Advances in Microbiology
Vol.4 No.1(2014), Article ID:42270,6 pages DOI:10.4236/aim.2014.41010
Molecular Detection and Association of QnrA, QnrB, QnrS and BlaCMY Resistance Genes among Clinical Isolates of Salmonella spp. in Iran
1Department of Microbiology, Pasteur Institute of Iran, Tehran, Iran
2Department of Stem Cells, Division of Nanobiomaterials and Tissue Engineering, National Institute of Genetic Engineering and Biotechnology, Tehran, Iran
3Research Institute for Endocrine Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
4Department of Mycobacteriology & Lung Research, Pasteur Institute of Iran, Tehran, Iran
5School of Management and Medical Informatics, Isfahan University of Medical Sciences, Isfahan, Iran
6Department of Veterinary Medicine, Kermanshah University of Medical Sciences, Kermanshah, Iran
7Department of Infectious Diseases, Islamic Azad University, Tehran Medical Branch, Tehran, Iran
8Department of Microbiology, Iranian References Laboratory, Research Center, Ministry of Health and Medical Education, Tehran, Iran
9Hope Generation Foundation, Tehran, Iran
Email: *d.siadat@gmail.com
Copyright © 2014 Raheleh Saboohi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Raheleh Saboohi et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received May 21, 2013; revised June 21, 2013; accepted July 8, 2013
KEYWORDS
QnrA; QnrB; QnrS; ESBL (Extended-Spectrum Beta-Lactamase); BlaCMY
ABSTRACT
Prevalence of three plasmid-mediated quinolone resistance determinant qnrA, qnrB, qnrS and extended spectrum Cephalosporins determinant blaCMY, among eighty-five isolates of Salmonella spp. collected in the community between 2008 and 2010 was determined by PCR. Not only qnr genes but also bla genes were positive in twenty-four different isolates. PCR assay detected that 22 of 85 (25.8%) Salmonella spp. carried the qnrA, 1 (1.17%) of 85 isolates harbored the qnrB, 1 (1.17%) of them contained the qnrS, 1 (1.17%) isolate carried all the three qnrA, qnrB, qnrS genes, 24 of 85 (28.2%) Salmonella carried blaCMY and 5 (5.88%) isolates carried qnrA and blaCMY. Antimicrobial susceptibility patterns of isolates were as follows: 49 (57.6%) exhibited resistance to Nalidixic acid and none of them to Ciprofloxacin. 33 (38.82%) isolates exhibited resistance to Cephalosporins and 2 (2.35%) of them exhibited ESBL phenotype and 12 (14.1%) isolates resistance to Ampicilin. These results were confirmed by MIC determination test as well. Having detected qnr and bla genes suggested that these genes spread antibiotic resistance among pathogenic bacteria.
1. Introduction
Salmonella spp. is an important cause of human infection worldwide. Resistance to flouroquinolone, quinolone and extended spectrum Cephalosporins (ESR) that are potentially lifesaving treatment is of great concern [1-3]. Resistance to extended-spectrum Cephalosporins was the result of blaCMY, a classC plasmid-encoded ampC gene [4]. Plasmid-mediated β-lactamases were classified into six genetic clusters and blaCMY was the most prevalent one. The blaCMY has been found on plasmid of sizes variable for 7 to 180 kb [5,6].
Plasmid-mediated quinolone resistance (PMQR) was first discovered in a clinical isolate of Kelebsiella penemoniae from Birmingham, Albama collected in 1998 by Martinez-Martinez. Since then, three major groups of qnr determinant were introduced. QnrA with 6 variants, QnrB with 19 variants and QnrS with 3 variants, differ from each other by 40% or more in nucleotide sequences [7]. Resistance to quinolone and flouroquinolone arises with mutation within the DNA gyrase (topoisomerase II) and topoisomerase IV gene, especially DNA gyrase and often with decreased expression of outer membrane proteins and overexpression of efflux pump. The qnr gene encodes a pentapeptide repeat protein that protects DNA gyrase against inhibition by quinolone and flouroquinolone [8-13].
Resistance to quinolone and flouroquinolone, is often associated with ESBL-producing organisms as well as association with integron carrying qnr. Sequencing is a good way to identify qnr and ESBL producing genes [8,14].
