Advances in Biological Chemistry
Vol.3 No.2(2013), Article ID:30815,11 pages DOI:10.4236/abc.2013.32029
Administration of grape seed extract alleviates age-associated decline in ubiquitin-proteasome system and cardiomyocyte apoptosis in rats
![]()
1ALM Post Graduate Institute of Basic Medical Sciences, Taramani Campus, University of Madras, Chennai, India
2Cardiology Division of the Department of Medicine, Gazes Cardiac Research Institute, Charleston, USA
Email: *cpanneerselvam@gmail.com
Copyright © 2013 Arun Kumar Vijayakumar et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 16 January 2013; revised 17 March 2013; accepted 19 April 2013
Keywords: Aging; Proteasome Dysfunction; Cardiac Muscle Cell Death
ABSTRACT
Effective clearance of oxidized, damaged, and/or misfolded proteins in the cell by the ubiquitin-proteasome system (UPS) is critical for cell homeostasis, survival and function. We hypothesized that in the aging heart, generation of free radicals could impair the UPS where the associated build-up of polyubiquitinated proteins could trigger programmed cell death. To test this, we used young (4 months old) and aged (24 months old) rats to analyze polyubiquitinated proteins, proteasome activity and programmed cell death in ventricular tissue samples. Our studies reveal excessive deposition of polyubiquitinated proteins in the ventricular tissue extracts of old rats when compared to younger rats. The increased ubiquitination was accompanied by a significant decrease in 20S proteasome activity. Since the loss of proteasome-mediated clearance of ubiquitinated proteins is linked to programmed cell death, we measured TUNEL activity in aged rat heart and compared with younger animals. Aged animal hearts showed a substantial increase in programmed cell death as evidenced by TUNEL positive nuclei and DNA fragmentation. Analyses of cell death/survival pathways support our findings in terms of age-associated increase in the nuclear localization of p53, Bax/Bcl2 ratio and cleaved (active) caspase-3 and decreased expression of cellular inhibitor of apoptosis (cIAP1). Administration of grape seed extract (GSE) as a source of antioxidants significantly reduced these ageassociated deleterious changes suggesting that free radicals primarily contribute to impaired UPS function and increased programmed cell death and that administration of antioxidants during aging could protect cardiac muscle cells and preserve ventricular function.
1. INTRODUCTION
It is well established that aging is accompanied by increases in the levels of oxygen-derived free radicals and pro-oxidants [1]. A number of studies investigated whether free radical production is causally related to functional defects in aging cells. A clear knowledge of the effect of age-related increase in the free radical levels could be gained by analyzing cells’ response to various forms of stresses. Of the various biological systems affected due to the increase in free radicals is the ubiquitin-proteasome system (UPS) that eliminates deleterious cellular proteins and plays a vital role in cellular homeostasis.
Degradation of proteins by the ubiquitin (Ub) proteasome pathway is critical in regulating the levels of several cellular proteins where UPS eliminates damaged and/or unwanted cellular proteins, such as misfolded proteins, oxidized proteins, and proapoptotic proteins [2-6]. However, the efficiency of the UPS to degrade cellular proteins declines with age, leading to accumulation of intracellular ubiquitinated proteins destined for degradation [7-9]. In the aging heart, studies have confirmed that accumulation of misfolded proteins, damaged or oxidized proteins and proapoptotic proteins in cardiomyocytes due to compromised UPS function contributes significantly to cardiac failure [10-12]. In support of these findings, ubiquitination of cellular proteins for their subsequent elimination was found to be critical for the compensatory mechanism of the stressed heart [13,14] .
Therefore, improving UPS function in the aging heart could be one potential mechanism to prevent age-associated cardiomyocyte loss and function.
Grape seed extract is a natural extract from the seeds of Vitis vinifera. Grape seeds contain high levels of phytochemicals, which have been correlated with a decreased risk of chronic diseases. When administered, GSE is known to function as an effective free radical scavenger that reduces lipid peroxidation [15]. Grape seeds and skins are common supplements known to be rich in phytochemicals such as gallic acid, catechin, flavan-3-ol epicatechin and procyanidinoligomers [16]. Over 8000 structural variants of phenolic and polyphenolic compounds have been identified [17]. GSE contains approximately 75% - 80% oligomeric proanthocyanidins and 3% - 5% monomeric proanthocyanidins [18]. In 1976, Langcake and Pryce discovered Resveratrol (3,5,40-trihydroxystilbene) as phytoalexin in grapes [19]. Catechins are abundantly present in GSE. The uptake of flavonoids and their in vivo metabolites is dependent on cell type [20]. GSE has been shown to mediate cell survival signaling through the action of pro-apoptotic transcription factors and expression of genes, such as JNK-1 and c-JUN [21]. In the aging heart, GSE has been shown to protect heart function and reduce infarct size in experimental cardiac ischemia [22].
Since several studies demonstrate that protein damage by oxygen-derived free radicals is high during aging [23], we analyzed whether polyubiquitinated proteins accumulate in the aging rat heart with a corresponding loss of proteasomal function and whether the administration of a nutraceutical supplement of grape seed extract (GSE), known to have significant antioxidant properties [24], to aged rats reverses these adverse effects and promotes cardiomyocyte survival.
