American Journal of Plant Sciences
Vol. 4 No. 2A (2013) , Article ID: 28471 , 10 pages DOI:10.4236/ajps.2013.42A057
Rapid In-Vitro and In-Vivo Detection of Chalara fraxinea by Means of Mass Spectrometric Techniques
![]()
1Institute of Ecology, Berlin University of Technology, Berlin, Germany; 2Institute for Forest Genetics (TI), Federal Research Institute for Rural Areas, Forestry and Fisheries, Waldsieversdorf, Germany; 3Leibniz Institute for Molecular Pharmacology, Berlin, Germany.
Email: *lamhuong.pham@tu-berlin.de
Received December 21st, 2012; revised January 22nd, 2013; accepted January 30th, 2013
Keywords: Chalara fraxinea; Rapid In-Vivo and In-Vitro Detection; MS Techniques; MALDI; LC-MS
ABSTRACT
For the first time, mass spectrometric (MS) techniques were employed to rapidly detect the pathogen Chalara fraxinea in-vitro and directly in-vivo in tissues of diseased ash trees caused by C. fraxinea, using a range of characteristic novel secondary metabolites of C. fraxinea as chemical markers for the presence of the pathogen. We have found an evident correlation between the presence and amount of these-only for C. fraxinea characteristic and novel-secondary metabolites (named chalarafraxinines) and the degree of disease of respective infected ash seedlings. As demonstrated in this work, the MS based high-throughput-screening approach constitute an alternative to the time consuming and expensive micro biological isolation procedures for detection of the pathogen C. fraxinea and furthermore, can be used to rapidly test ash genotypes for resistance/susceptibility to C. fraxinea infection.
1. Introduction
Since the discovery of the aggressive ascomycetous fungus Hymenoscyphus pseudoalbidus on common ash (Fraxinus excelsior L.) in Eastern and Northeastern Europe [1-3], the disease has widespread throughout 22 countries in Europe [4]. The outer symptoms include wilting, necrotic bark lesions of variable size, dieback and decline. Trees at various ages are affected, in forest stands as well as in landscape and in nurseries [5]. At first, the mitosporic ascomycete Chalara fraxinea Kowaslski has been identified as the causal agent of this disease in ash trees [6,7]. More recently, based on molecular genetic studies using inter-simple sequence repeat anchored PCR (ISSRPCR) fingerprinting and three sequence loci, the teleomorph Hymenoscyphus pseudoalbidus has been linked to C. fraxinea [8]. This species was separated from the formerly assigned discomycete Hymenoscyphus albidus, which in contrary is known to be a non-pathogenic litter decomposing organism in Europe since more than 150 years. This new species can be clearly distinguished by its pathogenicity to common ash. The isolation of the pathogen from ash tissue is difficult because of the abundance of secondary pathogens [9]. Therefore, the search for an objective evidence of C. fraxinea as the primary cause of ash decline is of interest and has been intensified lately. The exact and rapid in planta detection of C. fraxinea has to be carried out by real-time PCR assay using a dual-labelled probe [10]. From the genus Chalara, many secondary metabolites of several substance groups were isolated and identified, for examples the antifungal and antibacterial isofusidienols from Chalara sp. strain 6661—an endophytic fungus isolated from Artemisia vulgaris [11]—and the antifungal lipopeptides FR227673 and FR190293 from fermentation broth of the soil fungus Chalara sp. 22210 [12]. Recently, analyses of the anamorphic stage of Hymenoscyphus pseudoalbidus indicated also the presence of the phytotoxin viridiol and the fungistatic compound viridin as well as viridin-like steroids [13,14].
In this paper, we describe our work on the use of mass spectrometric techniques for rapid detection of the pathogen C. fraxinea via a range of its characteristic novel secondary metabolites of C. fraxinea (named chalara fraxinines). These metabolites have molecular weight mainly in the range from 900 to 1250 Da and can be easily detected by means of MS techniques directly from culture media or plant tissue extracts. Therefore, these metabolites are ideal to be used as chemical markers for indication of the presence of C. fraxinea [15,16]. A rapid mass spectrometric screening for the pathogen C. fraxinea in-vitro and in-vivo can be well achieved by using MALDI-TOF-MS. This technique is particularly suitable for detection of compounds with a molecular weight higher than 400 Da, such as the chalarafraxinines (especially when α-cyano-4-hydroxycinnamic acid is used as matrix), but also for peptides and proteins. Convincing advantages of MALDI-TOF-MS are its simplicity, high sensitivity, capability to provide data within minutes and low detection limits (pico to femtomole) [17-19].
The identification of secondary metabolites of C. fraxinea, especially of the chalarafraxinines itself can be conducted using also MALDI-TOF/TOF-MS but also LCESI-TOF-MS/MS or -MSn techniques. LC-MS and especially LC-MS/MS, such as LC-ESI-MS/MS, LC-Q-TOFMS/MS, LC-Ion-TRAP-MSn, and high-resolution LCOrbitrapTM-MS are powerful tools to investigate complex samples because they provide important information on the molecular mass and the presence of certain functional groups of the analyses. Furthermore, the determination of the amount and in particular the quantification of secondary metabolites such as the chalarafraxinines can be carried out via LC-MS and LC-MS/MS, too. High sensitivity and selectivity make these techniques well suited for high-throughput analysis [20-24].