ESBL was first explained in 1983. β-lactamases are bacterial enzymes that are inactive β-lactame antibiotic. These enzymes have the ability to hydrolyse oxyimino Cephalosporin but not Cephamycines or Carbapenems. From different parts of the world, ESBL has been introduced in Enterobacteriaceae and Pesedomonadaceaesis. These enzymes belong to the Ambler class A and D, β-lactamases. The activity of class A of β-lactamases is inhibited by β-lactamases inhibitor such as Clavulonic Acid, Sulbactam and tazobactam [15].
2. Material and Methods
2.1. Bacterial Isolates
Eighty-five isolates of Salmonella spp. were obtained from blood, stool, synovial fluid, abscess, urine and bone marrow during the years 2008 to 2010 in different provinces of Iran. Each strain obtained from a unique patient in different hospital. Stool isolates were the highest one of all the others. Biochemical and serological methods were used to identify each isolates [16].
2.2. Antimicrobial Susceptibility Test
Disk diffusion method was used to select resistant bacteria. All isolates tested and inoculated with nalidixic acid 30 µgml, Ciprofloxacin 30 µgml, Ceftazidim 30 µgml, Ceftriaxon 30 µgml, Cefepime 30 µgml, Cefpodoxime 30 µgml, Ampicilin 30 µgml by clinical and laboratory standard institute (CLSI). Disk prepared by Mast company [14,17]. Broth microdilution method [Sensitite, West Lake, OH, USA] was used to determine the minimum inhibitory concentration (MIC), confirmed by E test [AB Biodisk, Piscataway NJ, USA]. Kelebsiella pneumonia ATCC 700603 was used as quality control strain.
2.3. PCR Detection
The qnrA, qnrB, qnrS and blaCMY were detected by PCR in clinical isolates using the following primers. For qnrA_ up (GATAAAGTTTTTCAGCAAGAGG) and qnrA_ dw (ATCCAGATCCGCAAAGGTTA) to give a 700 bp product (Figure 1). Primer for qnrB_ up (ATGACGCCATTACTGTATAA) were qnrB_ dw (GATCGCAATGTGTGAAGTTT) generating a 120 bp fragment (Figure 2). Primer for qnrS_ up (ATGGAAACCTACAATCATAC) and qnrS_ dw (AAAAACACCTCGACTTAAGT) and amplicon size of qnrS was 280 bp and primers (Figure 3) for blaCMY_ up (ATGATGAAAAAATCGTTATGC) and blaCMY_ dw (TTGCAGCTTTTCAAGAATTCGC) with 800bp amplicon size (Figure 4) [4].
2.4. DNA Sequencing
The qnrA, qnrB, qnrS and blaCMY were sequenced by specific primers according to previous study. Analysis was performed using Bio Edit
(www.Mbio.ncsu.edu/Bioedit/bioedit.html). The BLAST

Figure 1. PCR products of qnrA and 50 bp DNA ladder.

Figure 2. PCR products of qnrB and 1 kb DNA ladder.

Figure 3. PCR products of qnrS and 100 bp DNA ladder.

Figure 4. PCR products of blaCMY-2 100 bp DNA ladder.
algorithm used to screen the gene bank database at the National Center of Biotechnology information website (http://www.ncbi.nlm. nih.gov/BLAST).
3. Results
3.1. Prevalence of Quinolone and Flouroquinolone and Ampicilin, Cephalosporins Resistance
Antimicrobial susceptibility pattern of 85 isolates of Salmonella were as a fallow: 9 (10.5%) isolates exhibited resistance to Ceftazidim. MIC range of these antibiotics were 0.25 - 128 µgml, 6 (7.05%) isolates to Ceftriaxone, 9 (10.5%) isolates to Cefotaxime, 6 (7.05%) isolates to Cefexime, 5 (5.88%) isolates to Cefepime, 4 (4.7%) isolates to Cefpodoxime, 12 (14.1%) isolates to ampicilin with MIC range of <4 - >2048 µgml. 49 (57.6%) isolates were resistance to Nalidixic acid and none of them were resistant to Ciprofloxacin with the MIC range of 0.64 0.125 µgml (Table 1). Among all the isolates only 2 isolates were resistant to all Cephalosporins that we tested. We showed that these two isolates were exhibited ESBL phenotype, but only one isolate had a blaCMY gene indicating that it may have the other β-lactamase gene.
