [25] [26] [27] .
Probe 2X was synthesized by GDH. Its precision for silencing and as a hybridization probe for mRNAs encoding resveratrol synthase, MD, PGM, PGlyM, MS, enolase, and PEPCase was phenomenal. An alternative method for hybridization probe design of probe 2X would be the algorithm approach. But the wide sequence span of the probe and some variable nucleotide residues would defy the assembly of its biological and physical properties in one molecule by computational language. Algorithm is yet to be adapted for the design of nucleic acid hybridization probes specific for monitoring the integration/discrimination regulation of carbon-nitrogen biochemical pathways.
Horticultural stress inductiveness by stoichiometric mineral salt mixes: The big picture (Table 1) provided the context for further discussion on the signaling properties of the optimized mineral ion mixes relative to peanut yields. GDH is a target site of mineral ion action [38] , crop yield being regulated by the resultant redox isomerization stresses [35] [36] [39] . The responses of yield (Table 1) to the mineral salt mixes illustrated the interplay of synergism and antagonism. Separate K, and N treatments induced about the same very low podded seed yields; but as stoichiometric KN treatment, their synergism doubled and maximized the yield. Conversely, PPN, and K treatments induced higher yields than NPPK treatment thus illustrating the operation of mineral ion antagonism. Similar examples of mineral ion synergism and antagonism abound in the Table.
Figure 4. Biosynthesis of resveratrol in peanut. [Abbreviations: PGM, phosphoglucomutase; ACCase, acetyl coenzyme A carboxylase; PKS, polyketide synthase; RS, resveratrol synthase].
Conversations on the physiology of mineral ion antagonism and synergism have centered on agronomy [31] [61] because commercial fertilizes are applied in weight ratios per unit area of land. The peanut yield responses to the stoichiometric mixes of mineral salts (Table 1) manifested a successive series of sigmoidal curves indicating that enzymes were the target sites of the mineral ion actions. Peanut yield did not plateau in response to the stoichiometric mixes of mineral salts because GDH is a target site of mineral nutrient action [38] [39] . Mineral ions acting as electrophiles and nucleophiles (signals) induce the isomerization of the enzyme to synthesize some RNA that silence the mRNAs that are homologous to them [36] [37] [41] . When biomass or a metabolite increased in accumulation as the result of the differential silencing of mRNAs, in K-, N-, and KN-treated peanuts (Table 1) then the outcome was synergism. Conversely, when biomass or a metabolite declined in accumulation as the result of the differential silencing of mRNAs, in PPN-, K-, and NPPK-treated peanuts (Table 1), then the outcome was antagonism. GDH under a mineral nutrient regiment integrates all the signals from the aggregate nucleophiles and electrophiles to give one single response in the synthesis of a unique set of RNA sequences, which then silence the corresponding set of homologous mRNAs. This is the signal integration-discrimination (synergism-antagonism) function of the enzyme [62] [63] . With respect to the RNAs synthesized by peanut GDH (Figure 3), the more the complementary RNA was synthesized, the more was the silencing of the resveratrol synthase mRNA, outcome of which was antagonism. Resveratrol accumulated the least in the KN-treated peanut (Figure 1). Conversely, the less the complementary RNA synthesized, the less was the silencing of resveratrol synthase mRNA, outcome of which was synergism. Resveratrol accumulated the highest in the NPPK-treated peanut. Additional physiological stress factors could amplify/modulate the RNA synthetic activity of GDH. Only the acidic isoenzymes of the GDH of NPPK-treated peanut synthesized RNAs that were homologous to the mRNA encoding resveratrol synthase (Figure 3). Acidic isoenzymes of GDH might signal threshold inability of the RNA to silence the resveratrol synthase mRNAs. The GDHs of the control, KN-, and NPKS-treated peanuts synthesized RNAs in the acidic, neutral and alkaline pI ranges (Figure 3) and were able to silence resveratrol synthase mRNA to different degrees (Figure 2). Therefore, mineral ion-induced stresses, antagonism, and synergism in agronomy have roots that stretch into the molecular and enzyme levels of metabolism. Optimized stoichiometric mixes of mineral nutrients are formulated to exploit the principles of antagonism and synergism and to manipulate crop plants to accumulate desirable metabolites beneficial to human health [34] [35] [36] .