In principle, the here presented approach can be used as alternative to normally applied standard biological and biochemical methods. Possible advantages are the simplicity of sample preparation, inherent high detection sensitivity and high sampling rate [15,16].
2. Material and Methods
2.1. Isolation of C. fraxinea
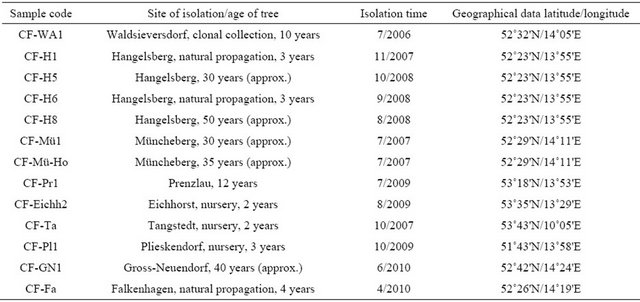
13 strains C. fraxinea were isolated from necrotic tissues of diseased ash trees from different forest sites and from nurseries located in the countries of Brandenburg and Schleswig-Holstein, Germany (see Table 1). They were identified according to Kowalski (2006) [6] via the typical phialidic conidiogenous cells and conidia formation, by sequencing of ITS1/2 regions of rDNA (18S rRNA gene partial, ITS1, 5.8S rRNA gene, ITS2 rRNA gene complete, 28S rRNA partial), and preserved at the Thuenen-Institute of Forest Genetics, Waldsieversdorf, Germany.
2.2. Preparation of C. fraxinea Cultures and Extracts for MS Experiments
C. fraxinea was cultured on several agar media MEA, PDA, mPDA, CMA, mSeed at the different temperatures 7˚C, 15˚C, 18˚C, 22˚C, and 28˚C.
Details on agar media: MEA (malt extract agar): malt extract (Roth, Karlsruhe, Germany) 1.7%, agar-agar 1.5%; PDA (potato dextrose agar): potato dextrose (Duchefa, Haarlem, The Netherlands) 3.9%; mPDA (modified potato dextrose agar): PDA supplemented with yeast extract (Duchefa) 0.15%; CMA (corn meal agar): corn meal (Serva, Heidelberg, Germany) 4%; mSeed (modified Seed agar medium, [25]): Pharmamedia (non-hydrolized globular protein from cotton seeds, Traders Inc. USA) 2%, sojapepton (Merck, Darmstadt, Germany) 0.2%, maltose (Merck) 0.4%, corn meal (Serva) 2.6%, MgSO4 × 7 H2O (Merck) 0.2%, NaCl (Merck) 0.2%, CaCO3 (Merck) 0.3%.
After 8 weeks of growing, the cultural characteristics were determined and extraction of secondary metabolites from C. fraxinea cultures for further investigations by means of MS techniques was conducted. The extraction was carried out from at 22˚C overgrown C. fraxinea culture plates and at room temperature. For that, each plate was separated in four equal parts and each quarter was taken and mixed with 10 ml of methanol. The suspensions were subsequently sonicated for 15 minutes at 15˚C and finally filtered. All extracts were stored at 4˚C.
Table 1. List of C. fraxinea isolates.
2.3. Cultivation and Preparation of Plant Materials for MS Experiments
The in-vivo detection of C. fraxinea in ash tissues via its secondary metabolites by means of MS techniques was carried out on young plants artificially and naturally infected, respectively.
33 two-year-old ash seedlings (selected propagation material, origin area 81,103, Germany) were used in artificial infection experiments. 26 thereof were inoculated with a predetermined level of the C. fraxinea strain WA1, potted into 3L-containers in autumn, and kept completely randomized during the experimental period in a cold greenhouse. Plant infection was performed via woundinoculation in next spring by brushing 0.1 ml of mycelium/conidia suspension on a vertical section 1 cm along shoot bark. Temperature and light conditions followed as close as possible natural fluctuations. During winter month, the greenhouse was kept at above zero degrees. To prevent heat accumulation by direct sunlight opening of windows and shading installation worked automatically. The plants were watered periodically. The rest of ash seedlings (7 seedlings) were grown under same conditions and used as negative control and therefore not inoculated with the pathogen C. fraxinea.
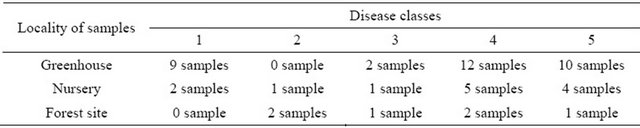
16 months later, the 33 aforementioned ash seedlings were regularly evaluated according to outer disease symptoms and classified by their degree of disease in five appropriate disease classes 1 to 5 (see Table 2, 1: without symptoms, healthy plant; 2: wilted leaves at the top; 3: necrosis in the upper shoot, wilted leaves; 4: extended necrosis and wilting downwards; 5: shoot complete dry, dead plant). Two of the inoculated plants were not affected by the pathogen and accordingly were assigned to disease class 1.