3.2. Screening of Qnr and Bla Genes by PCR
Twenty two (25.8%) isolates were found to be qnrA-positive among all of the quinolone resistance isolates, qnrS and qnrB genes were detected in one (1.17%) of them. blaCMY gene were identified in twenty four (28.23%) among all of the Cephalosporins resistance isolates. Moreover, 5 isolates were found to have blaCMY and qnrA genes (Table 2). Some isolates carrying qnrA, qnrB, qnrS and blaCMY genes were selected for sequencing. The comparison between sequence results and Gene bank databases revealed identity to sequence of the qnrA, qnrB and qnrS with the accession numbers No. AY906856.1, DQ303919.1, FJ4181530.1 respectively and sequence of blaCMY-2 with the accession number Nos. EU1132222.1, EU1132220.1, AB3658670.1.
4. Discussion
Three major plasmid-mediated quinolone resistance and
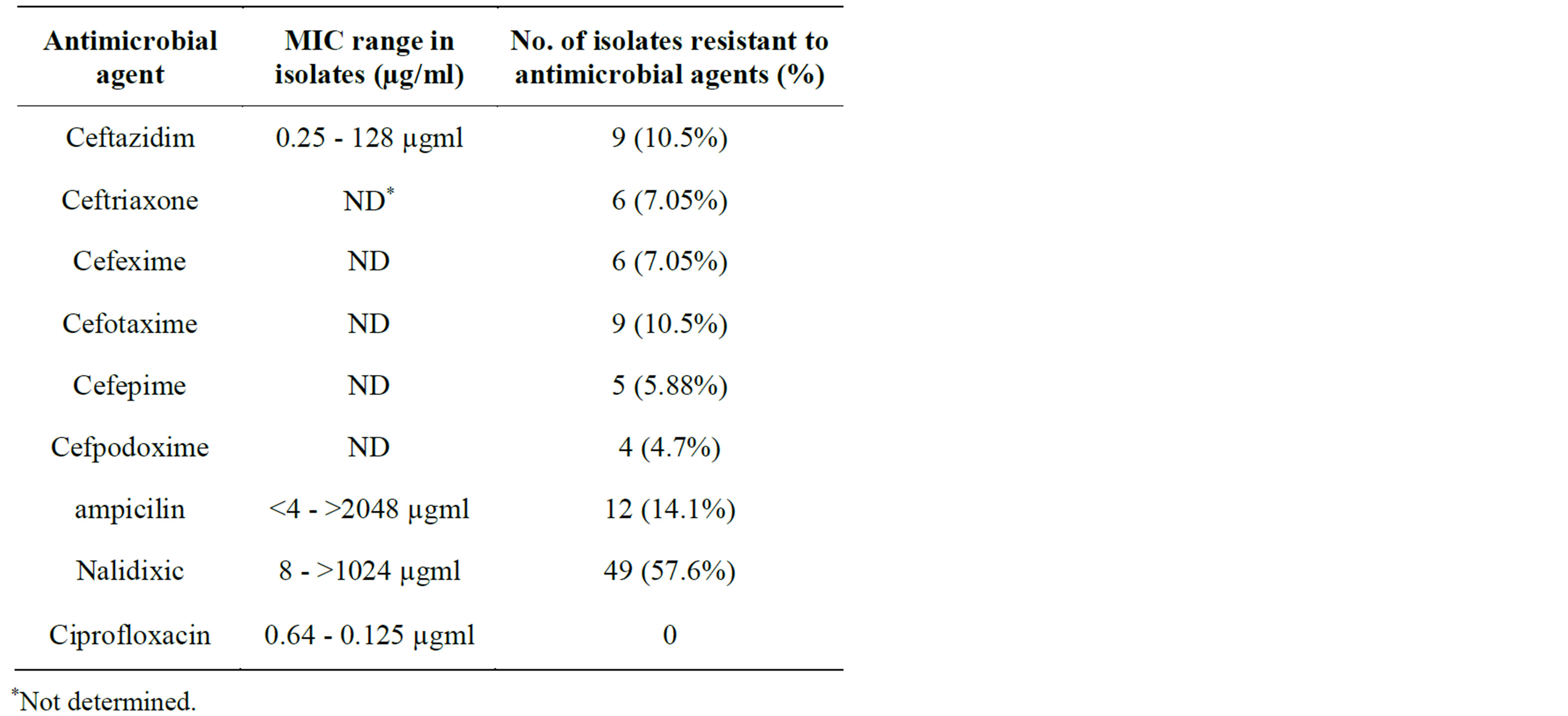
Table 1. Information about antimicrobial agents, MIC range and antimicrobial resistance percentage for 85 samples of Salmonella isolates obtained from stool, blood, bone marrow, synovial fluid, abscess, urine.