The control untreated peanut gave higher yield than the commercial fertilizer treatment, and many of the mineral ion-treated peanuts. Therefore, neither the control nor the commercial fertilizer- treated peanuts contained the control GDH. This is a common experimental problem encountered in plant GDH research [63] . It was the KN-treated peanut that contained the positive control GDH, and produced the highest yield of podded seeds; whilst the NPPK treatment induced the highest stress on GDH, and consequently the highest yield of resveratrol but the lowest podded seed yield. These are in agreement with the repeatedly published results that high resveratrol yields are induced by exertion of stresses on whole plants or plant tissue systems [4] [21] [22] [23] [27] [32] [33] [51] .
Other neutraceticals of peanut: Pterostilbene content was highest in the NPPK-treated peanut (81.3 µg. g−1) but was insignificant in the other peanuts (Figure 1). Similarly trans-resveratrol-3-D-glucoside content was highest in the NPPK-treated (6.6 µg∙g−1), and NPKS-treated (6.1 µg∙g−1) peanuts, but was insignificant in the other peanuts (Figure 1). Pterostilbene, is a methylated resveratrol compound present in blueberries and peanut hairy roots [10] . Significant concentrations of pterostilbene and resveratrol 3-O-β-D-glucoside in high resveratrol peanut is an added dietary benefit. Mineral ion regulation of the pathway is favorable for the sustainable cultivation of peanut.
4. Conclusion
Resveratrol naturally occurring antioxidant is associated with reduction of inflammatory cancer risks. Its levels in peanut are ultra-low and variable (0 to 26 µg∙g−1 fresh wt.). The aim of this project was to identify related reaction step(s) in glycolysis, citric-glyoxylic acid cycle that optimize resveratrol metabolism to maximum resveratrol accumulation. The GC-LC profiles showed that resveratrols were lowest in concentration in the KN-treated (9.0 µg∙g−1) and control untreated (6.0 µg∙g−1) peanuts; and were highest in the NPPK-treated (1.14 mg∙g−1) peanut. Fatty acid yields aligned in decreasing order from KN-treated (56.96 g∙kg−1), control (55.60 g∙kg−1), and to NPPK-treated (44.88 g∙kg−1) peanuts in converse order to the resveratrol contents because high resveratrol synthase activity (NPPK-treated peanut) implies diversion of malonyl CoA to resveratrol synthesis instead of fatty acid synthesis. The NPPK treatment gave the lowest ever yield of peanut due to the antagonistic stresses induced by the mineral ion mix. Regulatory enzymes in glycolysis, citric-glyoxylic acid cycles were functionally coupled to the resveratrol biosynthetic pathway thus inducing the ultra-high resveratrol accumulation in the NPPK-treated peanut.
Acknowledgements
Research was funded under Evans Allen Funds made to Prairie View A&M University by USDA-NIFA. Thanks and appreciation to Dr. Yoonsung Jung for biometric review of the peanut research proposal.
Competing Financial Interests
The authors declare no competing financial interests.
Cite this paper
Osuji, G.O., Johnson, P., Duffus, E., Woldesenbet, S. and Kirven, J.M. (2017) Horticultural Production of Ultra High Resveratrol Peanut. Agricultural Sciences, 8, 1173-1194. https://doi.org/10.4236/as.2017.810086
References
- 1. Sanders, T.H., McMichael, R.W. and Hendrix, K.W. (2000) Occurrence of Resveratrol in Edible Peanuts. Journal of Agricultural and Food Chemistry, 48, 1243-1246. https://doi.org/10.1021/jf990737b
- 2. Creasy, L.L. and Coffee, M. (1988) Phytoalexin Production Potential of Grape Berries. Journal of the American Society for Horticultural Science, 113, 230-234.
