American Journal of Plant Sciences
Vol.4 No.5(2013), Article ID:32137,9 pages DOI:10.4236/ajps.2013.45127
Bioassay and Characterization of Several Palmer Amaranth (Amaranthus palmeri) Biotypes with Varying Tolerances to Glyphosate
![]()
US Department of Agriculture-Agricultural Research Service (USDA-ARS), Crop Production Systems Research Unit, Stoneville, USA.
Email: bob.hoagland@ars.usda.gov
Copyright © 2013 Robert E. Hoagland et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 11th, 2013; revised April 27th, 2013; accepted May 10th, 2013
Keywords: Amaranthus palmeri; Betalain; Chlorophyll; EPSPS; Glyphosate-Resistant Weeds; Palmer Amaranth; Pigweed; Western Blot
ABSTRACT
The wide distribution of Palmer amaranth (Amaranthus palmeri) in the southern US became a serious weed control problem prior to the extensive use of glyphosate-resistant crops. Currently glyphosate-resistant populations of Palmer amaranth occur in many areas of this geographic region creating an even more serious threat to crop production. Investigations were undertaken using four biotypes (one glyphosate-sensitive, one resistant from Georgia and two of unknown tolerance from Mississippi) of Palmer amaranth to assess bioassay techniques for the rapid detection and level of resistance in populations of this weed. These plants were characterized with respect to chlorophyll, betalain, and protein levels and immunological responses to an antibody of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) the target site of glyphosate. Only slight differences were found in four biotypes grown under greenhouse conditions regarding extractable soluble protein and chlorophyll content, but one biotype was found to be devoid of the red pigment, betalain. Measurement of early growth (seedling shoot elongation) of seedlings was a useful detection tool to determine glyphosate resistance. A leaf disc bioassay (using visual ratings and/or chlorophyll analysis) and an assay for shikimate accumulation were effective methods for determining herbicide resistance levels. The two unknown biotypes were found to be resistant to this herbicide. Some differences were found in the protein profiles of the biotypes, and western blots demonstrated a weak labeling of antibody in the glyphosate-sensitive biotype, whereas strong labeling occurred in the resistant plants. This latter point supports research by others, that increased copy number of the EPSPS gene (and increased EPSPS protein levels) is the resistance mechanism in this species. Results indicate the utility of certain bioassays for the determination of resistance and provide useful comparative information on the levels of inherent constituents among closely related plants.
1. Introduction
Chemical management of weeds is a necessary and expensive challenge, and herbicides have been very effective for weed control in food and fiber crops and in other unwanted plant growth situations. However, herbicide use has also resulted in the development of resistant weeds and presently 397 unique cases of herbicide resistant weeds have been documented [1]. A major portion of the resistance problem has been exacerbated by the increasing use of glyphosate [N-(phosphonomethyl)glycine] to control weeds in genetically modified organisms (GMO) crops such as corn, cotton and soybeans [2,3]. Early resistance risk considerations assumed low probability for the development of resistant weeds using glyphosateresistant GMO crops [4]. Initially glyphosate controlled most weeds, but as early as 1997, glyphosate resistant weeds were reported and presently there are 24 documented cases of resistance [1]. These resistant weed biotypes have been reported from virtually all countries in the world where agrochemicals have been used for weed control and are causing major problems in rangelands, natural settings and rights-of-way.
In the southern US, glyphosate-resistant Palmer amaranth populations are widespread where this weed is an economically important detriment in row crop production.
Palmer amaranth was one of the most troublesome weeds of agronomic crops across the southern US prior to resistance issues [5]. The first glyphosate-resistant Palmer amaranth population was discovered in Georgia [6] and shortly thereafter, glyphosate resistant Palmer amaranth was confirmed in North Carolina, South Carolina, Tennessee, and Arkansas [7,8]. Palmer amaranth is a dioecious plant (either male or female) and due to crosspollination, genetic traits are exchanged each season. Palmer amaranth seed production can range between 60,000 [9] and 500,000 seed m−2 [10] depending on plant density. Its characteristics of a rapid growth rate and tall stature [11] make Palmer amaranth extremely competitive with all crops. A single glyphosate-resistant Palmer amaranth spaced every 7 m of row while emerging with the crop reduced yield 7% in cotton [12]. Palmer amaranth also impacts harvesting, and one Palmer amaranth plant every 3 m of row can reduce mechanical harvesting efficiency by 2.4% [13]. The economical management of glyphosate-resistant Palmer amaranth is very difficult with currently available herbicide technology in cotton [14-17]. Palmer amaranth also causes large yield reductions (17% to 68%) in soybean [9]. Palmer amaranth in the southeast has also evolved resistances to dinitroanilines and acetolactate synthase inhibitors [6] and multiple resistance in Palmer amaranth to glyphosate and pyrithiobac was recently confirmed in Georgia [18].
The molecular mode of action of glyphosate in plants is competitive inhibition of the enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS; E.C. 2.5.1.19), which catalyzes the penultimate step of the pre-chorismate portion of the plastid-localized shikimate pathway [19-21]. As much as 20% of the total fixed carbon in a plant is directed through the shikimate pathway resulting in biosynthesis of aromatic amino acids (Tyr, Phe, and Trp), and other products (vitamin K, vitamin E, tetrahydrofolate, and ubiquinone) [20,22]. These aromatic amino acids, are precursors for many secondary metabolites including lignin, flavonoids, and alkaloids [23,24]. In glyphosate-susceptible plants, shikimate levels increase after glyphosate application due to inhibition of EPSPS [25].
Recently, glyphosate-resistant Palmer amaranth was shown to possess an extraordinarily high copy number of the EPSPS gene compared to glyphosate-susceptible plants [26]. This high EPSPS gene copy number results in production of sufficient EPSPS to support aromatic amino acid synthesis even after treatment with glyphosate. The increased EPSPS gene copy number is a heritable trait when plants are cross-bred [26]. This genetic basis for glyphosate resistance in Palmer amaranth allows rapid spread of the trait over any distance that the pollen may travel. Movement of resistance traits via pollen for distances up to 1000 feet from resistant male plants to susceptible female plants has been reported [27].