2. MATERIALS AND METHODS
2.1. Chemicals
The following antibodies were obtained commercially: ubiquitin for Western analysis, and ubiquitin for microscopy were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Caspase-3, cleaved caspase-3 was purchased from Cell Signaling, (Beverly, MA). GAPDH was purchased from Fitzgerald (Concord, MA). Horseradish peroxidase-labeled secondary antibodies were purchased from Promega (Madison, WI). Actin antibody was purchased from Sigma (St. Louis, MO). cIAP1 was obtained from R & D Systems (Minneapolis, MN), and Alexa Fluor secondary antibodies was purchased from Invitrogen (Carlsbad, CA). ApopTag Fluorescein in situ Apoptosis Detection Kit for TUNEL assays was obtained from Chemicon, (Billerica, MA). 20S proteasome fluorometric (AMC) assay kit WAS OBTAINED from EMD Millipore (Millipore Corporation, MA). For nuclear and cytoplasmic extrac-tion from ventricular tissue samples, Pierce (Thermo Scientific, Rockford, IL) NE-PER extraction kit was used. DNA and RNA extractions were performed using Qiagen (Valencia, CA) extraction kits.
All other chemicals used were of analytical grade and were obtained from Sigma (St. Louis, MO), SRL (Sisco Research Laboratories Pvt. Ltd, India) and CDH (Central Drug House Pvt. Ltd., Mumbai, India).
2.2. Preparation of grape seed extract
Grapes, as large clusters with red berries, were bought from a local supermarket in Chennai and identified as Vitis vinifera. Grape seeds were removed from the grapes, air dried for 1 week and milled to a particle size of <0.4 mm. The grape seed powder (100 g) was macerated for 12 h at room temperature three times with 800 ml of 100 mM acetate buffer, pH 4.8, in water/acetone (30:70, v/v), each time. The three macerates were combined and concentrated until no acetone was left using a rotary evaporator under reduced pressure and a water bath temperature < 35˚C. The concentrated solution was extracted four times with 200 ml of ethyl acetate each time. The extracts were combined, evaporated to remove ethyl acetate and grape seed polyphenols was obtained as a lyophilized powder [25].
2.3. Animals
All experiments were conducted in accordance with guidelines approved by the Institutional Animal Ethical Committee (IAEC. No 02/024/09). Young (4-month-old) and aged (24-month-old) male Wistar albino rats were used throughout the study. Since the average life span of the Wistar albino rat is 2(1/2) to 3(1/2) years and the aged animals were defined as those that had achieved the age at which one half of the population ordinarily die (median survival time), we chose 24 months rats for the older group. Rats were divided into four groups and each group consisted of six animals: Group I, young control rats; Group II, young rats supplemented with grape seed extract; Group III, aged control rats; Group IV, aged rats supplemented with grape seed extract.
Grape seed extract (50 mg/kg body weight/day) dissolved in physiological saline was administered orally using an intragastric canula for 30 days as described previously [25]. Control rats received physiological saline alone. On completion of the experimental period, rats were sacrificed by cervical decapitation under isoflurane anesthesia. Hearts were excised, weighed, and part of the tissue sample was used immediately, for the isolation of RNA followed by gene expression studies.
2.4. Preparation of Tissue Lysates
Heart tissue was excised, weighed and immediately transferred to ice-cold physiological saline. After several washes, 10% homogenate was prepared in ice-cold 100 mM Tris-HCL buffer, pH 7.4. The heart homogenate was used for the biochemical analysis. Triton X-100 soluble and insoluble fractions were prepared from fresh tissue samples as before [26]. Briefly, 50 mg of tissue was homogenized in ice-cold Tris/Triton X-100 extraction buffer (100 mM Tris-HCl, pH 7.4, 10 mM EGTA, and 2% Triton X-100, 1:100 dilution of Sigma protease inhibitor cocktail (P8340) and Sigma phosphatase inhibitor cocktails I and II (P2850 and P5726), containing 4-(2- aminoethyl) benzenesulfonyl fluoride (AEBSF), aprotinin, leupeptin, bestatin, pepstatin-A, E-64 (trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane), cantharidin, bromotetr-amisole, microcystin, sodium orthovanadate, sodium molybdate, sodium tartrate, and imidazole. After homogenization and centrifugation at 14,000× g, the supernatant was mixed with an equal volume of 2X SDS sample buffer (soluble fraction). The pellet (insoluble) was re-extracted with extraction buffer to remove any remaining detergent-soluble proteins, and was again pelleted, resuspended, and boiled in 0.5 ml of 1X SDS sample buffer to obtain the Triton-insoluble cytoskeletal-associated protein fraction. All samples mixed with SDS-sample buffer were boiled for 5 min and used for gel electrophoresis.
2.5. Nuclear Preparations in Tissue
Nuclear isolation was performed as per the manufacturer’s (Pierce) protocol with minor modifications. Briefly, 50 mg of heart tissue was homogenized in 1 ml PBS with protease and phosphatase inhibitor cocktails. The homogenate was centrifuged at 1000× g for 8 min. To the pellet, 500 μl of cytoplasmic extraction buffer I (CERI) with inhibitors was added. After suspending the pellet, the solution was incubated on ice for 15 min, followed by the addition of 27 μl CERII. The contents were incubated on ice for 2 min, centrifuged at 16,000× g for 15 min, and the supernatant (cytoplasmic proteins) was mixed with an equal volume of 2X SDS sample buffer. The pellet was washed with PBS, suspended in 250 μl of nuclear extraction buffer with inhibitors and incubated on ice for 40 min, then vortexed for 10 sec every 10 min. After centrifugation at 16,000× g for 15 min at 4˚C, the supernatant, consisting of nuclear proteins, was mixed with an equal volume of 2X SDS sample buffer.