- 3. Schroder, G., Brown, J.W. and Schroder, J. (1998) Molecular Analysis of Resveratrol Synthase cDNA, Genomic Clones and Relationship with Chalcone Synthase. European Journal of Biochemistry, 172, 161-169. https://doi.org/10.1111/j.1432-1033.1988.tb13868.x
- 4. Giovinazzo, G., Ingrosso, I., Paradiso, A., De Gara, L. and Santino, A. (2012) Resveratrol Biosynthesis: Plant Metabolic Engineering for Nutritional Improvement of Food. Plant Foods for Human Nutrition, 67, 191-199. https://doi.org/10.1007/s11130-012-0299-8
- 5. Jang, M., Cai, G.O., Udeani, K.V., Slowing, C.F., et al. (1997) Cancer Chemopreventive Activity of Resveratrol, a Natural Product Derived from Grapes. Science, 275, 218-220. https://doi.org/10.1126/science.275.5297.218
- 6. Wu, J.M., Wang, Z.R., Hsieh, T.C., Bruder, J.L., et al. (2001) Mechanism of Cardioprotection by Resveratrol, a Phenolic Antioxidant Present in Red Wine. International Journal of Molecular Medicine, 8, 3-17.
- 7. Nolan, C.J., Damm, P. and Prentki, M. (2011) Type 2 Diabetes across Generations: From Pathophysiology to Prevention and Management. The Lancet, 378, 169-181.
- 8. Beak, S.-H., Shin, W.-C., Ryu, H.-S., Lee, D.-W., Moon, E., et al. (2013) Creation of Resveratrol-Enriched Rice for the Treatment of Metabolic Syndrome and Related Diseases. PLoS ONE, 8, e57930. https://doi.org/10.1371/journal.pone.0057930
- 9. Guarente, L. (2006) Sirtuins as Potential Targets for Metabolic Syndrome. Nature, 444, 868-874. https://doi.org/10.1038/nature05486
- 10. Szajdek, A. and Borowska, E.J. (2009) Bioactive Compounds and Health-Promoting Properties of Berry Fruits: A Review. Plant Foods for Human Nutrition, 63, 147-156. https://doi.org/10.1007/s11130-008-0097-5
- 11. Rimando, A.M., Nagmani, R., Feller, D.R. and Yokoyama, W. (2005) Pterostilbene, a New Argonist for the Peroxizome Proliferator-Activated Receptor α-Isoform, Lowers Plasma Lipoproteins and Cholesterol in Hypercholoesteromic Hamsters. Journal of Agricultural and Food Chemistry, 53, 3403-3407. https://doi.org/10.1021/jf0580364
- 12. McCormack, D. and McFadden, D. (2013) A Review of Pterostilbene Antioxidant Activity and Disease Modification. Oxidative Medicine and Cell Longevity. https://doi.org/10.1155/2013/575482
- 13. Wang, L., Heredia, A., Song, H., Zhang, Z., et al. (2004) Resveratrol Glucuronides as the Metabolites of Resveratrol in Humans: Characterization, Synthesis, and Anti-HIV Activity. Journal of Pharmaceutical Sciences, 93, 2448-2454. https://doi.org/10.1002/jps.20156
- 14. Jungong, C.S. and Novikov, A.V. (2012) Practical Preparation of Resveratrol 3-O-β-D-Glucuronide. Synthetic Communications: An International Journal for Rapid Communication of Synthetic Organic Chemistry, 42, 3589-3597. https://doi.org/10.1080/00397911.2011.585733
- 15. McMurtrey, K.D., Minn, J., Pobanz, K. and Schultz, T.P. (1994) Analysis of Wines for Resveratrol using Direct Injection High-Pressure Liquid Chromatography with Electrochemical Detection. Journal of Agricultural and Food Chemistry, 42, 1997-2000. https://doi.org/10.1021/jf00046a001
- 16. Bioscience Technology (2014) Red Wine May Not Prevent Heart Disease, Prolong Life. http://www.biosciencetechnology.com
- 17. Wang, K.H., Lai, Y.H., Chang, J.C., et al. (2005) Germination of Peanut Kernels to Enhance Resveratrol Biosynthesis and Prepare Sprouts as a Functional Vegetable. Journal of Agricultural and Food Chemistry, 53, 242-246.
- 18. Chung, I., Park, M.R., Chun, J.C. and Yun, S.J. (2003) Resveratrol Accumulation and Resveratrol Synthase Gene Expression in Response to Abiotic Stress and Hormones in Peanut Plants. Plant Science, 164, 103-109.
- 19. Sobolev, V.S. and Cole, R.J. (1999) Trans-Resveratrol Content in Commercial Peanuts and Peanut Products. Journal of Agricultural and Food Chemistry, 47, 1435-1439. https://doi.org/10.1021/jf9809885
- 20. Fletcher, S.M. (2009) Evaluation and Results Report on the National Peanut Board’s Domestic Peanut Market Program 1st quarter 2005-3rd quarter 2009.