Individual plants of a given weed species may appear similar, but at the molecular level there may be vast genetic variability within a population from a localized area or among populations from diverse locations. Conversely, plants of the same species may possess obvious outward physical differences. Since Palmer amaranth covers a large geographic area there could be major differences (morphological and molecular) among plant sources or biotypes. Therefore characterization of growth characteristics and molecular traits among these populations are important in the full understanding of the development, competitiveness, spread, and control of problematic weeds. Using leaf disc bioassays, we have screened numerous Palmer amaranth plants for possible resistance to glyphosate, and found a wide range of tolerance levels to this herbicide. Our first objective in this study was to examine several bioassays to establish glyphosate resistance (and level of resistance) in four Palmer amaranth biotypes using seed germination and early growth, leaf disc injury, and shikimate accumulation as assay tools. Secondly, we wanted to characterize four biotypes from different locations with respect to their pigmentation, chlorophyll content, protein profiles and immunological responses to an EPSPS protein antibody and possibly correlate this information with tolerance levels to glyphosate.
2. Materials and Methods
2.1. Sources of Seeds and Chemicals
All chemicals were of reagent grade or higher purity. Glyphosate (99.8% pure, free acid) was obtained from Chem Service (Chem Service, Inc. 660 Tower Lane P.O. Box 599, West Chester, PA). Seeds were obtained from field-grown plants as follows: Source C3 (glyphosate sensitive) from Decatur County, GA, source C7 (glyphosate resistant) from Macon County, GA (both kindly provided by Dr. Stanley Culpepper); and R1 and G1 from Washington County, MS. R1 and G1 were found growing in close proximity to each other and R1 possessed red stems, while visibly there was no red pigmentation in G1.
2.2. Culture of Plant Seedlings
Seeds of each Palmer amaranth population were planted in a commercial potting mix Jiffy-mix (Jiffy Products of America, Inc., Batavia, IL) contained in plastic trays. The small trays were placed in sub-irrigated trays that were placed on greenhouse benches. Greenhouse temperatures ranged from 28˚C - 32˚C with 40% - 90% relative humidity (RH). The photoperiod was 12 - 14 h with 1650 µEm−2·s−1 photosynthetically active radiation as measured at midday with a light meter (LI-COR, Inc., Lincoln, NE). After germination and emergence the plants were thinned to 6 - 8 per tray. The young seedlings were supplemented with a controlled-release (14:14:14, NPK) fertilizer. When seedlings of each population were in the 8 - 10 leaf growth stage, they were used as tissue sources for the various tests described below.
2.3. Seed Germination and Growth Bioassay for Tolerance to Glyphosate
Fifty seeds of each Palmer amaranth population were placed in a petri dish (100 × 15 mm) on a filter paper moistened with 1.5 ml deionized H2O, or solutions of various concentrations of glyphosate (0.1 - 1.0 mM). Dishes were incubated in a growth chamber (30˚C - 32) in the light (150 µEm−2·s−1) and examined for germination and growth at 24 h intervals over a period of 5 - 7 days after treatment. At the end of the incubation period, shoot length of individual seedlings was determined. Tests were conducted in triplicate and repeated.
2.4. Leaf Disc Bioassay for Tolerance to Glyphosate
Discs were cut from leaves of 3 - 4 plants from each population using a cork borer (4 mm) and placed in 12- well microtiter plates (4 discs per well) with each well containing 2.5 ml deionized H2O, or solutions of various concentrations of glyphosate. The plates were incubated under continuous light (150 µEm−2·s−1) in an environmental chamber at 28˚C for 96 h. The test was visually assessed for chlorosis at various times after treatment, and at 96 h the chlorophyll from discs was extracted using dimethyl sulfoxide (DMSO) and quantitatively determined spectrophotometrically (Microplate Reader, Model SIAFRT, Bio-Tek Instruments, Inc., Winooski, VT) [28,29]. The tests were replicated 2 to 3 times. The visual rating utilized a rating scale from 0 - 10, where 10 = no injury or chlorosis, and 0 = total chlorosis/bleaching (mortality).
2.5. Chlorophyll Extraction and Quantification
The biotypes were compared on the basis of chlorophyll content by extracting leaf tissue from 3 - 4 plants of each population using DMSO and quantitatively measured spectrophotometrically as described above [28,29]. The tests were replicated 2 - 3 times. This method was also used to measure injury (chlorosis/bleaching) in the leaf disc bioassays caused by exposure to glyphosate at various concentrations and times.
2.6. Betalain Extraction and Quantification
Weighed amounts of Palmer amaranth stem tissue from 3 - 4 plants from each population were homogenized in 15 ml acidic methanol (1% HCl, v/v) at 0˚C - 4˚C. Homogenates were clarified (20,000 g, 15 min, 0˚C) and betalain content in the supernatants was quantified as A549 - A622 g−1 fresh weight in 15 ml of extraction medium, modified and outlined previously for anthocyanin measurement [30].
2.7. Shikimate Assay
The glyphosate dose-response assay was performed in 20 ml borosilicate scintillation vials as adapted from [31]. Ten leaf discs (4 mm) from 3 - 4 plants of each biotype were placed in separate vials and 1.0 ml 10 mM ammonium phosphate plus 0.1% (v/v) Tween 80 surfactant containing 700, 300, 180, 75, 25, 5 or 0 µM glyphosate was added. The vials were shaken to ensure that all leafdiscs were wetted and submersed. The vials were placed under fluorescent lights (120 µEm−2·s−1) for 24 h. Following incubation the vials were immediately frozen and stored at −20˚C pending shikimate assay. Vials were removed from the freezer, thawed at room temperature, and 0.25 ml of 1.25 M HCl was added followed by incubation (60˚C, 15 min).