Additionally, 13 containerized plants of the same provenience grown in the nursery in the same time and six 3- to 5-year-old seedlings obtained from natural regeneration of a common ash stand near the site of Falkenhagen (Germany) were chosen for this study. All these plants showed also clear symptoms of C. fraxinea infection by natural ways with different degrees of disease (see Table 2).
In total, 52 plants of five disease classes (see Table 2) were used for further investigations on the secondary metabolites by means of MS techniques.
To prepare the samples for the direct detection of C. fraxinea in plant tissue, each selected ash seedling was divided in three sections: shoot apex, central section and shoot basis with collar. The upper section of the shoot apex comprised predominantly young tissues from the last vegetation period, the segments subjacent consisted of wood from the last two and three years, respectively. The section samples were cut to small pieces of 3 to 8 mm and extracted with methanol (2 ml methanol per 5 cm ash sample). The obtained suspensions were sonicated for 15 minutes at 15˚C and subsequently filtered. All extracts were stored at 4˚C.
2.4. MS Experiments
For rapid detection of C. fraxinea secondary metabolites, a Voyager MALDI-TOF-MS spectrometer (PerSeptive Biosystems) was used. For structure elucidation of secondary metabolites and for verification of rapid detection of the pathogen C. fraxinea, a MALDI-TOF/TOF-MS spectrometer AB SCIEX 5800 TOF/TOF System (Applied Biosystems) and the 4700 Proteomics Analyzer (Applied Biosystems) as well as the LC-MS equipment such as LC-ESI-TOF-MS/MS Q-Tof micro and Q-Tof Ultima API (Micromass/Waters) as well as LTQ Orbitrap XL (Thermo Fisher) were used. The obtained MS data were compared with international databases, SciFinder of CAS and dnp.chemnetbase.
2.4.1. Conditions of MALDI-MS Experiments
MALDI-TOF-MS spectrometer: Voyager-DE STR BioSpectrometry workstation (Perseptive Biosystems, Framingham, MA, USA), N2 laser, 337 nm; MALDI-TOF/ TOF-MS spectrometers: AB SCIEX TOF/TOF 5800 (Applied Biosystems, Framingham, MA, USA) Neodymiumdoped yttrium lithium fluoride laser (Nd: YLF) mit 349 nm laser repetition rate of 400 Hz in MS and 1000 Hz in MS/MS mode; 4700 Proteomics analyzer (Applied Biosystems, Framingham, MA, USA), Nd: YAG laser (355 nm), operating at a frequency of 200 Hz; Matrix: α-cyano-4-hydroxycinnamic acid (CHCA): 5 mg of matrix dissolved in 1 mL of 0.3% TFA in acetonitrile/water (3:7, v/v).
Table 2. List of selected ash seedling samples for MS experiments.
1: without symptoms, healthy plant; 2: wilted leaves at the top; 3: necrosis in the upper shoot, wilted leaves; 4: extended necrosis and wilting downwards; 5: shoot complete dry, dead plant.
2.4.2. Conditions of LC-MS Experiments
LC-ESI-TOF-MS/MS Q-Tof micro (HPLC Waters Alliance 2695, detectors Waters 2996 PDA and Waters-Micromass Q-TOF micro mass spectrometer): MS spectra with m/z ranging from 200 to 2000 Da; 5000 nominal resolution; using the lock spray for accurate mass measurements; 5 - 8 ppm mass error; cone gas flow: 75 L/h; desolvation gas flow: 450 L/h; interface temperature: 250˚C; source temperature: 120˚C; cone voltage: maintained at 28 V for all transitions of positive and negative modes; column: Gemini NX C18 (150 × 2 mm, 3 µm), precolumn: Gemini NX C18 (10 × 3 mm); gradient: solvent B from 45% to 80% in 47 min (solvent A: 0.1% formic acid in water/acetonitrile: 95/5; solvent B: 0.1% formic acid in acetonitrile).
LTQ-Orbitrap XL with Nano LC (Eksigent Nano 2D Ultra): MS spectra with m/z 750 to 2000 Da; 100,000 nominal resolution; using the lock mass for accurate mass measurements; 5 ppm mass error; column: C18 PepMap (3 µm, 75 µm × 250 mm, Dionex); precolumn: C18 PepMap (5 µm, 0.3 × 5 mm, Dionex); flow: 300 nl/min; gradient: solvent B from 50% to 80% in 40 min, then from 80% to 85% in 5 min, and kept by 85% in 60 min (solvent A: 0.1% formic acid in water; solvent B: 0.1% formic acid in acetonitrile); Samples were diluted 1/25 (1 µl orginal sample added to 24 µl 50% acetonitrile 0.1% formic acid).
LC-ESI-TOF-MS/MS Q-Tof Ultima API (Eksigent Nano 2D Ultra): MS spectra with m/z 200 to 2000 Da; 20,000 nominal resolution.