2.6. Western Blot Analysis
Amount of nuclear level of p53, cytosolic level of Bcl2, Bax, cIAP1, and Caspase-3, ubiquitin were assessed by Western blot using 4% - 12% SDS-polyacrylamide gels. The gels were blotted on to PVDF membrane. The membranes were then blocked in 10% nonfat milk in Trisbuffered saline with 0.2% Tween 20 (TBS-T) at room temperature for 1 h, and probed with the following primary antibodies diluted in TBS-T: anti-ubiquitin, Bcl-2, Bax, p53 and cIAP1 mouse monoclonal antibodies (1:300 dilution), and anti-pro-/cleaved caspase-3 rabbit polyclonal antibodies (1:2000 dilution) for 3 h at room temperature. Following incubation with secondary antimouse or rabbit IgG linked to horseradish peroxidase at a 1:5000 dilution for 45 min, the band was visualized using enhanced chemiluminescence and X-ray film. Band intensity was measured by using NIH-ImageJ. Quantification for each protein of interest was accomplished by first normalizing the protein band in each ventricle to a control endogenous protein (either GAPDH or actin).
2.7. Measurement of 20S Proteasome Activity
The 20S subunits were purified as described [27]. Briefly, 50 mg of heart tissue were homogenized in 1.5 volumes buffer A (20 mM Tris-HCl, 20 mM KCl, 10 mM magnesium acetate, 2 mM DTT, and 10% glycerol, pH 7.6). The soluble protein fraction was isolated by centrifugation at 30,000 g for 30 min at 4˚C. The pellet was discarded, and the supernatant of this first centrifugation step was centrifuged again at 100,000 g for 6 h at 4˚C. The pellet of this second centrifugation step was washed twice and resuspended in 200 μl fresh buffer A. The protein concentrations were determined using the BCA Protein Assay Kit (Pierce, Rockford, IL).
The proteasome assay was performed using 20S proteasome fluorometric (AMC) assay kit (Millipore Corporation). The degradation of the fluoropeptide Suc-LeuLeu-Val-Tyr-AMC (chymotrypsin like) was measured after addition of the substrate to the tissue homogenates. 10 μl of heart tissue homogenates were incubated with 985 μl of reaction buffer (500 mM HEPES, pH 7.6 and 10 mM EDTA) for 5 minutes. 5 μl of 200× (2 mM of substrate solution diluted in reaction buffer) and 10 μl of 3% SDS were added. The mixture was incubated for 60 minutes at 37˚C, and the reaction was then stopped by addition of an equal volume of ice-cold 96% ethanol. The fluorescence determination was performed at 380 nm excitation and 460 nm emission using free AMC as a standard, and the measurements were corrected by subtracting the background fluorescence (without SucLLVY-AMC).
2.8. Confocal Microscopy
Fresh frozen tissue samples were placed in OCT freezing solution and left at -80˚C. Tissue sections (12 μm thick) were fixed with 2% paraformaldehyde, permeabilized in 2% SDS for 5 min at room temperature, and blocked with 10% donkey serum for 1 h at room temperature.
Primary anti-ubiquitin antibody (1:100, Dako) was added overnight at 4˚C. Sections were washed in PBS and 200 μl of DAPI was added. Slides were then incubated with Alexa Fluor secondary antibodies for 2 h. After the secondary antibody incubation, the sections were washed with PBS, mounted in Anti-Fade, and subjected to laser scanning confocal microscopy (Olympus IX81, Japan) at 40× magnification. The images were processed with Adobe Photoshop.
2.9. TUNEL Assay
TUNEL (TdT-mediated dUTP Nick End Labeling) assay was performed as per manufacturer’s protocol with slight modifications. Briefly, fresh frozen tissue samples were used for TUNEL studies. Tissue sections (12 μm thick) were prepared and incubated in 1% PFA for 10 min at room temperature and washed twice with PBS each for 5 min. Then, the slides were incubated at -20˚C for 5 min in ethanol:acetic acid solution (2:1 dilution) for permeabilization and washed subsequently with PBS and then with 75 ml of equilibration buffer for 1 min. The enzymatic reaction was performed by incubating the tissue sections for 1 hour at 37˚C with 55 μl of terminal deoxynucleotidyl transferase (Reaction Buffer and TdT). After incubation, the tissues were soaked in wash buffer at room temperature for 10 min. The slides were washed three times each for 1 min and incubated in 65 ml of anti-digoxigenin fluorescein. Followed by 3 washes in PBS, the sections were stained for specific proteins or nucleus and mounted using anti-Fade and analyzed by confocal microscopy.
2.10. RT-PCR Analysis
Total RNA extraction from the heart muscle was performed using Qiagen’s RNeasy Minikit. Total RNA was solubilized in RNase-free H2O, and quantified in duplicate by measuring the optical density (OD) at 260 nm. Purity of RNA was assured by examining the OD260/ OD280 ratio. Total RNA (100 - 150 ng) was subjected to a total volume of 50 μl RT-PCR containing 0.6 μM final concentrations of forward and reverse primers for cIAP1 and β-actin. Thermal cycling conditions were started with RT reaction for 1 h at 50˚C followed by initial activation of Taq DNA polymerase for 10 min at 94˚C. cIAP1 and β-actin cDNA amplification was started with initial thermo-cycle at 94˚C for 30 s and then at 59˚C for 45 s for primer annealing. Extension was done at 72˚C for 45 s and the final extension was at 72˚C for 10 min. PCR was performed using a programmed thermocycler (Thermocycler gradient; Eppendorf, Germany). The primer pairs used for RT-PCR amplification are as follows: cIAP1: 5’-TCCCTGTCATCTCACCATGA-3’ and 5’-TGTCTAGCATCAGGCCACAG-3’; β-actin: 5’-GCCATGTACGTAGCCATCCA-3’ and 5’-GAACCGCTCAT-TGCCGATAG-3’. After amplification, the RTPCR products were electrophoresed on 2% agarose gels and stained with ethidium bromide. Images were captured and subjected to densitometric analysis.