A 25 µL aliquot of the 0.25 M HCl extract was pipetted into each of three wells of a microtiter plate so that three replicate extract samples were assayed per vial. Then 100 µL 0.25% periodate/0.25% meta-periodate was as added to each well and the plate incubated at 37˚C, 40 min. The reaction was quenched by addition of 100 µl of 0.6 M NaOH/0.22 M Na2SO4 solution to each well and the optical density of each well measured at 380 ηm using a plate reader as described above. Shikimate standards were prepared by adding known amounts of shikimate to vials containing leaf discs of a glyphosate-sensitive biotype that had not been exposed to glyphosate. Shikimate levels are reported as µg shikimate ml−1 of HCl solution. Data were analyzed by analysis of variance of fixed effects with differences presented as Least Squares Means (Software, Statistical Analysis Systems®, Version 9.1, SAS Institute, Inc., SAS Campus Drive, Cary, NC).
2.8. Accumulation of Shikimate
The vial method as outlined above was used to quantify and establish the linearity of the shikimate extracted from a tissue assay using glyphosate-sensitive Palmer amaranth (biotype C3) leaf discs. Known amounts of shikimate to provide a range of concentrations (0 - 180 µM) were added to each vial containing 10 leaf discs from 3 - 4 plants of each population (4 mm diameter). The assay protocol was immediately initiated and the recovered shikimate in each vial determined using the assay described above.
2.9. Protein Extraction, SDS-PAGE, and Protein Electrophoresis
Generally, leaf tissue was excised, immediately weighed, and 0.5 g tissue was ground with 2.0 ml extraction buffer in a chilled mortar and pestle. Samples consisted of tissue from 3 - 4 plants of each biotype and were replicated at least 3 times. Extraction buffer consisted of 100 mM 3-(N-morpholino) propanesulfonic acid (Mops), 5 mM EDTA, 5% polyvinylpyrrolidone, 1% Triton-X 100, 10% glycerin, 50 mM KCl, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 100 µM iodoacetamide as modified [26]. The homogenate was centrifuged (15,000 ×g at 4˚C for 15 min) and the supernatant was retained for protein analysis. Protein levels in samples were quantified (Lowry-based Bio-Rad RC DC protein assay). Aliquots of the samples were boiled 10 min in Laemmli sample buffer containing 2-mercaptoethanol and 25 µg total protein of each sample were fractionated by SDS polyacrylamide gel electrophoresis on a 12.5% Tris-HCl Criterion gel (Bio-Rad Corp., Van Nuys, CA). The gel was run for 45 min at a constant voltage of 200 V. Gels were stained using Biosafe Coomassie stain (Bio-Rad Corp., Van Nuys, CA) and photographed. Molecular weight marker proteins were electrophoresed in separate lanes as size standards (Bio-Rad Corp., Van Nuys, CA).
2.10. Immunoblotting of EPSP Synthase
Two sets of the four Palmer amaranth protein extract samples (25 µg) were loaded on a 12.5% Tris-HCl gels (Bio-Rad, Van Nuys, CA). A set of standard protein markers was also added to separate lanes and electrophoresed for 45 min at constant voltage of 200 V. One half of the gel was stained with Biosafe Coomassie stain as described and the other half was processed for immunoblotting of EPSP synthase by western blot analysis. Proteins on the unstained half of the gel were transferred to a PVDF membrane in a semi-dry Trans-blot apparatus (Bio-Rad, Van Nuys, CA) for 55 min at constant voltage of 15 V. The blots were then blocked for non-specific binding 12 h at 4˚C with 5% Blotto [5% dry milk in 10 mM Tris, 150 mM NaCl, 0.1% Tween-20 (pH 8.0 TTBS)] washed in TTBS. The blocked blot was then incubated 1 h at room temperature with an antibody produced (Agrisera AB, Vännäs, Sweden) against a sequence of residues (numbers 193 - 206) of Palmer amaranth EPSPS (GenBank accession number FJ861242) [26]. The antibody was diluted 1:2000 in 0.4% Blotto [0.4% dry milk in 10 mM Tris, 150 mM NaCl, 0.1% Tween-20 (pH 8.0 TTBS)]. Following incubation, the membrane was washed in TTBS and incubated 1 h at room temperature with an anti-rabbit secondary antibody-conjugated alkaline phosphatase diluted 1:20,000 in 0.4% Blotto. Bands were detected with 5-chloro-2-methoxybenzenediazonium chloride hemi (zinc chloride) salt (Fast Red RC, Sigma Aldrich, St Louis, MO) in naphthol AS-TR Phosphate substrate (Sigma Aldrich). The blot was photographed on a Versa doc molecular imager (Bio-Rad, Van Nuys, CA) and analyzed with Carestream Molecular Imaging Software (Carestream Health, Inc., Rochester, NY).
3. Results and Discussion
3.1. Comparison of General Traits of Four Palmer Amaranth Biotypes
Seeds of the four biotypes grown under greenhouse conditions produced seedlings with a generally equal appearance except one biotype lacked red pigmentation (betalain) (Figure 1). Analysis of betalain content in the four biotypes showed some variation in pigment content, but the green stemmed biotype was void of betalain (Figure 2). Betalains are nitrogen-containing, water-soluble, yellow-to-red pigment compounds found only in a limited number of plants. Although derived from tyrosine, the biosynthetic pathways of betalains and the associated enzymes and genes are not as well characterized as those of flavonoids and carotenoids [32]. Inherent chlorophyll content in leaves of the four biotypes in greenhousegrown seedlings was not significantly different (Figure 3). No obvious or major differences in growth characteristics among the biotypes were noted, however this parameter was not part of the objectives or the experimental design.