3. Results and Discussion
3.1. Characteristics of C. fraxinea Cultures
All 13 strains of C. fraxinea were slowly growing and showed similar growth patterns of mycelia depending on media. The best radial growth was observed on CMA, mPDA and mSeed agar at temperatures between 18˚C and 22˚C. All strains grew rather restricted below 15˚C and over 22˚C and no growth was observed at 28˚C. The mycelium on MEA was restricted flat, velvety, whitish, and developed later dark grey-black stromatic dots between 18˚C and 22˚C. Under the same temperature conditions, the mycelium on CMA was formed velvety, at first whitish-grey, later orange-brown up to dark grey with cottony white pulvinations, whereas on mPDA a velvety, yellowish-orange coloured mycelium with whitish areas was typically. On mSeed agar, at first a whitish-grey mycelium was formed, which changed soon to dark brown or black and pseudostromatic areas with a hard surface were developed.
3.2. Investigation of Pure Cultures of C. fraxinea Using Mass Spectrometric Techniques
The investigation of the methanol extracts of cells of all 13 C. fraxinea pure isolates, grown on several agar media, by means of MALDI-TOF-MS or LC-ESI-TOF-MS in positive and negative modes yielded a range of novel nonpolar secondary metabolites further referred as the chalarafraxinines A, B, C, and D (CF-A, -B, -C, and -D) with molecular weight from 800 to 1250 Da as shown in Table 3. The production of secondary metabolites types from the fungus depended on culture media. The structure elucidation of the chalarafraxinines would be beyond the scope of this presented work and will be reported in a future work.
As shown in Figure 1, the growth of C. fraxinea depends on the medium and on the applied temperature. Accordingly, the metabolite production of C. fraxinea is
Table 3. Found chalarafraxinines in methanol extracts of C. fraxinea grown on different agar culture media.
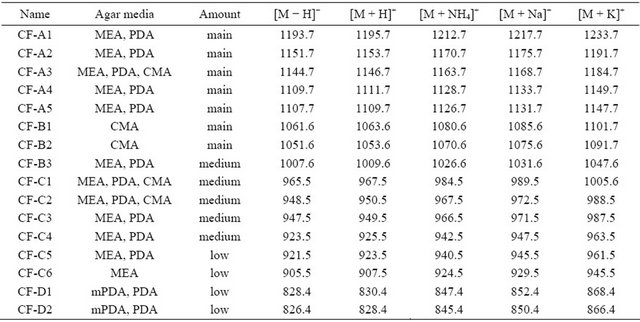
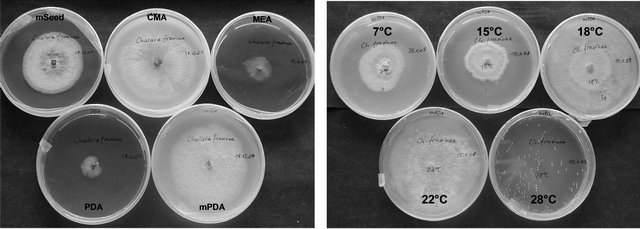
Figure 1. Growth of Chalara fraxinea on different media and at different temperatures.
therefore also depending on the substrate, too.
In cell extracts of C. fraxinea grown on MEA and PDA agar media, the chalarafraxinines CF-A (including CF-A1 to CF-A5 with molecular weight from 1100 to 1200 Da) and the chalarafraxinenes CF-C (including CF-C1 to CF-C6 with molecular weight from 900 to 1000 Da) were mainly detected (see Table 3, Figures 2 and 3). The investigation on these cell extracts by means of LC-MS revealed a range of more than three isomers of CF-A1 with [M + Na]+ at m/z 1217.7, four isomers of CF-A2 with [M + Na]+ at m/z 1175.7, and three isomers of CF-A4 with [M + Na]+ at m/z 1133.7 (for example: see LC-MS chromatograms in Figure 7, several peaks with different retention time, but of the same mass).
In cell extract of C. fraxinea grown on CMA, the chalarafraxinines CF-B (CF-B1 to CF-B3 with molecular weight from 1000 to 1100 Da) were mainly found (see Table 3 and Figure 4).
In cell extract of C. fraxinea grown on mPDA (modified potato dextrose agar with 0.15% yeast extract), the chalarafraxinines CF-A, -B and -C were not found. Only chalarafraxinines CF-D with molecular weight from 800 to 900 Da were detected (see Table 3 and Figure 5).
In cell extract of C. fraxinea grown on mSeed (with 2% non-hydrolized globular protein from cotton seeds and 0.2% sojapepton), no characteristic chalarafraxinines (CF-A, -B, -C, and -D) were found.
Obviously, the chalarafraxinines CF-A, -B and -C are produced only on culture media with carbohydrates. The media containing amino acids and proteins clearly inhibit the production of chalarafraxinines with higher molecular weight, especially CF-A.