2.11. Analysis of DNA Fragmentation
The level of DNA fragmentation was measured as described previously [28]. For this, the heart tissue was homogenized with four volumes of lysis buffer (10 mM Tris-HCl, 10 mM EDTA and 0.5% Triton X-100, pH 10) and centrifuged at 16,000 rpm for 20 min to separate intact DNA from fragmented DNA. After transfer of the supernatant to a separate tube, 0.2 ml of lysis buffer was added to the pellet. To both fragmented and intact DNA fractions, 0.2 ml of perchloric acid was added. After maintaining the mixture at 4˚C for 30 min, the samples were centrifuged at 16,000 rpm for 20 min and the supernatant was removed. The pellets were dissolved in 0.05 ml of perchloric acid and heated at 70˚C for 20 min. Agarose gel electrophoresis was carried out for the analysis of DNA fragmentation [28]. For this, the DNA samples (1 mg) were electrophoresed on 1.4% agarose gel using TBE buffer at 40 V for 5 h. Then the gel was stained with ethidium bromide and viewed under UVtransilluminator and photographed.
2.12. Statistics
Differences in the proteasome activity between the groups were compared by one-way ANOVA followed by a Tukey test for multiple comparisons. Statistical significance was defined as P < 0.05.
3. RESULTS
3.1. GSE Administration Decreases Age-Associated Build-Up of Ubiquitinated Proteins
In the normal heart, polyubiquitination marks the damaged proteins for their subsequent effective elimination by the 26S proteasome. However, since the volume of damaged/oxidized proteins in the aged heart is substantially increased due to excessive generation of reactive oxygen species, it is possible that the ubiquitinated protein level increases in the aged heart with a corresponding decrease in the proteasome function. Furthermore, such an intracellular buildup of polyubiquitinated proteins due to the ineffective clearance by the UPS could eventually lead to programmed cell death. Therefore, to gain insight into potential consequences of age-dependent increase in the polyubiquitinated protein level in the heart, we performed immunohistochemical studies to establish whether aged rat hearts exhibit enhanced protein ubiquitination accompanied by increased programed cell death (Figure 1(a)). Our data showed the presence of several ubiquitin-enriched cells only in the old rat heart samples but not young rat hearts. To explore whether the increased ubiquitination in aged rat hearts was associated with cell death, TUNEL staining was also performed in these tissue samples. The data indicated that the accumulation of ubiqutinated proteins in the untreated aged rat hearts was accompanied by increased levels of TUNEL positive cells (Figure 1(a)). Quantification of TUNEL positive cells was performed by counting TUNEL positive nuclei relative to the total number of nuclei, and the ratio was estimated as 1.85% for the aged rat hearts (data not shown). We then performed a similar staining protocol in GSE treated young and old rat hearts. WITH the GSE-treated old rat hearts both ubiquitin-enrichment and TUNEL reactivity were found to be absent. As expected, in young rats, ubiquitin and TUNEL positive cells were undetected in both control and GSE treated groups.
We performed additional biochemical experiments to confirm our immunohistochemical studies. To show that the polyubiquitinated protein level was increased in old rats that could be reverted with GSE treatment, immunoblots using anti-ubiquitin antibody was performed in Triton X-100 soluble and insoluble samples prepared from young and old rat hearts treated with ±GSE (Figure 1(b)).Consistent with confocal data, old heart samples when compared with young heart samples showed an overall increase in the polyubiquitinated protein levels.
Administration of GSE was found to partially reverse this trend in protein ubiquitination. Finally to confirm that the old heart samples had higher levels of programmed cell death that could be controlled with GSE treatment, DNA fragmentation was analyzed. DNA fragmentation in the aged rat hearts was high when compared to young rat heart samples. However, in GSE fed rats, DNA fragmentation was substantially attenuated and the DNA migration pattern resembled similar to young rat hearts (Figure 1(c)).
Accumulation of polyubiquitinated proteins in the aging rat heart could be due to excessive generation of damaged proteins with their subsequent ubiquitination and/or decreased clearance of polyubiquitinated proteins due to less efficient proteasome function. Therefore, we next measured proteasome activity in young and old rat hearts and then explored if administration of GSE could improve proteasome function. 20S proteasome activity in the heart extracts was measured using a fluorescence conjugated peptide substrate [29] as described in the Methods section.
As shown in Figure 1(d), a significant loss (53%) of proteasome activity was observed in older heart tissue samples when compared to younger rat hearts. Importantly, administration of grape seed extract showed improved (72%) proteasome activity in aged rat hearts while there was no major effect in younger rats. These data suggest that decreased proteasome function could be one possible mechanism for the accumulation of polyubiquitinated proteins and the associated programmed cell death which could be controlled by the administration of GSE.
3.2. GSE Controls Changes in Cell Survival/Death Regulators in Aged Rat Heart
Enhanced programmed cell death in older hearts indicates changes in the regulators of cell survival and programmed cell death. Therefore, we next analyzed known key components regulating programmed cell death. For this, we measured the cellular levels of the following parameters in old and young rat hearts after GSE administration: The transcription factor p53 is a tumor suppressor protein that functions by controlling cell cycle progression and/or promoting apoptosis under stress conditions. Normally, the nuclear level of the tumor suppressor protein p53 is maintained at low levels and accumulates in response to various types of stress. Nuclear localization of p53 has been shown to activate mitochondrial pathways for apoptosis by increasing Bax/Bcl2 ratio [30,31]. We observed an elevated level of p53 in the nuclear and cytoplasmic fractions of aged rat hearts and compared them with young rat heart samples (Figure 2(a)). Compared to young hearts, treated or untreated, old rat hearts showed enhanced levels of p53 both in the nuclear and cytoplasmic fractions. Histone-H4 and GAPDH were present only in their known fractions, namely nuclear and cytoplasmic, respectively. Importantly, the deleterious increase of p53 in the older rat hearts was found to be reverted with GSE treatment, although such treatment did not cause any significant change in young rat hearts. The increased level of p53 in older heart samples and its reversal towards the levels of young hearts were also confirmed at the mRNA level (data not shown).