expended spectrum Cephalosporins were studied. Although resistance to quinolone and flouroquinolone with qnr gene is rare, most of the qnrA positive clinical isolates were found to have high level quinolone resistance in the present study which was usually detected by resistance to Nalidixic acid. The ability of these genes to supplement resistance is due to mutation in DNA gyrase and topoisomerase IV, porin or efflux mutations and qnrB seems to be even more potent than QnrA in blocking the action of Ciprofloxacin. These data show that qnrA presents a potential problem for the spread of quinolone resistance. Spread of these gene and bla gene is depending on the geographical region. The high prevalence of qnr among Salmonella spp. isolates and extended spectrum Cephalosporin-resistant or ESBL producing isolates has also been described in several countries. For example, the qnrB gene has been found in Senegal, USA and Korea, while the qnrS gene has been found in Enterobacteriacea in Germany, USA, Taiwan, Vietnam, France, Sothern and eastern Asia, North and south American, Europe and blaCMY has been reported in France, Turkey, Greek, USA, Mexico and United Kingdom [8,12,18,19]. Cephalosporins resistance is quite important due to its rapid spread not only among clinical isolates but also among Community in other countries [20]. The frequent association of qnr resistance gene with bla has been noted in several studies. It is Noteworthy that perception of qnrA, qnrB, qnrS in the overall quinolone and flouroquinolone was noticeable. Nonetheless, association between qnr and blaCMY was more important [10]. In both India and United States qnrB has been found on plasmid that encoding blaESBL, CTX-M-15 in India and SHV-12 in United States [13]. The presence of qnr and
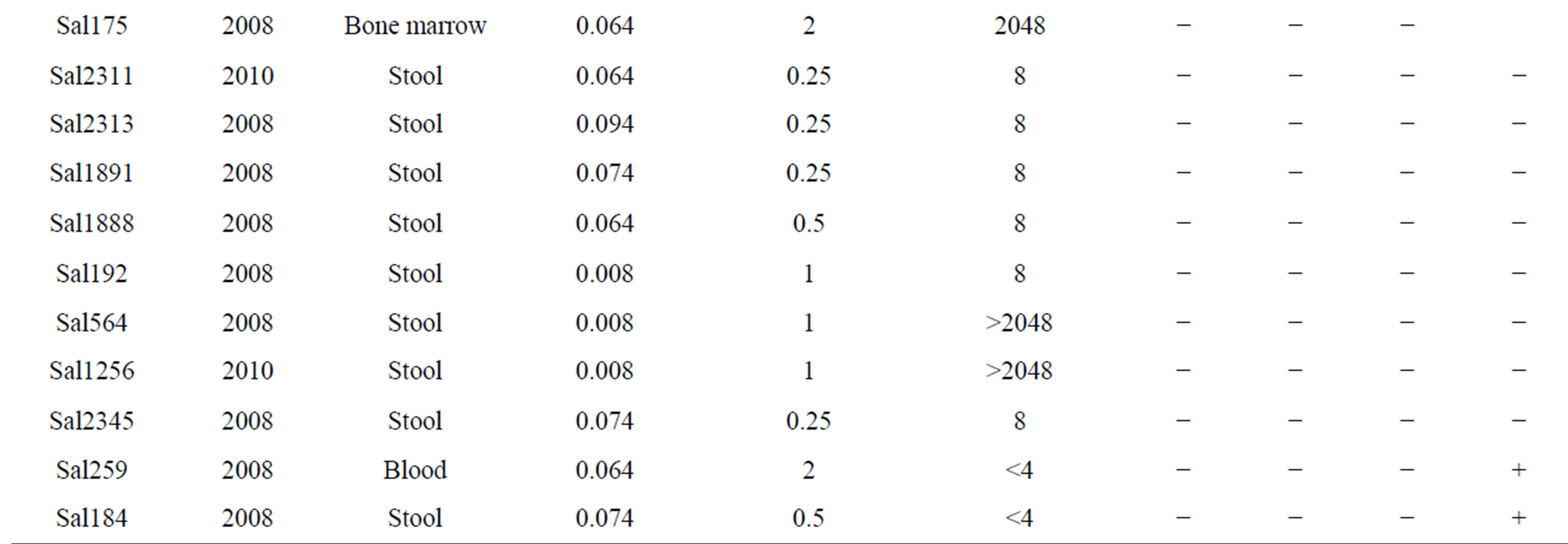
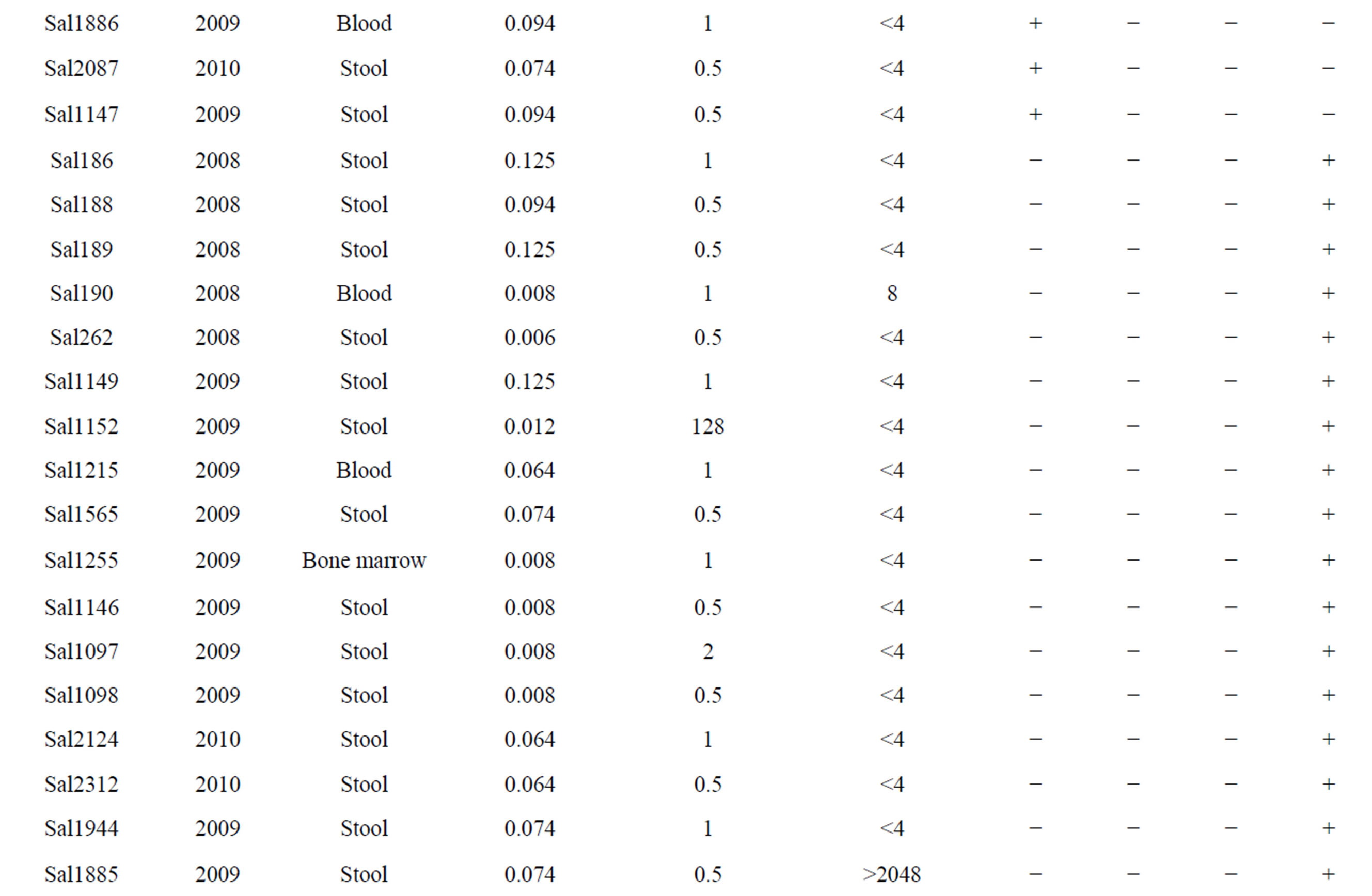
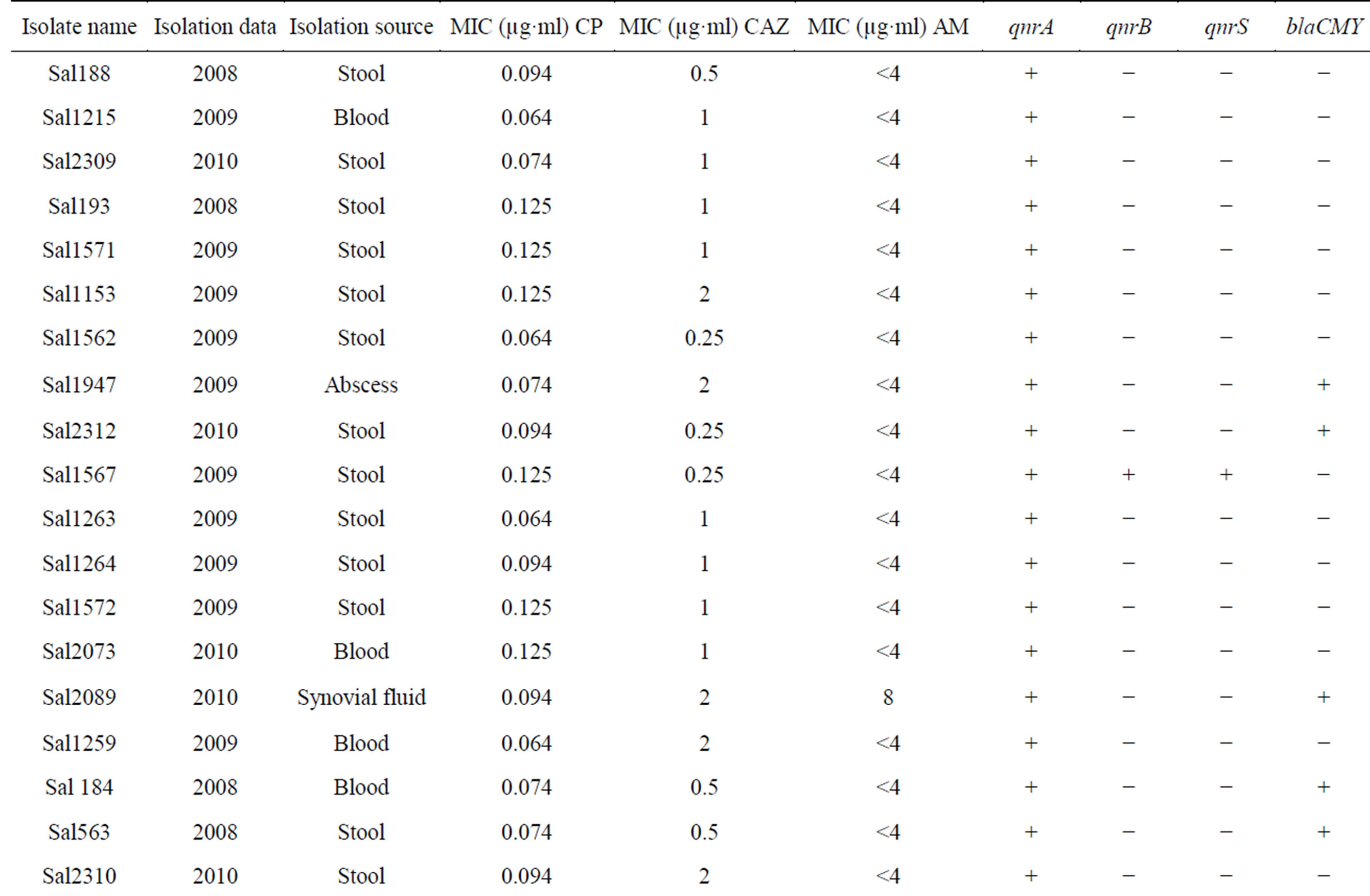
Table 2. The information of BlaCMY-2 and qnrA, qnrB, qnrS positive in Salmonella spp.
bla genes on the same plasmid is one of the several possible explanations. In this study, qnrA gene coresistance with blaCMY in some isolates may be on the same plasmid. qnrA was as common as blaCMY in these community but there is no qnr gene with ESBL in the same isolates. The similar results were acquired at Turkey in 2008 [20]. All our qnr positive isolates couldn’t carry blaCMY-2. The qnrA gene was more prevalent than the qnrB and qnrs. In 85 clinical isolates of Salmonella, 57.6% of isolates were resistant to quinolone, 38.82% to Cephalosporins and 14.1% to ampicillin. If the plasmids carrying both the qnrA and blaCMY genes spread rapidly, an important ally like the quinolone could be lost in the near future. Exploration about the events leads to co resistance of quinolone and extended spectrum Cephalosporins. In another study, we have shown that class 2 integron carrying gene cassettes which confer resistance to different classes of antibiotics such as Aminoglycosides and Trimethoprim are prevalent in Salmonella serovars isolated in Iran [21]. Presence of plasmid-mediated resistance, as well as association of class 2 integron, in Salmonella serovars demonstrates that antimicrobial selection pressure is widespread in our clinical settings. Regarding this issue and to avoid distribution of multidrug resistance, the limited use of antibiotics in clinics would be recommended.