It should be noted that in MALDI-MS, the prior found compounds, e.g. viridol or viridian [13,14], cannot be observed since there molecular masses are smaller than 400 Da and thus, lie in the range of matrix region. Thus, in our work we mainly focused on the mass range above 500 Da.
3.3. Detection of Secondary Metabolites of the Pathogen C. fraxinea in Plant Tissue
A total of 52 seedlings were investigated using MS techniques, from which 33 plants were cultivated under greenhouse conditions, 13 plants had been grown in the nursery, and 6 plants were selected from a natural forest site. 41 plants of all three samples (79%) showed visible symptoms of C. fraxinea infection. 38 plants (73%) out of these 41 had severe infections ranged into disease classes 3 to 5 (Table 4).
The chalarafraxinines, especially CF-A (CF-A1 to CFA5), which had been already demonstrated as a characteristic secondary metabolite complex of cell extracts of C. fraxinea grown on MEA and PDA (Figures 2 and 3), were also found in the methanol extracts of wooden part of the most diseased ash plants (Figure 6).
In fact, the CF-A were only detected in seedlings with severe symptoms of disease classes 3 to 5 with a frequency of 89%. For all plants of the group 5, the CF-A were found (100%). In contrast, in all healthy seedlings without symptoms of C. fraxinea infection, no CF-A were found, as well as in the small group of plants showing only weak symptoms (disease class 2) (see Table 4). Therefore, it could be concluded that the occurrence of CF-A is correlated with the degree of disease of infected trees. This can be understood in such a way that a high CF-content is also equal to a high presence of C. fraxinea.
The distribution of chalarafraxinines CF-A between the different shoot sections was not uniformly. The compounds were largely found in the wooden part of middle shoot segments (76%) and even more in the section of shoot basis and root collar (88%) (see Table 4). The highest concentration could be predominantly detected in lower parts of the shoots, whereas the shoot apex of infected plants was free of CF. Only in 23.5% of analysed shoot tops, the metabolites were evident, but only in
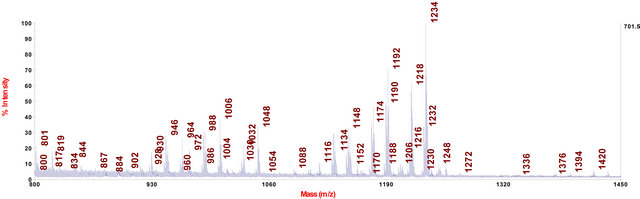
Figure 2. MALDI-TOF-MS spectrum of C. fraxinea cell grown on MEA medium.
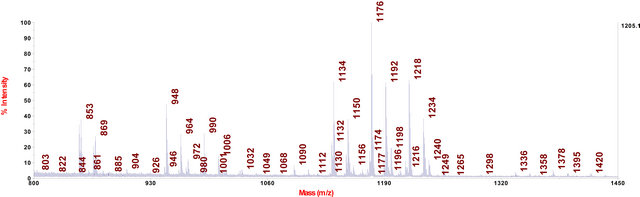
Figure 3. MALDI-TOF-MS spectrum of C. fraxinea cells grown on PDA medium.
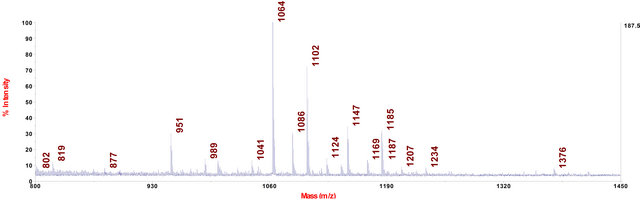
Figure 4. MALDI-TOF-MS spectrum of C. fraxinea cells grown on CMA medium.
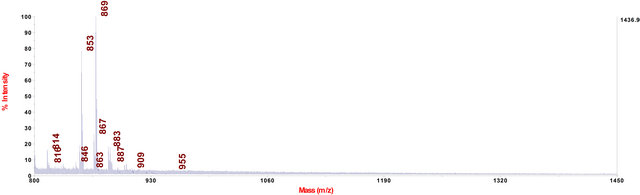
Figure 5. MALDI-TOF-MS spectrum of C. fraxinea cells grown on mPDA medium.
Table 4. Detection of chalarafraxinines and in particular the CF-A in shoots from common ash seedlings: Distribution of the occurrence of chalarafraxinines CF-A as a function disease classes and shoot sections, respectively.
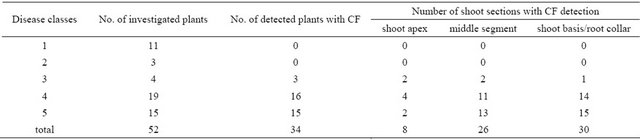
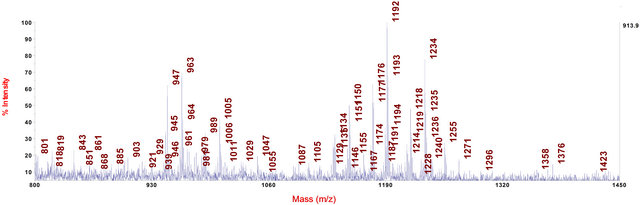
Figure 6. MALDI-TOF-MS spectrum of the methanol extract of wood from shoot basis and root collar of an infected common ash seedling ranged into disease class 5.
small concentration.