Since p53 has been known for its role as a transcriptional factor for the expression of Bax and other proapoptotic factors [30,31], we next measured the mRNA and protein levels of Bax, a proapoptotic factor, and Bcl-2, a prosurvival factor in aged and young hearts treated with ±GSE. In untreated rats, the Bax level in the aged animal group when compared to younger animals, showed a significant (>2.5 fold) increase at the protein (Figure 2(b)) and mRNA (data not shown) levels when compared to control young rats. Oral administration of GSE to aged rats significantly decreased the Bax levels
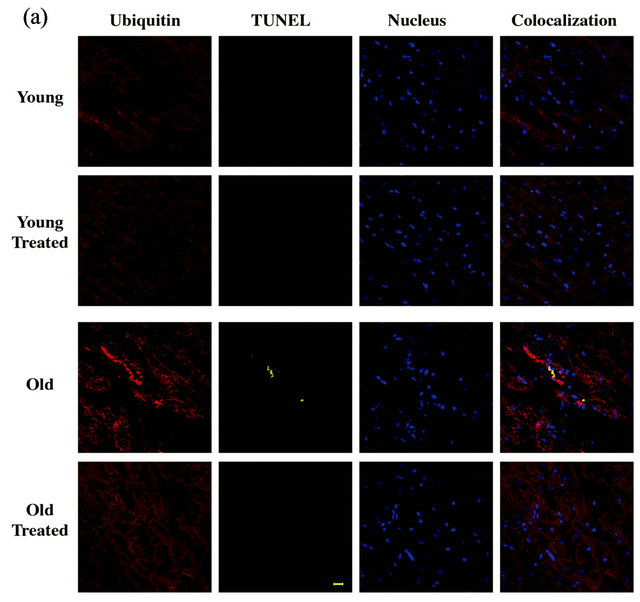
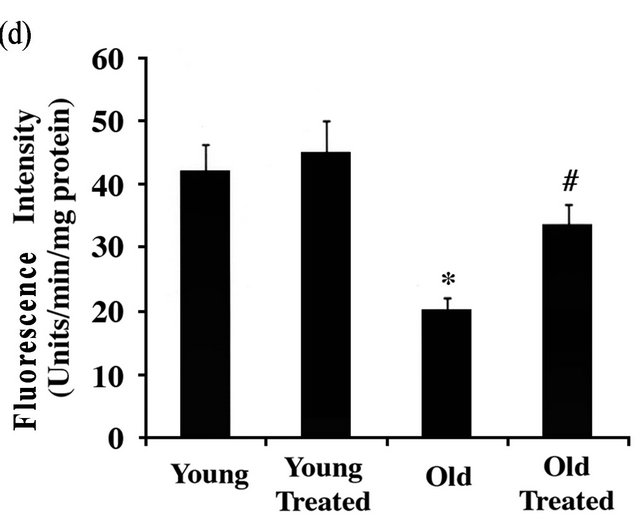
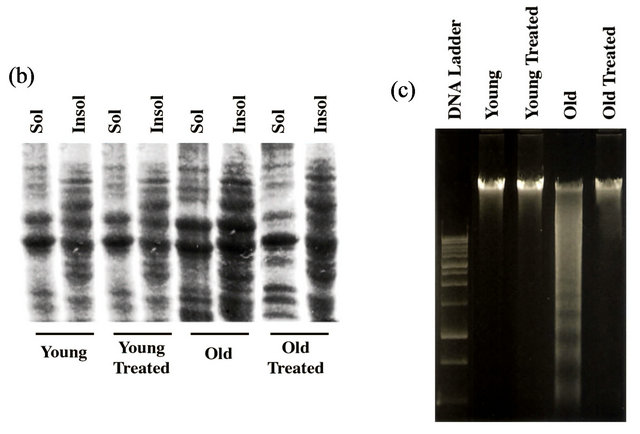
Figure 1. GSE effect on the age-associated changes in ubiquitin-proteasome system (UPS) and programmed cell death: (a) Ventricular tissue samples from young and old rats fed with ±GSE were fixed in 4% paraformaldehyde. Tissue sections (12 mm) AND were used for ubiquitin immunostaining (decorated in red with Alexa Fluor 568), TUNEL staining (decorated in green with Alexa Fluor 488) and nuclear staining (decorated in blue with DAPI) as outlined under Methods. Sections were analyzed using confocal microscope. Scale bar, 10 mm. (b) Heart tissue samples were lysed in Triton X-100 buffer and the soluble and insoluble protein fractions were prepared as described under Methods. Samples were run 12% SDS-PAGE and transferred to PVDF membrane and Western blotted using anti-ubiquitin antibody. (c) Genomic DNA extracted from ventricular rat heart ventricular samples of control and experimental animals were electrophoresed on 1% agarose gel and visualized by ethidium bromide staining. All experimental results were confirmed in at least three independent rat samples for each group. (d) Cytosolic extracts were prepared from the ventricular tissue samples of control and experimental groups. The 20S proteasome activity was measured by monitoring the release of free AMC (aminomethylcoumarin) from the flurogenic peptide Suc-LeuVal-Tyr-AMC. Values are expressed as mean ± SD for six rats in each group. *P < 0.05 vs. young control; #P < 0.05 vs. old.
towards that of young rats, while no change by GSE was observed in young rate hearts. Similarly, the level of Bcl-2 as a pro-survival marker was assessed. Immunoblot analyses also showed a notable decrease in the Bcl-2 level in the aged rat heart (Figure 2(b)). Oral administration of GSE to aged rats significantly increased the Bcl-2 level to that was observed with young rats. These results indicate the cell survival effects of GSE by lowering Bax/Bcl-2 ratio.