- 21. Aleynova, O.A., Dubrovina, A.S., Manyakhin, A.Y., Keratin, Y.A. and Kiselev, K.V. (2015) Regulation of Resveratrol Production in Vitis amurensis Cell Cultures by Calcium-Dependent Protein Kinases. Applied Biochemistry and Biotechnology, 175, 1460-1476. https://doi.org/10.1007/s12010-014-1384-2
- 22. Resurreccion, V., Anna, A., Rudolf, J.L., Phillips, R.D. and Chinnan, M. (2005) Method for Enhancing Content of Peanut Compositions (Patent No. US 766455).
- 23. Fritzemeier, K.H., Rofls, C.H., Pfau, J. and Kindl, H. (1983) Action of Ultraviolet-C on Stilbene Formation in Callus of Arachis hypogaea. Planta, 159, 25-29. https://doi.org/10.1007/BF00998810
- 24. Paiva, N.L. and Hipskind, J.D. (2005) Transgenic Legume Plants Modified to Produce Resveratrol Glucoside and Uses Thereof. US Patent 6974895.
- 25. Farina, A., Ferranti, C. and Marra, C. (2006) An Improved Synthesis of Resveratrol. Natural Product Research, 20, 247-252. https://doi.org/10.1080/14786410500059532
- 26. Trants, E., Panopoulos, N. and Ververidis, F. (2009) Metabolic Engineering of the Complete Pathway Leading to Heterologous Biosynthesis of Various Flavonoids and Stilbenoids in Saccharomyces cerevisiae. Metabolic Engineering, 11, 355-366.
- 27. Wang, H., Liu, L., Guo, Y.X., et al. (2007) Biotransformation of Piceid in Polygonum cuspidatum to Resveratrol by Aspergillus oryzea. Applied Microbiology and Biotechnology, 75, 763-768.
- 28. Hasan, M.M., Cha, M., Bajpai, V.K. and Baek, K. (2013) Production of a Major Stilbene Phytoalexin, Resveratrol in Peanut (Arachis hypogaea) and Peanut Products: A Mini Review. Reviews in Environmental Science and Biotechnology, 12, 209-221. https://doi.org/10.1007/s11157-012-9294-7
- 29. Yu, O. and Jez, J. (2008) Nature’s Assembly Line: Biosynthesis of Simple Phenylpropanoids and Polyketides. The Plant Journal, 54, 750-763. https://doi.org/10.1111/j.1365-313X.2008.03436.x
- 30. Osuji, G.O., Duffus, E., Johnson, P., Woldesenbet, S., Weerasooriya, A., Ampim, P.A., Carson, L., Jung, Y., South, S., Idan, E., Johnson, D., Clarke, D., Lawton, B., Parks, A., Fares, A. and Johnson, A. (2015) Enhancement of the Essential Amino Acid Composition of Food Crop Proteins through Biotechnology. American Journal Plant Sciences, 6, 3091-3108. https://doi.org/10.4236/ajps.2015.619302
- 31. Jeandet, P., Bessis, R., Adrian, M., Yvin, J. and Joubert, J. (2000) Use of Aluminum Chloride as a Resveratrol Synthesis Elicitor. US Patent Number US6080701A. http://www.google.com/patents/US6080701
- 32. Kiselev, K.V., Shumakova, O.A. and Manyakhin, A.Y. (2013) Effect of Plant Stilbene Precursors on the Biosynthesis of Resveratrol in Vitis amurensis Rupr. Cell Cultures. Applied Biochemistry and Microbiology, 49, 53-58. https://doi.org/10.1134/S0003683813010079
- 33. Rietra, R.P.J.J., Heinen, M., Dimkpa, C. and Bindraban, P.S. (2015) Effects of Nutrient Antagonism and Synergism on Fertilizer Use Efficiency. VFRC Report 2015/5. Virtual Fertilizer Research Center, Washington DC, 42 p.