Figure 7 shows the partition of samples (7 samples from the shoot apex to root of ash seedling) for the LCMS investigation and the LC-ESI-Q-TOF-MS chromatograms of tissue extracts of ash samples affected by C. fraxinea and of cell extract of C. fraxinea grown on MEA culture medium. The chromatogram of cell extract of C. fraxinea shows clearly the peaks of chalarafraxinines CF-A1 (with [M + Na]+ at m/z 1217.7), CF-A2 (with [M + Na]+ at m/z 1175.7), and CF-A4 (with [M + Na]+ at m/z 1133.7) (see first chromatogram, left above). The chromatograms of samples No. 1, 2, and 3 of ash tissues above the inoculation spot shows no peaks of chalarafraxinines CF-A. The selected chromatograms at m/z 1217.7 for CF-A1 of the samples No. 1, 2, and 3 reveals only noise signals, no CF-A compounds (see selected chromatograms of the samples No. 1-3 in Figure 7). The chromatograms of samples No. 4, 5, 6, and 7 (samples at and under the inoculation spot) showed again the peaks of chalarafraxinines CF-A1, CF-A2, and CF-A3. The highest concentration of CF-A can be seen in sample No. 5 (shoot basis, direct under the inoculation spot) and was equal to the concentration of CF-A in cell extract of C. fraxinea grown on MEA culture medium (see Figure 7, blue row: peak intensity). In sample No. 4 (inoculation spot) and No. 6 (root collar), the concentration of CF-A was lower than in the sample No. 5.
The disease of the pathogen has a typical development: The natural infection is induced by free spores of the teleomorph H. pseudoalbidus starting from top or from upper leaves and proceeded with wilting and increasing formation of shoot necroses to stem basis of the seedlings. In case of artificially infection, the disease distributed from the inoculation point with similar symptoms downwards. The incidence of high concentration of CF-A in the wood of the stem basis and collar, higher than in the upper parts of infected shoots, confirmed that the fungus was expanded from top to bottom. This observation is only made for plants with severe damage (see Table 4, disease classes 4 and 5). This can be rationalized in three different ways:
1) It is possible that CF-A is transported along with other assimilation products downwards into the root system. However, experiments with fluid broth fermentations of C. fraxinea showed that no chalarafraxinines CF-A were found in fluid culture filtrates, only in cell suspensions of C. fraxinea, meaning that CF-A are produced only in cells of C. fraxinea and not released from the cell. Thus, it could be concluded that the CF-A cannot be transported with other assimilation products downwards into the root system.
2) Also possible is that the pathogen C. fraxinea may spread out from the inoculation spot to the stem basis and root system. The concentration of CF-A is in this case
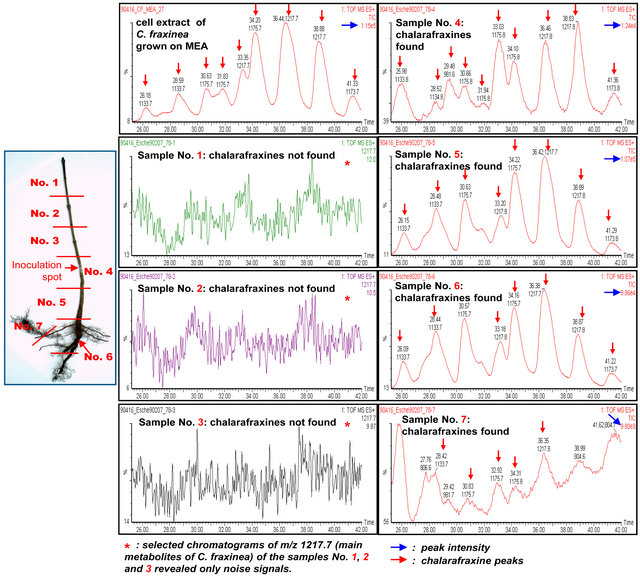
Figure 7. Right: Sample partition and LC-ESI-Q-TOF-MS chromatograms of tissue extracts of ash samples affected by C. fraxinea and of cell extract of C. fraxinea grown on MEA culture medium. Left: Division of investigated seedlings in 7 different sections. Details can be found in the text.
proportional to the amount C. fraxinea cell material. This cannot be proven.
3) Finally, the fungus could be spread over the complete ash seedling. However, it could be that the substrate conditions at different parts of the plant may promote the production of CF-A differently. As shown in section 3.1 and 3.2, the production of CF-A is strongly dependent from the substrate, i.e. medium. For example, although on mSeed and mPDA best cell growth of C. fraxinea was observed (see Figure 1), CF-A was in fact not produced. In contrast, highest CF-A concentrations were found for MEA and PDA culture media (see Figures 2 and 3), where only weak cell growth of C. fraxinea was observed (see Figure 1).