3.2. Leaf Disc Bioassay for Tolerance to Glyphosate
In this bioassay, two methods for evaluating tolerance to glyphosate were used, i.e., a visual rating of injury over time, and chlorophyll extraction and quantization at the termination of the bioassay. Visualization and rating of injury to leaf discs caused by exposure to several concentrations of glyphosate provided a qualitative estimate of relative injury compared to control discs exposed to H2O (Table 1). Biotype C3 was more sensitive to glyphosate at all concentrations tested, whereas biotypes C7, R1 and G1 were resistant after 96 h exposure. The high glyphosate concentration (1 mM) severely damaged the susceptible biotype. Visual analysis allowed non-destructive comparison of glyphosate injury to a given biotype, as well as comparison of injury among biotypes at

Figure 1. Visual comparison of seedlings of four A. palmeri biotypes used in this study. Top photo (whole seedlings; four biotypes); bottom photo (close-up of lower stems of two biotypes from Washington County, MS).
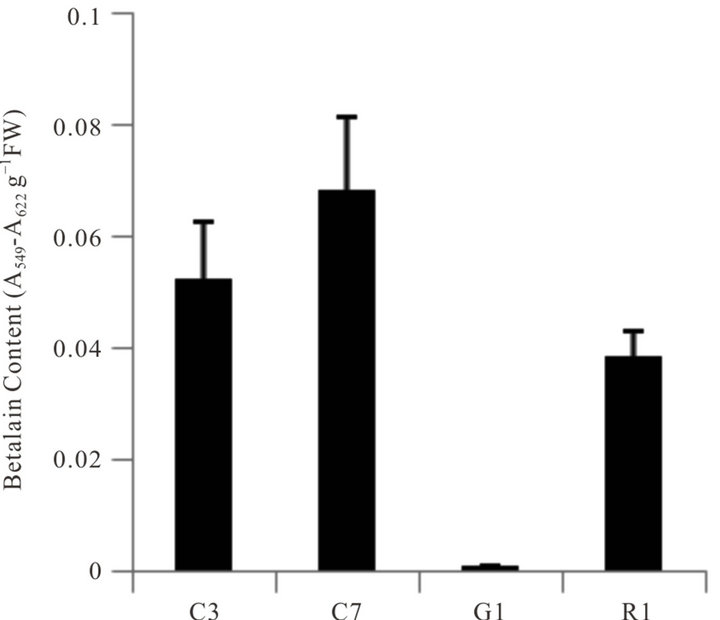
Figure 2Comparison of betalain content in stem tissues of four A. palmeri biotypes. Error bars represent ± 1 SEM.
any time point during the bioassay. Chlorophyll analysis at the termination of the bioassay provided a quantitative measure of injury and the reduction in chlorophyll content paralleled that of the visual ratings (data not shown). Similar bioassays for detecting injury caused by herbicidal and allelopathic activity have been discussed and
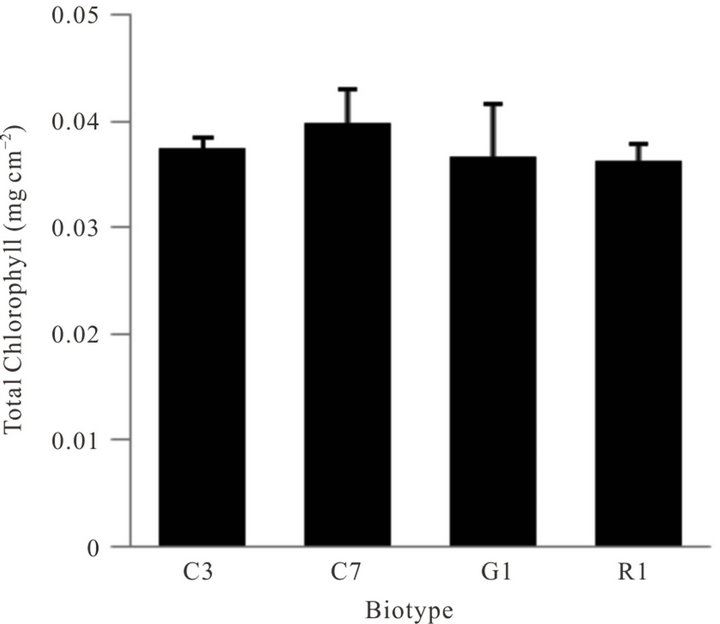
Figure 3. Comparison of total chlorophyll content in leaves of four biotypes of A. palmeri grown under greenhouse conditions. Error bars represent ± 1 SEM.
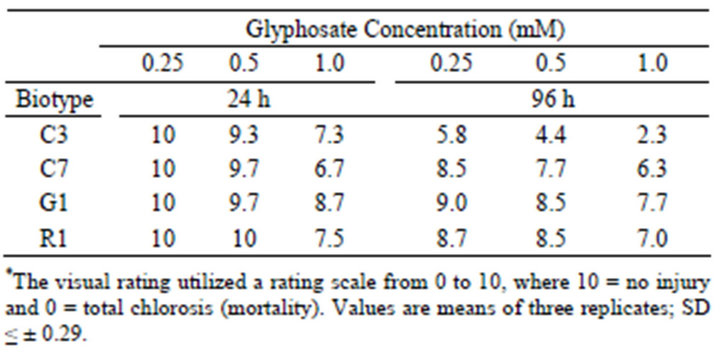
Table 1. Visual injury rating of leaf discs of four Palmer amaranth biotypes exposed to glyphosate at several concentrations under continuous light over a 96 h time course.*
reviewed [33-36]. Furthermore, glyphosate (1.0 mM) spray application to plants of these four biotypes corroborated the bioassay results in that C7, R1 and G1 were glyphosate-resistant and C3 was susceptible to glyphosate when treated at the 4-5 leaf growth stage (data not shown).