- 34. Osuji, G.O., Brown, T.K., South, S.M., Johnson, D. and Hyllam, S. (2012) Molecular Modeling of Metabolism for Allergen-Free Low Linoleic Acid Peanuts. Applied Biochemistry and Biotechnology, 168, 805-823. https://doi.org/10.1007/s12010-012-9821-6
- 35. Osuji, G.O., Brown, T.K., South, S.M., Duncan, J.C. and Johnson, D. (2011) Doubling of Crop Yield through Permutation of Metabolic Pathways. Advances in Bioscience and Biotechnology, 2, 364-379. https://doi.org/10.4236/abb.2011.25054
- 36. Osuji, G.O., Brown, T.K. and South, S.M. (2010) Optimized Fat and Cellulosic Biomass Accumulation in Peanut through Biotechnology. International Journal of Biotechnology & Biochemistry, 6, 451-472.
- 37. Osuji, G.O., Konan, J. and M’Mbijjewe, G. (2004) RNA Synthetic Activity of Glutamate Dehydrogenase. Applied Biochemistry and Biotechnology, 119, 209-228. https://doi.org/10.1007/s12010-004-0003-z
- 38. Osuji, G.O., Reyes, J.C. and Mangaroo, A.S. (1998) Glutamate Dehydrogenase Isomerization: A Simple Method for Diagnosing Nitrogen, Phosphorus, and Potassium Sufficiency in Maize (Zea mays L.). Journal of Agricultural and Food Chemistry, 46, 2395-2401. https://doi.org/10.1021/jf971065x
- 39. Osuji, G.O., Mangaroo, A.S., Reyes, J. and Wright, V. (2003) Biomass Enhancement in Maize and Soybean in Response to Glutamate Dehydrogenase Isomerization. Biologia Plantarum, 47, 45-52.
- 40. Osuji, G.O., Brown, T.K. and South, S.M. (2008) Discovery of the RNA Synthetic Activity of GDH and Its Application in Drug Metabolism Research. The Open Drug Metabolism Journal, 2, 1-13. https://doi.org/10.2174/1874073100802010001
- 41. Osuji, G.O., Brown, T.K. and South, S.M. (2009) Nucleotide-Dependent Reprogramming of mRNAs Encoding Acetyl Coenzyme a Carboxylase and Lipoxygenase in Relation to the Fat Contents of Peanut. Journal of Botany, 2009, Article ID: 278324. https://doi.org/10.1155/2009/278324
- 42. Osuji, G.O. and Brown, T. (2007) Environment-Wide Reprogramming of mRNAs Encoding Phosphate Translocator and Glucosyltransferase in Relation to Cellulosic Biomass Accumulation in Peanut. ICFAI Journal of Biotechnology, 1, 35-47.
- 43. Streb, S., Elgi, B., Eicke, S. and Zeeman, S. (2009) The Debate on the Pathway of Starch Synthesis: A Closer Look on Low-Starch Mutants Lacking Plastidial Phosphoglucomutase Supports the Chloroplast Localized Pathway. Plant Physiology, 151, 1769-1777. https://doi.org/10.1104/pp.109.144931
- 44. Hattenbach, A. and Heineke, D. (1999) On the Role of Chloroplastic Phosphoglucomutase in the Regulation of Starch Turn Over. Planta, 207, 527-532. https://doi.org/10.1007/s004250050513
- 45. Manjunath, S., Lee, C.H.K., VanWinkle, P. and Bailey-Serres, J. (1998) Molecular and Biochemical Characterization of Cytosolic Phosphoglucomutase in Maize Expression during Development and in Response to Oxygen Deprivation. Plant Physiology, 117, 997-1006. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3649683/
- 46. Periappuram, C., Steinhauer, L., Barton, D.L., Taylor, D.C., Chatson, B. and Zou, J. (2000) The Plastidic Phosphoglucomutase from Arabidipsis. A Reversible Enzyme Reaction with an Important Role in Metabolic Control. Plant Physiology, 122, 1193-1199. https://doi.org/10.1104/pp.122.4.1193
- 47. Harrison, C.J., Mould, R.M., Leech, M.K., Johnson, S.A., Turner, L., Schreck, S.L., Baird, K.M., Jack, P.L., Rawsthorne, S., Hedley, C.L. and Wang, T.L. (2000) The rug3 of Pea Encodes Plastidial Phosphoglucomutase. Plant Physiology, 122, 1187-1192. https://doi.org/10.1104/pp.122.4.1187
- 48. Rose, D.T., Scherf, U., Eisen, M.B., Perou, C.M., Reese, C., Spellman, P., et al. (2000) Systematic Variation in Gene Expression Patterns in Human Cancer Cell Lines. Nature Genetics, 24, 227-235. https://doi.org/10.1038/73432
- 49. Grierson. D., Slater, J. and Tucker, G.A. (1985) The Appearance of Polygalacturonase mRNA in Tomatoes. Planta, 163, 263-271. https://doi.org/10.1007/BF00393517
- 50. Cammaerts, D. and Jacobs, D. (1983) A Study of the Polymorphism and the Genetic Control of the Glutamate Dehydrogenase Isoenzymes in Arabidopsis Thaliana. Plant Science Letters, 31, 67-73.