3.3. GSE Treatment Reduces Active Caspase-3 in Old Rat Heart
Finally, we measured the level of cellular inhibitor of apoptosis (cIAP1), a member of E3 ubiquitin ligase that promotes cell survival through ubiquitin-mediated elimination of caspases. Measurement of cIAP1 at the mRNA expression level was significantly decreased in aged rat hearts (Figure 3(a)), indicating higher level of caspase
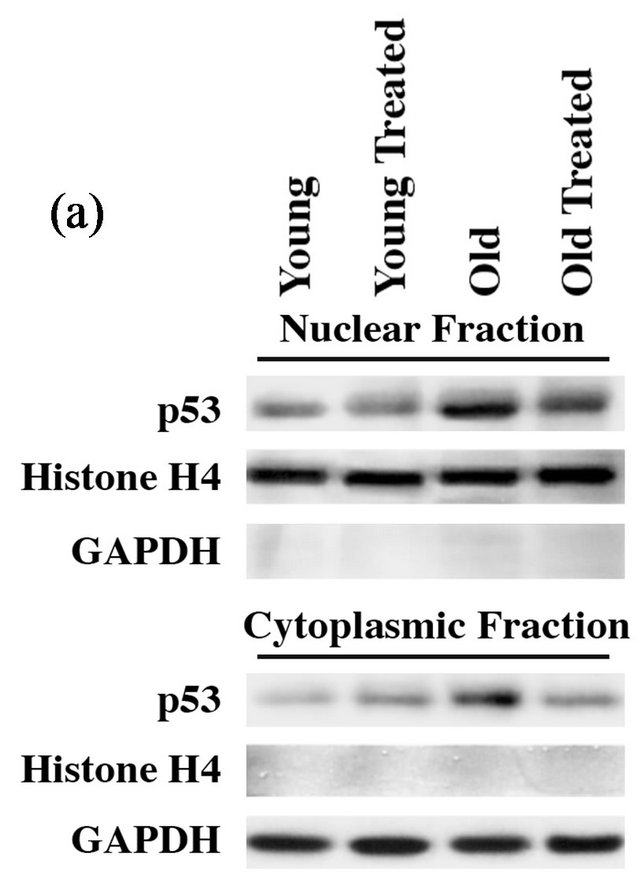
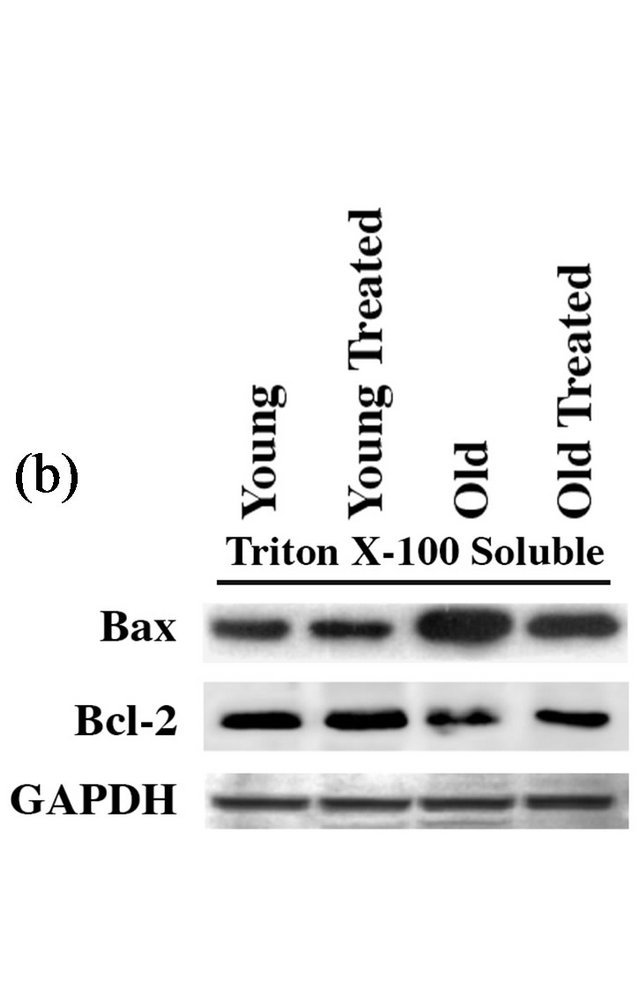
Figure 2. GSE administration reverses aging-associated changes in proapoptotic and prosurvival factors: (a) Nuclear and cytoplasmic extracts were prepared from ventricular tissue samples of control and experimental rats. Proteins were resolved on 12% SDS-PAGE, transferred to PVDF membrane and Western blotted with anti-p53, histone-H4 and GAPDH antibodies. (b) Triton X-100 soluble fractions were prepared from ventricular tissue samples of control and experimental rats. Proteins were resolved on 12% SDS-PAGE, transferred to PVDF membrane and Western blotted with anti-Bax, Bcl-2 and GAPDH antibodies. Results were confirmed in at least three independent rat samples for each group.