3.3. Seed Germination and Early Growth Bioassays
To carry out an effective seed germination bioassay, two important criteria must be met. The seed source should be of sufficient viability to provide relatively high germination percentages, and the seed stock should be ample enough to use statistically significant seed numbers set-up in triplicate or more replications. Although seed germination bioassays at various glyphosate concentrations were conducted, most of the seed stocks were either insufficient in quantity or lacked proper viability thus proper statistical analysis of data (e.g. transformation using an arc sin-square root transformation) was not possible. Thus the seed germination bioassays to detect glyphosate resistance in these tests were unsuccessful or inconclusive (data not shown). However, measurement of the early growth of seedlings after seeds imbibed and grew in various glyphosate concentrations in this bioassay system was useful to measure tolerance to glyphosate by these plants. Shoot growth of such seedlings demonstrated that biotypes C7, R1 and G1 were resistant to glyphosate, whereas biotype C1 was very susceptible to low levels of glyphosate (Figure 4).
3.4. Accumulation of Shikimate as a Function of Time after Exposure to Glyphosate
Quantification of the shikimate extracted from leaf disc tissue of glyphosate-sensitive Palmer amaranth (biotype C3) leaf discs showed that shikimate recovery was linear (data not shown). The accumulation of shikimate over a 48 h time-course in leaf discs of biotype C3 exposed to glyphosate (250 µM) was also linear (Figure 5). Profiles of shikimate accumulation in these four biotypes strongly correlated with their resistance levels to glyphosate (Figure 6). Shikimate accumulated as exposure time increased in the sensitive biotype C3, but no shikimate build-up occurred in the glyphosate resistant biotypes C7, R1 and G1 indicating that these latter three biotypes are resistant to glyphosate.
3.5. Protein Extraction, SDS-PAGE, and Protein Electrophoresis
There was a high degree of similarity of the profiles of
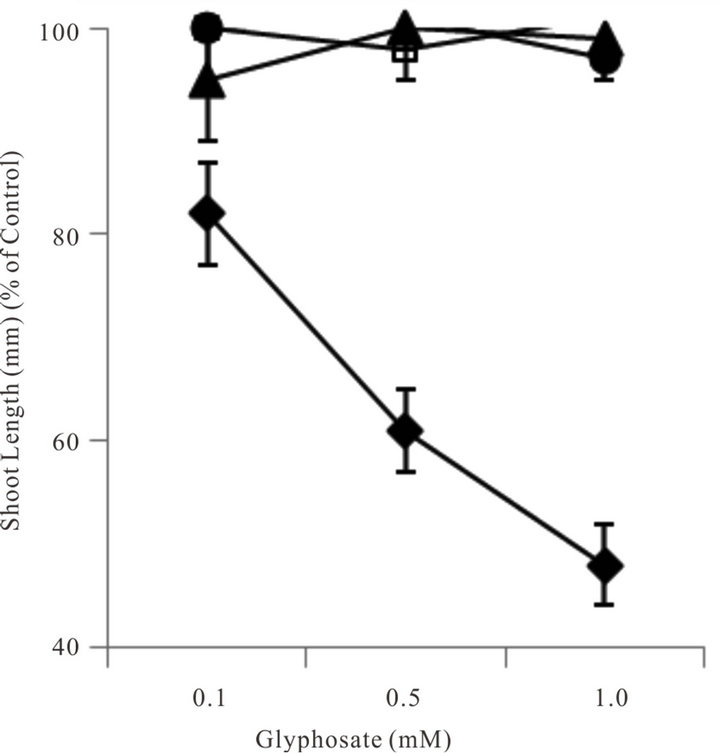
Figure 4. Comparative growth (shoot length) of seedlings of four A. palmeri sources 120 h after seed imbibition and growth in several glyphosate concentrations in the light. Data points are defined as; ♦ = C3; □ = C7; ▲= G1 and ● = R1. Error bars represent ± 1 SEM.

Figure 5. Accumulation of shikimate with time in leaf discs of a glyphosate-sensitive (C3) A. palmeri biotype exposed to 250 μM glyphosate and light over a 48 h-time course. The relationship between shikimate accumulation with time is best described by the equation Y = 0.422X – 2.021; R2 = 0.91.

Figure 6. Accumulation of shikimate in leaf discs of four A. palmeri biotypes exposed to various concentrations of glyphosate and light for 24 h. Data points are defined as; ♦ = C3; □ = C7; ▲= G1 and ● = R1. Error bars represent ± 1 SEM.
extracted, denatured protein in the four biotypes (Figure 7). However, the relative levels of some of the bands differed when the biotypes were compared. Distinct bands, but with different intensities, were apparent in all biotypes corresponding to an estimated molecular weight of ~47 - 50 kD, a region closely matching that of the molecular weight of the EPSPS protein. Immuno-blotting with antibody for EPSP synthase (western blot) gave strong immuno-precipitation reactions at an estimated 47.2 kD in the biotypes shown to be resistant to glyphosate (C7, R1 and G1) and a very weak response in the
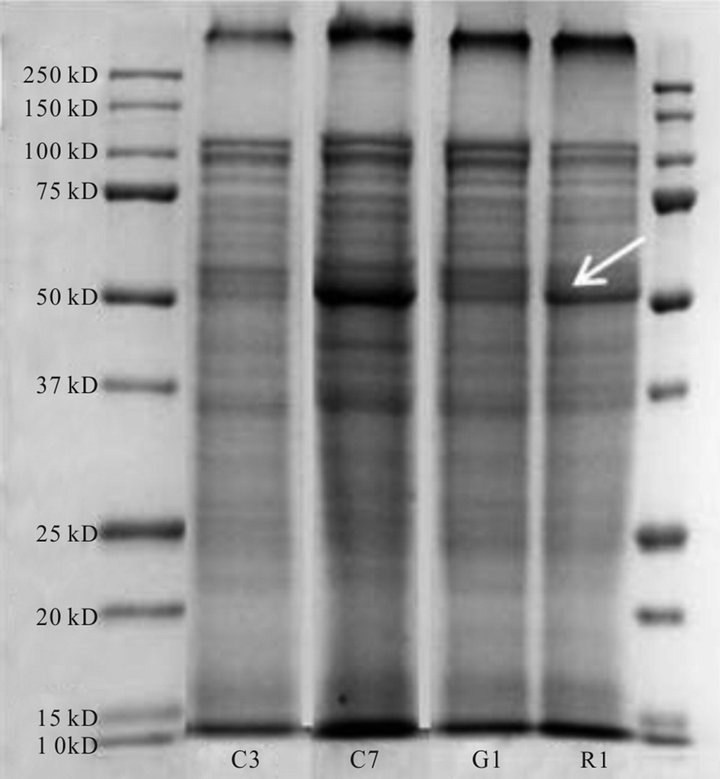
Figure 7. Protein profiles of four A. palmeri biotypes after gel electrophoresis and staining with Coomassie stain.