- 51. Schoppner, A. and Kindl, H. (1984) Purification and Properties of a Stilbene Synthase from Induced Cell Suspension Cultures of Peanut. The Journal of Biological Chemistry, 259, 6806-6811.
- 52. Law, R.D. and Plaxton, W.C. (1995) Purification and Characterization of a Novel Phosphoenolpyruvate Carboxylase from Banana Fruit. Biochemical Journal, 307, 807-816. https://doi.org/10.1042/bj3070807
- 53. Singal, H.R. and Singh, R. (1986) Purification and Properties of Phosphoenolpyruvate Carboxylase from Immature Pods of Chickpea (Cecer arietium L). Plant Physiology, 80, 369-373. https://doi.org/10.1104/pp.80.2.369
- 54. Rothermel, B.A. and Nelson, T. (1989) Primary Structure of Maize NADP-Dependent Malic Enzyme. The Journal of Biological Chemistry, 246, 19587-19592.
- 55. Schearer, H.L., Turpin, D.H. and Dennis, D.T. (2004) Characterization of NADP-Dependent Malic Enzyme from Developing Castor Oil Seed Endosperm. Archives of Biochemistry and Biophysics, 429, 134-144.
- 56. Beeching, J.R. and Northcote, D.H. (1987) Nucleic Acid (cDNA) and Amino Acid Sequences of Isocitrate Lyase from Castor Bean. Plant Molecular Biology, 8, 471-475. https://doi.org/10.1007/BF00017992
- 57. Comai, L., Diewtrich, R.A., Maslyar, D.J., Baden, C.S. and Harada, J.J. (1989) Coordinate Expression of Transcriptionally Regulated Isocitrate Lyase and Malate Synthase Genes in Brassica napus L. The Plant Cell, 1, 293-300.
- 58. Mano, S., Hayashi, M., Kond, S. and Nishimura, M. (1996) cDNA Cloning and Expression of a Gene for Isocitrate Lyase in Pumpkin Cotyledons. Plant and Cell Physiology, 37, 941-948. https://doi.org/10.1093/oxfordjournals.pcp.a029043
- 59. Grana, X., Broceno, C., Garriga, J., de la Ossa, P.P. and Climent, F. (1993) Phosphoglycerate Mutase Activity and mRNA Levels during Germination of Maize Embryos. Plant Science, 89, 147-151.
- 60. Huang, Y., Blakeley, S.D., McAleese, S.M., Fothergill-Gilmore, L.A. and Dennis, D.T. (1993) Higher Plant Cofactor-Independent Phosphoglyceromutase: Purification, Molecular Characterization and Expression. Plant Molecular Biology, 23, 1039-1053. https://doi.org/10.1007/BF00021818
- 61. Osvalde, A. (2011) Optimization of Plant Mineral Nutrition Revisited: The Roles of Plant Requirements, Nutrient Interactions, and Soil Properties in Fertilization Management. Environmental and Experimental Biology, 9, 1-8.
- 62. Osuji, G.O. and Breathwaite, C. (1999) Signaling by Glutamate Dehydrogenase in Response to Pesticide Treatment and Nitrogen Fertilization of Peanut. Journal of Agricultural and Food Chemistry, 47, 3332-3344. https://doi.org/10.1021/jf9805303
- 63. Osuji, G.O. and Madu, W.C. (2015) Glutamate Dehydrogenase. In: D’Mello, J.P.F., Ed., Amino Acids in Higher Plants, Chapter 1, CABI Publishers, Wallingford.