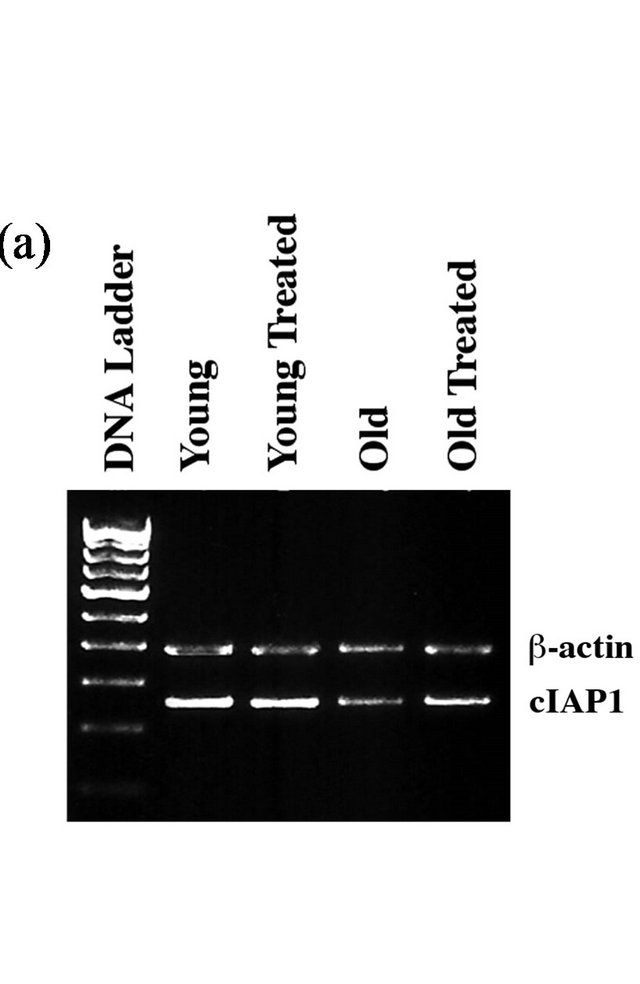
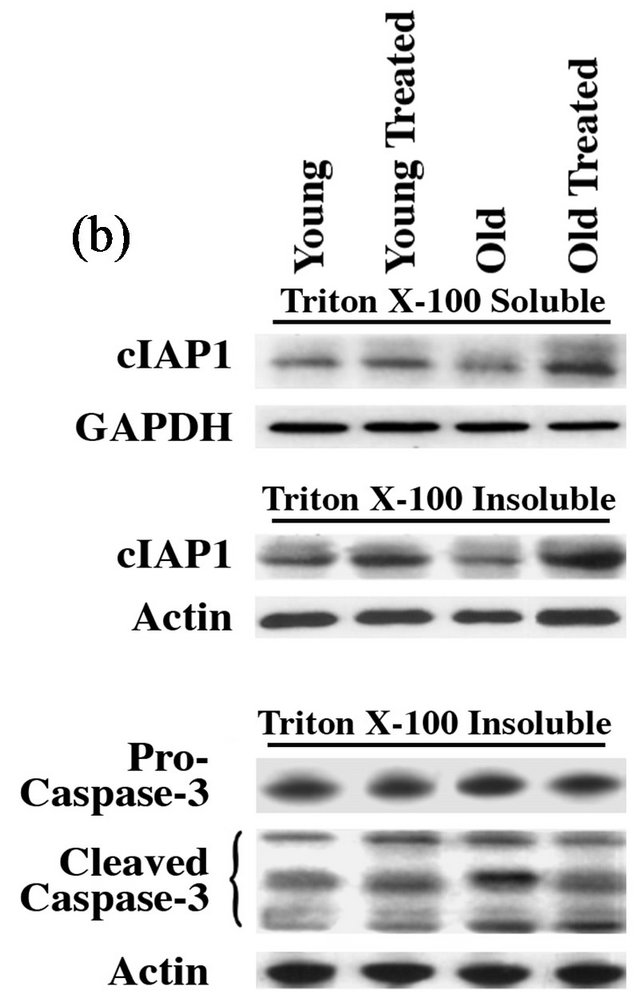
Figure 3. GSE administration corrects aging-associated changes in cIAP1 and caspase-3: (a) Total RNA extracted from rat heart ventricular samples was used as template. RT-PCR products were amplified from cDNA corresponding to cIAP1 and b-actin (internal control) using respective primer pairs as described in the Methods section. The amplified products shown in the figure correspond to 374 bp and 264 bp and they represent cIAP1 and b-actin, respectively. (b) Heart tissue samples were lysed in Triton X-100 buffer and the soluble and insoluble protein fractions were prepared as described in the Methods. Soluble proteins were used for Western blot analysis using anti-cIAP1 and GAPDH antibodies while the insoluble protein samples were used for Western blot with anti-cIAP1 and GAPDH antibodies. (c) The insoluble protein fraction prepared above was used for Western blot analysis using anti-procaspase-3 (full-length caspase-3), active caspase-3 (cleaved caspase-3), and actin antibodies. The active caspase-3 antibody recognizes cleaved fragments of caspase-3, thus representing the active forms of caspase-3. All experimental results were confirmed in at least three independent rat samples for each group.
activity and the associated increase in programmed cell death. This was also reflected at the protein level as evidenced from the immunoblot analysis that showed a substantial decrease in cIAP1 level in the aged heart when compared to young rat hearts (Figure 3(b)). This trend in aged rats could be significantly restored to that of younger animal by the oral administration of GSE.