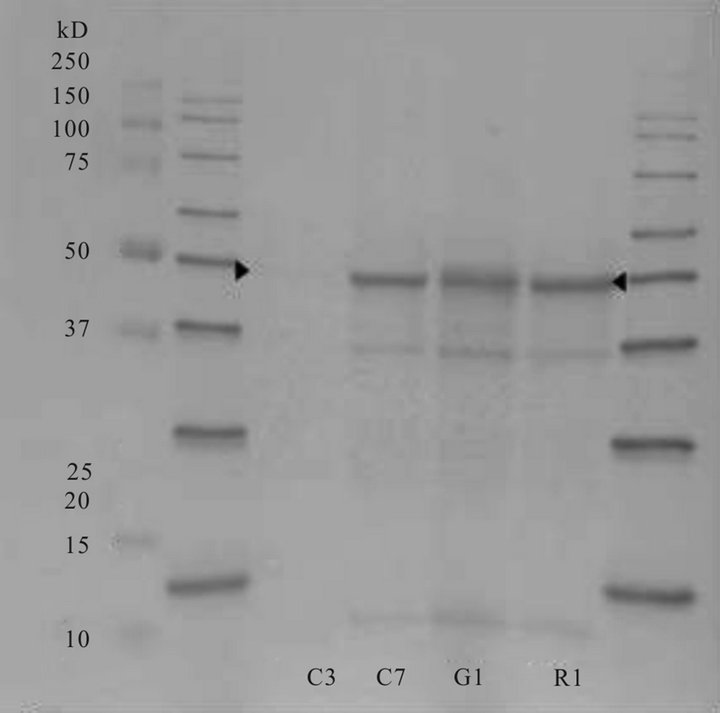
Figure 8. Immunological localization (western blot) of EPSPS in four A. palmeri biotypes using an EPSPS antibody. Standard protein molecular weight markers appear in the extreme left and right lanes.
sensitive biotype (C3) (Figure 8). Some extraneous staining also occurred in the resistant biotypes at ~38.5 and 18.1 kD. These stained areas could be fragments or artifacts. Further, an explanation of some of the positive reactions on the blots may be due to the fact that our antibody was in the form of serum, and not used as a purified antibody. However, in preliminary tests we showed that the pre-immune serum produced no bands in western blots using Palmer amaranth protein extracts (data not shown).
Alternatively, early research on EPSPS and glyphosate on cell cultures of Corydalis sempervirens using immuno-precipitation reported that the protein is synthesized as an extra-plastidic precursor at a molecular weight of 53.9, compared to 45.5 kD for the mature enzyme [37]. We are unaware of reports on precursor versus mature enzyme studies in Palmer amaranth. Our immunological results support reports [26] that resistance in this species is due to gene amplification allowing more than ample amounts EPSPS to be synthesized. Further studies using two-dimensional gels followed by western blotting will be needed to characterize specific protein differences among these biotypes and to identify EPSPS and possible isozymes.
Knowledge of the distribution of herbicide-resistant weeds is important to provide best management practices and to potentially reduce spread to non-infested areas. Overall, the use of bioassays to detect resistance of plant to herbicides can allow rapid screening of many populations when compared to spray applications of herbicide to greenhouse-grown or to field plants. The early growth after seed imbibition of glyphosate from solutions and the leaf disc bioassays used here can depict overall injury caused by most phytotoxic agents including effects caused by glyphosate (EPSPS inhibition). In contrast, the assay for shikimate accumulation following glyphosate treatment is applicable only to EPSPS inhibition and is therefore herbicide-specific (glyphosate) since it is the only herbicidal compound possessing this mode of action. Results using the leaf disc bioassay, followed by chlorophyll quantification, were supported by visual ratings of injury caused by glyphosate exposure. Furthermore, results from the leaf disc bioassay also corroborated results obtained from the shikimate accumulation assay. These various bioassays indicated that the two Washington County, MS biotypes tested from were resistant to glyphosate. Western blot analysis depicted some differences in intensity that support resistance due to variation in copy number as reported earlier [26].
4. Conclusions
Although the G1 biotype (void of the red pigment betalain) was resistant to glyphosate, future studies will be needed to determine the nature of this mutation and how (or if) it is in any way related to glyphosate resistance. Since the biosynthesis of this pigment is derived from an amino acid (tyrosine) [32] whose synthesis is inhibited by glyphosate, further investigation of this will be of interest.
Growth and physiological attributes of weeds (and herbicide-resistant weeds) are important to determine competitiveness, and some information has been provided on Amaranth species [10,11]. However, the growth characteristics and competitiveness of these Palmer amaranth biotypes and others under controlled environmental conditions and/or after exposure to chemical or biological stresses has not been accessed, but is an important subject for future research. Also further examination of the proteomic aspects of these and other biotypes will be undertaken.