Authors’ Contributions
This work was carried out in collaboration between all authors. Authors Raheleh Saboohi and Seyed Davar Siadat with the same contributions, designed the study, managed the literature searches, performed the analysis of data and wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This study is supported by Pasteur Institute of Iran. Kind cooperation of my colleagues in Department of Microbiology, Pasteur Institute of Iran, is highly appreciated. Special thanks to Dr. Seyed Mehdi Sadat, Mr. Alireza Azizi and Amin Arsang.
REFERENCES
- Z. Shahina, M. D. Jahdul Islam, J. Abedin, A. I. Chowdhury and M. D. Arifuzzaman, “A Study of Antimicrobial Susceptibility and Resistance Pattern of Ecoli Causing Urinary Tract Infection in Chitagong Bangladesh,” Asia Journal of Biological Sciences, Vol. 4, No. 7, 2011, pp. 548-555. http://dx.doi.org/10.3923/ajbs.2011.548.555
- F. Mansouri, F. Norouzi, M. Moradi and N. Nakaee, “Comparison of Virulence Factors among Clinical Isolates of Psoedomonase and Non-Producing Extended Spectrum β-Lactamases,” Current Research in Bacteriology, Vol. 4, No. 3, 2011, pp. 85-93. http://dx.doi.org/10.3923/crb.2011.85.93
- T. N. Farivar, M. Naderi, A. H. Mohagheghi Fard, H. O. Oweist and B. Sharifi Moud, “Drug Resistance of Mycobacterium tuberculosis Strains Isolated from Patients with Pulmonary Tuberculosis in South Eastern of Iran,” Journal of Medical Science, Vol. 6, No. 2, 2006, pp. 275-278. http://dx.doi.org/10.3923/jms.2006.275.278
- J. M. Whichard, K. Gay, J. E. Stevenson, K. J. Joyce, K. L. Cooper, M. Omondi, F. Medella, G. A. Jacoby and T. J. Barrett, “Human Salmonella and Concurrent Decrease Susceptibility to Quinolones and Extended-Spectrum Cephalosporins,” Journal of Antimicrobial Chemotherapy, Vol. 13, No. 11, 2007, pp. 1681-1688.
- A. Carattoli, F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs and F. J. Angulo, “Characterization of Plasmids Carrying CMY-2 from Expanded-Spectrum Cephalosporin-Resistant Salmonella Strains Isolated in the United States between 1996-1998,” Antimicrob Agents Chemother, Vol. 46, No. 5, 2002, pp. 1269-1272. http://dx.doi.org/10.1128/AAC.46.5.1269-1272.2002
- T. M. Coque, F. Baquero and R. Canton, “Increasing Prevalence of ESBL-Producing Enterobacteiaceae in Europe,” Eurosurveillance, Vol. 13, No. 47, 2008, pp. 1-10.