In ash seedlings with slight disease symptoms, the CF-A detection was negative. There are also other possible reasons for absence of CF like the mode of sampling, the extraction method from wood, or in general another biotic or abiotic cause than C. fraxinea that had damaged the seedlings. For MS based screening for C. fraxinea, samples should be taken from stem basis or root section, where the CF-A concentration is maximal.
Possible extensions of the collected data, that may help to clarify the origin of the disease, may include experiments with higher quantities of seedlings damaged by the pathogen C. fraxinea in a different degree and different duration in which the plant was subjected to the pathogenic influence of the fungus. Furthermore, the investigation of other species of the genus Hymenoscyphus regarding production of secondary metabolites, especially strains of the related H. albidus as a saprophytic litter decomposer of ash are from interest.
4. Conclusions
This is the first report on the indirect detection of C. fraxinea in damaged ash seedlings via its pathogen specific secondary metabolites by means of MS techniques. Although several other fungi beside C. fraxinea can cause necrotic ash lesions, the chalarafraxinines, in particular the CF-A, indicated exclusively the presence of C. fraxinea and are therefore ideal to be used as chemical markers in MS experiments.
Using these markers, it could be shown that the occurrence of the chalarafraxinines is correlated with the degree of disease of the tested seedlings. Moreover, by analyzing different parts of infected seedlings separately, the distribution of the metabolite compounds could be monitored, which in turn may allow conclusion on the propagation of C. fraxinea infections.
The here described MS based high-throughput-screening approach is shown to be suitable to detect in-vitro and in-vivo the pathogen C. fraxinea and may be extended to be used to rapidly test ash genotypes for resistance/susceptibility to C. fraxinea infections. In comparison with standard microbial and biochemical detection methods, the use of MS techniques, especially MALDIMS, exhibits several advantages, such as simplicity of sample preparation, inherent high sensitivity and high sampling rate, capability to provide data in minutes. Therefore, our presented approach may be used as an alternative to the standard detection methods.
5. Acknowledgements
We thank Mrs. Thanh Dam Huynh (Institute for Ecology, Berlin University of Technology) and Mrs. Heidrun Mattauch (Thuenen Institute for Forest Genetics, Waldsieversdorf) for their help with the experiments.
REFERENCES
- A. Juodvalkis and A. Vasiliauskas, “The Extent and Possible Causes of Dieback of Ash Stands in Lithuania,” LŽUU Mokslo Darbai, Biomedicinos Mokslai, Vol. 56, 2002, pp. 17-22.
- K. Przybyl, “Fungi Associated with Necrotic Apical Parts of Fraxinus excelsior Shoots,” Forest Pathology, Vol. 32, No. 6, 2002, pp. 387-394. doi:10.1046/j.1439-0329.2002.00301.x
- R. Bakys, R. Vasaitis, P. Barklund, K. Ihrmark and J. Stenlid, “Investigations Concerning the Role of Chalara fraxinea in Declining Fraxinus excelsior,” Plant Pathology, Vol. 58, No. 2, 2009, pp. 284-292. doi:10.1111/j.1365-3059.2008.01977.x
- V. Timmermann, I. Borja, A. M. Hietala, T. Kirisits and H. Solheim, “Ash Dieback: Pathogen Spread and Diurnal Patterns of Ascospore Dispersal, with Special Emphasis on Norway,” EPPO Bulletin, Vol. 41, No. 1, 2011, pp. 14-20. doi:10.1111/j.1365-2338.2010.02429.x
- J. Schumacher, R. Kehr and S. Leonhard, “Mycological and Histological Investigations of Fraxinus excelsior Nursery Saplings Naturally Infected by Chalara fraxinea,” Forest Pathology, Vol. 40, No. 5, 2010, pp. 419-429.
- T. Kowalski, “Chalara fraxinea sp. nov. Associated with Dieback of Ash (Fraxinus excelsior) in Poland,” Forest Pathology, Vol. 36, No. 4, 2006, pp. 264-270. doi:10.1111/j.1439-0329.2006.00453.x
- T. Kowalski and O. Holdenrieder, “The Teleomorph of Chalara fraxinea, the Causal Agent of Ash Dieback,” Forest Pathology, Vol. 39, No. 5, 2009, pp. 304-308.
- V. Queloz, C. R. Grünig, R. Berndt, T. Kowalski, T. N. Sieber and O. Holdenrieder, “Cryptic Specification in Hymenoscyphus albidus,” Forest Pathology, Vol. 41, No. 2, 2010, pp. 133-142.
- R. Vasiliauskas, R. Bakys, V. Lygis, P. Barklund, K. Ihrmark and J. Stenlid, “Fungi Associated with Crown Dieback of Fraxinus excelsior,” In: T. Oszako and S. Woodward, Eds., Possible Limitation of Dieback Phenomena in Broadleaved Stands, Forest Research Institute, Warsaw, 2006, pp. 45-53.