Under high selection pressure, resistant plants may dominate a field population relatively rapidly (four to five generations) once plant genes are selected that confer resistance to the selection pressure of a herbicide [38]. Thus, an increase in the spread of resistance to glyphosate among populations of Palmer amaranth will cause greater crop yield losses due to weed competition. Bioassays could provide the means for a cost-effective, nondestructive, rapid and high throughput identification of resistant weeds from field or greenhouse samples.
5. Acknowledgements
The contribution of Palmer amaranth seeds (a sensitive and a resistant population) collected in Georgia from Dr. Stanley Culpepper is greatly appreciated. We thank Dr. Rick Turley for helpful discussions during this project.
REFERENCES
- Heap, “International Survey of Herbicide-Resistant Weeds,” 2013. http://www.weedscience.org/in.asp
- Anonymous, “New Report Reveals Dramatic Rise in Pesticide Use on Genetically Engineered (GE) Crops Due to the Spread of Resistant Weeds,” 2009. http://wwwprnewswire.com/news-release
- B. G. Young, “Changes in Herbicide Use Patterns and Production Practices Resulting from Glyphosate-Resistant Crops,” Weed Technology, Vol. 20, No. 2, 2006, pp. 301- 307. doi:10.1614/WT-04-189.1
- L. D. Bradshaw, S. R. Padgette, S. L. Kimball and B. H. Wells, “Perspectives on Glyphosate Resistance,” Weed Technology, Vol. 11, No. 1, 1997, pp. 189-198.
- T. M. Webster, “Weed Survey-Southern States: Broadleaf Crops Subsection,” Proceedings of the Southern Weed Science Society, Vol. 58, 2005, pp. 291-294.
- S. Culpepper, T. L. Grey, W. K. Vencill, J. M. Kichler, T. M. Webster, S. M. Brown, A. C. York, J. W. Davis and W. W. Hanna, “Glyphosate-Resistant Palmer Amaranth (Amaranthus palmeri) Confirmed in Georgia,” Weed Science, Vol. 54, No. 4, 2006, pp. 620-626. doi:10.1614/WS-06-001R.1
- R. C. Scott, L. E. Steckel, K. L. Smith, T. Mueller, L. R. Oliver and J. Norsworthy, “Glyphosate-Resistant Palmer Amaranth in the Southeastern United States,” In: W. K. Vencill, Ed., Proceedings of the 60th Southern Weed Science Society, Nashville, 22-24 January 2007.
- C. York, J. R. Whitaker, A. S. Culpepper and C. L. Main, “Glyphosate-Resistant Palmer Amaranth in the Southeastern United States,” Proceedings of the Southern Weed Science Society, Vol. 60, 2007, p. 225.
- N. Bensch, M. J. Horak and D. Peterson, “Interference of Redroot Pigweed (Amaranthus retroflexus), Palmer Amaranth (A. palmeri), and Common Waterhemp (A. rudis) in Soybean,” Weed Science, Vol. 51, No. 1, 2003, pp. 37-43. doi:10.1614/0043-1745(2003)051[0037:IORPAR]2.0.CO;2
- A. Sellers, R. J. Smeda, W. G. Johnson, A. J. Kendig and M. R. Ellersieck, “Comparative Growth of Six Amaranthus Species in Missouri,” Weed Science, Vol. 51, No. 3, 2003, pp. 329-333. doi:10.1614/0043-1745(2003)051[0329:CGOSAS]2.0.CO;2
- M. J. Horak and T. M. Loughin, “Growth Analysis of Four Amaranthus Species,” Weed Science, Vol. 48, No. 3, 2000, pp. 347-355. doi:10.1614/0043-1745(2000)048[0347:GAOFAS]2.0.CO;2
- W. MacRae, A. S. Culpepper, T. M. Webster and J. M. Kichler, “The Effect of Glyphosate-Resistant Palmer Amaranth Density and Time of Establishment on Yield of Cotton,” Proceedings of the Southern Weed Science Society, Vol. 60, 2007, p. 228.
- D. T. Smith, R. V. Baker and G. L. Steele, “Palmer Amaranth (Amaranthus palmeri) Impacts on Yield, Harvesting, and Ginning in Dryland Cotton,” Weed Technology, Vol. 14, No. 1, 2000, pp. 122-126. doi:10.1614/0890-037X(2000)014[0122:PAAPIO]2.0.CO;2
- S. Culpepper, A. W. MacRae, A. C. York and J. Kichler, “Managing Glyphosate-Resistant Palmer Amaranth in Conventional and Strip-till Roundup Ready Cotton,” Proceedings of the Southern Weed Science Society, Vol. 61, 2008, p. 62.
- A. S. Culpepper, A. C. York and J. Kichler, ‘University of Georgia Herbicide Programs for Controlling Glyphosate-Resistant Palmer Amaranth in 2008 Cotton,” Circular No. 924, Georgia Cooperative Extension Service, Tifton, 2008.
- S. Culpepper, A. C. York, J. M. Kichler and A. W. MacRae, “Glyphosate-Resistant Palmer Amaranth Response to Weed Management Programs in Roundup Ready and Liberty Link Cotton,” Proceedings of the Beltwide Cotton Conference, Nashville, 10 January 2008, pp. 1689-1690.
- J. R. Whitaker, “Distribution, Biology, and Management of Glyphosate-Resistant Palmer Amaranth in North Carolina,” Ph.D. Dissertation, North Carolina State University, Raleigh, 2009, http://repository.lib.ncsu.edu/ir/bitstream/1840.16/3130/1/etd.pdf
- L. M. Sosnoskie, J. M. Kichler, R. D. Wallace and A. S. Culpepper, “Multiple Resistance in Palmer Amaranth to Glyphosate and Pyrithiobac Confirmed in Georgia,” Weed Science, Vol. 59, No. 3, 2011, pp. 321-325. doi:10.1614/WS-D-10-00132.1
- H. C. Steinrücken and N. Amrhein, “The Herbicide Glyphosate is a Potent Inhibitor of 5-Enolpyruvyl-Shikimic Acid-3-Phosphate Synthase,” Biochemical and Biophysical Research Communications, Vol. 94, No. 4, 1980, pp. 1207-1212. doi:10.1016/0006-291X(80)90547-1
- J. E. Franz, M. K. Mao and J. A. Sikorski, “Glyphosate’s Molecular Mode of Action,” In: Glyphosate: A Unique Global Herbicide, American Chemical Society, Washington DC, 1997, pp. 521-615.