- O. Vasilaki, E. Ntokou, A. Ikonomidis, D. Sofianou, F. Frantzidou, et al., “Emergence of the Plasmid-Mediated Quinolone Resistance Gene qnrS1 in Esherichia coli Isolates in Greece,” Antimicrobial Agents and Chemotherapy, Vol. 52, No. 8, 2008, pp. 2966-2997. http://dx.doi.org/10.1128/AAC.00325-08
- K. L. Hopkins, L. Wootton, M. R. Day and E. J. Threlfall, “Plasmid-Mediated Quinolone Resistance Determinant qnrs1 Found in Salmonella enterica Strains Isolated in the UK,” Journal of Antimicrobial Chemotherapy, Vol. 59, No. 6, 2007, pp. 1071-1075. http://dx.doi.org/10.1093/jac/dkm081
- K. Veldman, W. V. Pelt and D. Mevius, “First Reports of qnr Genes in Salmonella in the Netherland,” Journal of Antimicrobial Chemotherapy, Vol. 61, No. 2, 2008, pp. 452-453. http://dx.doi.org/10.1093/jac/dkm495
- T. K. M. Cheung, Y. W. Chu, M. Y. Ha, M. A. C. Chu, R. W. H. Yung and K. M. Kam, “Plasmid-Mediated Resistance to Ciprofloxacin and Cefotaxime in Clinical Isolates of Salmonella enterica Serotype Enteritidis in Hong Kong,” Journal of Antimicrobial Chemotherapy, Vol. 56, No. 3, 2005, pp. 586-589. http://dx.doi.org/10.1093/jac/dki250
- G. A. Jacoby, N. Chow and K. B. Waits, “Prevalence of Plasmid-Mediate Quinolone Resistance,” Antimicrobial Agents and Chemotherapy, Vol. 47, No. 2, 2003, pp. 559- 562. http://dx.doi.org/10.1128/AAC.47.2.559-562.2003
- Y. Jiang, Z. Zhou, Y. Qian, Z. Wei, Y. Yu, S. Hu and L. Li, “Plasmid-Mediated Quinolone Resistance Determinates qnr and acc (6) Ib-Cr Extended-Spectrum β-Lactamace—Producing Escherichia coli and Klebsiella Pneumonia in China,” Journal of Antimicrobial Chemotherapy, Vol. 61, No. 5, 2008, pp. 1003-1006. http://dx.doi.org/10.1093/jac/dkn063
- G. A. Jacoby, K. E. Walsh, D. M. Mills, V. J. Walker, H. Oh, A. Robisk and D. C. Hooper, “qnrB Another Plasmid-Mediated Gene for Quniolone Resistance,” Antimicrobial Agents and Chemotherapy, Vol. 50, No. 4, 2006, pp. 1178-1182. http://dx.doi.org/10.1128/AAC.50.4.1178-1182.2006
- A. Sharma, C. R. Bora and R. K. Chaursia, “Antibiotic Suseptibility and Genetic Analysis of Vibrio Species Isolated from Reverina Environment,” Current Research in Bacteriology, Vol. 2, No. 2, 2009, pp. 36-49. http://dx.doi.org/10.3923/crb.2009.36.49
- J. D. D. Pitout, P. Nordmann, K. B. Laupland and L. Poirel, “Emergence of Enterabacteriaceac Producing Extended-Spectrum β-lactamases of Enterobacteriaceac Producing Extended-Spectrum β-Lactamases (ESBLS) in the Community,” Antimicobial Chemotherapy, Vol. 56, No. 1, 2005, pp. 52-59.
- A. M. Ahmed, E. E. A. Younis, Y. Ishida and T. Shimamoto, “Genetic Basis of Multidrug Resistance in Salmonella enterica Serovars Enteritidis and Typhimurium Isolated from Diarrheic Calves in Egypt,” Acta Tropica, Vol. 111, No. 2, 2009, pp. 144-149. http://dx.doi.org/10.1016/j.actatropica.2009.04.004
- CLSI, “Performance Standards for Antimicrobial Susceptibility Testing; Nineteenth Informational Supplement,” M100-S19, CLSI, Wayne, PA, 2009.
- M. B. Zaidi, V. Leon, C. Canche, C. Perez, S. Zhao, S. K. Hubert, J. Abbott, K. Blickenstaff and P. F. Mcdermott, “Rapid and Widespread Dissemination of Multidrug-Resistance blaCMY-2 salmonella Typhimurium in Mexico,” Antimicrobial Chemotherapy, Vol. 60, No. 2, 2007, pp. 398- 401.
- L. H. Su, T. L. Wu, J. H. Chia, C. Chu and A. J. Kuo, “Increasing Ceftriaxon Resistance in salmonella Isolates from a University Hospital in Taiwan,” Antimicrobial Chemotherapy, Vol. 55, No. 6, 2005, pp. 846-852.
- M. A. Oktem, Z. Gulay, M. Bicmen and D. Gur, “qnrA Prevalence in Extended-Spectrum β-Lactamase-Positive Enterobacteriaceae Isolates from Turkey,” Japanese Journal of Infectious Diseases, Vol. 61, No. 1, 2008, pp. 13- 17.
- B. Rajaei, S. D. Siadat, N. S. Rad, F. Badmasti, M. R. Razavi, M. R. Aghasadeghi, R. Saboohi, T. Rajaei, A. Moshiri, M. Nejati and A. R. Bahremand, “Molecular Detection of Antimicrobial Resistance Gene Cassettes Associated with Class 2 Integron in Salmonella Serovars Isolated in Iran,” British Microbiology Research Journal, Vol. 4, No. 1, 2014, pp. 132-141.
NOTES
*Corresponding author.