- R. Ioos, T. Kowalski, C. Husson and O. Holdenrieder, “Rapid in Planta Detection of Chalara fraxinea by a Real-Time PCR Assay Using a Dual-Labelled Probe,” European Journal of Plant Pathology, Vol. 125, No. 2, 2009, pp. 329-335. doi:10.1007/s10658-009-9471-x
- S. Loesgen, J. Magull, B. Schulz, S. Draeger and A. Zeeck, “Isofusidienol: Novel Chromone-3-Oxepines Produced by the Endophytic Fungus Chalara sp.,” European Journal of Organic Chemistry, Vol. 2008, No. 4, 2008, pp. 698-703. doi:10.1002/ejoc.200700839
- R. Kanasaki, M. Kobayashi, K. Fujine, I. Sato, M. Hashimoto, S. Takase, Y. Tsurumi, A. Fujie, M. Hino, S. Hahimoto and Y. Hori, “FR227673 and FR190293, Novel Antifungal Lipopeptides from Chalara sp. No. 22210 and Tolypocladium parasiticum No. 16616,” The Journal of Antibiotics, Vol. 59, No. 3, 2006, pp. 158-167. doi:10.1038/ja.2006.23
- P. F. Andersson, S. B. K. Johansson, J. Stenlid and A. Broberg, “Isolation, Identification and Necrotic Activity of Viridiol from Chalara fraxinea, the Fungus Responsible for Dieback of Ash,” Forest Pathology, Vol. 40, No. 1, 2010, pp. 43-46.
- P. F. Andersson, S. Bengtsson, M. Cleary, J. Stenlid and A. Broberg, “Viridin-Like Steroids from Hymenoscyphus pseudoalbidus,” Phytochemistry, Vol. 86, 2013, pp. 195- 200.
- T. L. H. Pham, H. Weisshoff, C. Muegge, E. Krause, W. Rotard, A. Preiss and I. Zaspel, “Non-Target-Analytik in der Ökologie,” Umweltchemie und Ökotoxikologie, Vol. 16, No. 1, 2010, pp. 2-9.
- T. L. H. Pham and I. Zaspel, “Studies on Chalara fraxinea Infection Process of Ash Plants—Direct and Rapid Detection of the Pathogen Chalara fraxinea in Plant Tissue by Means of Mass Spectrometric Techniques,” Proceedings of the Annual Conference 2011 of the VAAM, Karlsruhe, 3-6 April 2011, p. 203.
- J. O. Lay, “MALDI-TOF Mass Spectrometry of Bacteria,” Mass Spectrometry Reviews, Vol. 20, No. 4, 2001, pp. 172-194. doi:10.1002/mas.10003
- Y. G. Shin and R. B. Van Breemen, “Analysis and Screening of Combinatorial Libraries Using Mass Spectrometry,” Biopharmaceutics & Drug Disposition, Vol. 22, No. 7-8, 2001, pp. 353-372. doi:10.1002/bdd.278
- R. Giebel, C. Worden, S. M. Rust, G. T. Kleinheinz, M. Robbins and T. R. Sandrin, “Chapter 6—Microbial Finger Printing Using Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOFMS): Applications and Challenges,” Advances in Applied Microbiology, Vol. 71, 2010, pp. 149-184. doi:10.1016/S0065-2164(10)71006-6
- M. S. Lee and E. H. Kerns, “LC/MS Applications in Drug Development,” Mass Spectrometry Reviews, Vol. 18, No. 3-4, 1999, pp. 187-279. doi:10.1002/(SICI)1098-2787(1999)18:3/4<187::AID-MAS2>3.0.CO;2-K
- M. Jemal, “High-Throughput Quantitative Bioanalysis by LC/MS/MS,” Biomedical Chromatography, Vol. 14, No. 6, 2000, pp. 422-429.
- A. W. Korfmacher, “Foundation Review: Principles and Applications of LC-MS in New Drug Discovery,” Drug Discovery Today, Vol. 10, No. 20, 2005, pp. 1357-1367. doi:10.1016/S1359-6446(05)03620-2
- L. H. Pham, J. Vater, W. Rotard and C. Muegge, “ Identification of Secondary Metabolites from Streptomyces violaceoruber TÜ22 by Means of On-Flow LC-NMR and LC-DAD-MS,” In: K. Albert, Ed., Hyphenated NMR Techniques, Magnetic Resonance in Chemistry, Vol. 43, No. 9, 2005, pp. 710-723. doi:10.1002/mrc.1633
- K.-W. Cheng, F. Chen and M. Wang, “Liquid Chromatography-Mass Spectrometry In Natural Product Research,” In: S. M. Colegate and R. J. Molyneux, Ed., Bioactive Natural Products: Detection, Isolation, and Structural Determination, CRC Press, Boca Raton, 2008, pp. 245-261.
- K. Yanai, N. Sumida, K. Okakura, T. Moriya, M. Watanabe and T. Murakami, “Para-Position Derivatives of Fungal Antihelmintic Cyclodepsipeptides Engineered with Streptomyces venezuelae Antibiotic Biosynthetic Genes,” Nature Biotechnology, Vol. 22, No. 7, 2004, pp. 848-355. doi:10.1038/nbt978
NOTES
*Corresponding author.