- K. J. Gruys and J. A. Sikorski, “Inhibitors of Tryptophan, Phenylalanine and Tyrosine Biosynthesis as Herbicides,” In: B. K. Singh, Ed., Plant Amino Acids: Biochemistry and Biotechnology, CRC Press, Boca Raton, 1999, pp. 357-384.
- E. Haslam, “Shikimic Acid: Metabolism and Metabolites,” John Wiley Sons, Chichester, 1993, 400 Pages.
- K. M. Herrmann, “The Shikimate Pathway: Early Steps in the Biosynthesis of Aromatic Compounds,” Plant Cell, Vol. 7, No. 7, 1995, pp. 907-919.
- K. M. Herrmann, “The Shikimate Pathway as an Entry to Aromatic Secondary Metabolism,” Plant Physiology, Vol. 107, No. 1, 1995, pp. 7-12. doi:10.1104/pp.107.1.7
- N. Amrhein, B. Deus, P. Gehrke and H. C. Steinrüchen, “The Site of the Inhibition of the Shikimate Pathway by Glyphosate. II. Interference of Glyphosate with Chorismate Formation in Vivo and in Vitro,” Plant Physiology, Vol. 66, No. 5, 1980, pp. 830-834. doi:10.1104/pp.66.5.830
- T. A. Gaines, W. Zhang, D. Wang, B. Bukun, S. T. Chisholm, D. L. Shaner, S. J. Nissen, W. L. Patzoldt, P. J. Tranel, A. S. Culpepper, T. L. Grey, T. M. Webster, W. K. Vencill, R. D. Sammons, J. Jiang, C. Preston, J. E. Leach and P. Westra, “Gene Amplification Confers Glyphosate Resistance in Amaranthus palmeri,” Proceedings of the National Academy of Sciences, Vol. 107, No. 3, 2010, pp. 1029-1034. doi:10.1073/pnas.0906649107
- L. M. Sosnoskie, T. M. Webster, J. M. Kichler, A. W. MacRae and A. S. Culpepper, “Preliminary Estimates of Glyphosate-Resistant Amaranthus palmeri Pollen Dispersal Distances,” Proceedings of the Beltwide Cotton Conference, New Orleans, 11 January 2007, p. 1228.
- J. D. Barnes, L. Balaguer, E. Manriques, S. Elvira and A. W. Davison, “A Reappraisal of the Use of DMSO for the Extraction and Determination of Chlorophylls a and b in Lichens and Higher Plants,” Environmental and Experimental Botany, Vol. 32, No. 2, 1992, pp. 85-100. doi:10.1016/0098-8472(92)90034-Y
- J. D. Hiscox and G. F. Israelstam. “A Method for the Extraction of Chlorophyll from Leaf Tissue without Maceration,” Canadian Journal of Botany, Vol. 57, No. 12, 1979, pp. 1332-1334. doi:10.1139/b79-163
- R. E. Hoagland, “Effects of Glyphosate on Metabolism of Phenolic Compounds: VI. Effects of Glyphosine and Glyphosate Metabolites on Phenylalanine Ammonialyase Activity, Growth and Protein, Chlorophyll and Anthocyanin Levels in Soybean (Glycine max) Seedlings,” Weed Science, Vol. 28, No. 4, 1980, pp. 393-400.
- D. L. Shaner, T. Nadler-Hassar, W. B. Henry and C. H. Koger, “A Rapid in Vivo Shikimate Accumulation Assay with Excised Leaf Discs,” Weed Science, Vol. 53, No. 6, 2005, pp. 769-774. doi:10.1614/WS-05-009R.1
- Y. Tanaka, N. Sasaki and A. Ohmiya, “Biosynthesis of Plant Pigments: Anthocyanins, Betalains and Carotenoids,” The Plant Journal, Vol. 54, No. 4, 2008, pp. 733-749. doi:10.1111/j.1365-313X.2008.03447.x
- H. J. Beckie, I. M. Heap, R. J. Smeda and L. M. Hall, “Screening for Herbicide Resistance in Weeds,” Weed Technology, Vol. 14, No. 2, 2000, pp. 428-445. doi:10.1614/0890-037X(2000)014[0428:SFHRIW]2.0.CO;2
- R. E. Hoagland and R. D. Williams, “Bioassays: Useful Tools for the Study of Allelopathy,” In: F. Macias, J. Galindo, J., Molinillo and H. Cutler, Eds., Allelopathy, CRC Press, 2004, pp. 315-351.
- P. Boger and G. Sandmann, “Target Assays for Modern Herbicides and Related Phytotoxic Compounds,” Lewis Publishers, Boca Raton, 1993, 299 pages.
- J. C. Streibig, “Herbicide Bioassay,” Weed Research, Vol. 28, No. 6, 1988, pp. 479-484. doi:10.1111/j.1365-3180.1988.tb00831.x
- H. Hollander-Czytko and N. Amrhein, “5-EnolpyruvylShikimate 3-Phosphate Synthase, the Target Enzyme of the Herbicide Glyphosate, Is Synthesized as a Precursor in a Higher Plant,” Plant Physiology, Vol. 83, No. 2, 1987, pp. 229-231. doi:10.1104/pp.83.2.229
- M. Jasieniuk, A. L. Brule-Babel and I. N. Morrison, “The Evolution and Genetics of Herbicide Resistance in Weeds,” Weed Science, Vol. 44, No. 1, 1996, pp. 176-193.