4. DISCUSSION
The UPS has been shown as the main nonlysosomal degradation mechanism for the removal of defective proteins, which have often undergone free radical-derived modifications. The data in the present study demonstrate changes associated with aging and its reversal by the administration of GSE that promotes proteasome-mediated degradation of deleterious proteins in the heart. Additionally, this study shows an age-dependent decline in the peptidase activity of 20S proteasome in the heart which could be corrected by the administration of GSE (Figure 1(D)). The loss of proteasome function in the aged heart could be due to the effect of: 1) increased production of damaged or misfolded proteins, 2) decreased overall proteasome content, 3) alterations in subunit composition, and/or 4) occurrence of posttranslational modifications. Recent studies indicate that mild oxidative stress rapidly inactivates 26S proteasome function, although 20S proteasome function was found to be unaffected [32]. However, during severe oxidative stress or aging, the 20S proteasome function also declines more steadily due to the generation of cross-linked oxidized protein aggregates that bind and inhibit 20S proteasome function. Since our parallel studies in these animals (data not shown) demonstrate significant reduction in the generation of oxygen-derived free radicals following GSE administration, it is possible that the intracellular accumulation of protein aggregates is effectively reduced in GSE treated animals, leading to improved 20S proteasome function. In support of these findings, previous studies show the stimulatory impact of naturally occurring phenolic compounds on proteasome activities [33]. Also, the present data corroborates with an earlier study, which suggests that antioxidants enhance proteasome activity [34].
Although the decreased proteasome function in the aging heart could be partly due to the excessive generation of oxidatively damaged proteins, other independent changes in the proteasome composition, as part of the aging process, might also contribute to the overall loss of proteasomal function. Additional studies to measure the level, composition and posttranslational modifications of proteasome machinery will help decipher the mechanism of age-associated changes in the proteasome function. In support of this idea, an earlier study indicates that the natural antioxidants have ability to enhance the expression of proteasome subunits, thus offering increased protection against various oxidants [35] . Therefore, enhancing the biogenesis of proteasome subunits in order to increase the proteasome capacity could be an additional mechanism for the increased peptidase activity observed in GSE-administered aged rats. In aged rat hearts, whatever might be the reason for the observed loss of proteasomal function, the present study strongly suggests that administration of GSE improves the proteasome function, indicating that GSE administration would facilitate the clearance of damaged protein in order to improve the survival of aging cells.
Our study was also designed to explore whether the build-up of damaged macromolecules during aging is associated with changes in cell survival/death mechanism and increased programmed cell death. Comparing young vs. older rats, the major findings in the aging rat heart include: 1) transcriptional activation of p53, and the associated increase in the Bax/Bcl-2 ratio, 2) age-dependent decline in the level of cellular inhibitor of apoptosis (cIAP1) with the resulting activation of caspases, and 3) reversal of all these age-associated changes and cardiomyocyte loss following treatment with GSE.
For cellular homeostasis, clearance of damaged proteins, such as oxidized proteins, mis-folded proteins, pro-apoptotic proteins, etc is critical, and the UPS serves as a major mechanism for their removal. Therefore, loss of proteasomal function as observed in the aging heart could lead to accumulation of ubiquitinated proteins meant for degradation, and this condition triggers programmed cell death in the form of either apoptotic or autophagic cell death. In this context, previous studies showed that cardiomyocyte apoptosis increases in parallel with proteasome activity depression [36]. In the present study, IHC studies showed enhanced polyubiquitination in cardiomyocytes of the aged heart relative to young rat heart samples. Furthermore, this ubiquitination pattern appears to be throughout the cell, suggesting global ubiquitination of one or more cellular proteins [36]. Importantly, confocal results for TUNEL assay show that cardiomyocytes positive for TUNEL reactivity also exhibit enhanced polyubiquitination in the aged rat heart (Figure 1(a)). Previous studies have shown that apoptotic cell death of cardiomyocytes is increased in patients with heart failure and has been reported to contribute to the progression of heart failure [37]. Similarly, inhibition of proteasome function alone results in the accumulation of the pro-apoptotic factors [5]. Finally, in addition to regulating programmed cell death, the cardiac UPS is integral in regulating key cellular processes, including cellular mass regulation, sarcomere quality control, and cardiac ischemia/reperfusion homeostasis [38]. Therefore, promoting proteasomal function in the aging heart can have beneficial effects by improving survival and function of cardiomyocytes. In support of this idea, the current study clearly demonstrates that GSE treatment in aged rats could improve proteasomal function, clear off ubiquitinated proteins and decrease programmed cell death.
It has been demonstrated that the anti-oxidant proanthocyanidins present in the GSE could function as an anti-apoptotic regulator by down-regulating pro-apoptotic genes [23]. Therefore, in addition to improving proteasome function, the anti-oxidants present in the GSE regulate cell death/survival pathways, thus contributing to the cardioprotective effect. In this context, it has been reported that age associated decline in the UPS leads to nuclear translocation and accumulation of p53, resulting in the transcription of pro-apoptotic factors and programmed cell death. In the current study, the nuclear level of p53 was markedly increased in parallel with the up-regulation of Bax in aged rat heart (Figure 2). In support of these findings, several studies report that down-regulation of the proteasome function in cells leads to accumulation of p53 [39,40].
p53 has been shown to regulate apoptosis by inhibiting the anti-apoptotic effects of Bcl-2 homologues while inducing the expression of pro-apoptotic genes such as PUMA, Bax, and Bak that mediate the release of cytochrome c from mitochondria and induce cell death [41-43]. Therefore, accumulation of p53 due to proteasome insufficiency and the associated changes in the pro-apoptotic gene products could be responsible for the increased cell death that was observed in the aged heart. The proteins of the Bcl-2 family have been suggested to play a pivotal role as intracellular checkpoint in the apoptotic signal transduction [44]. In particular, among the entire Bcl-2 family, Bax and Bcl-2 have been well examined and designated to be the main protagonists in the regulation of apoptosis [45]. Our studies show that the increased level of TUNEL activity in the aged hearts was accompanied by the increased Bax/Bcl-2 ratio. Previous studies have also demonstrated that the up-regulation of p53 may contribute to part of the apoptotic signaling during muscle atrophy [1,46]) . Importantly, administration of GSE to aged rats caused a significant decrease in p53 level, which might be responsible for the decreased amount of cell death through the regulation of proand anti-apoptotic Bcl-2 family proteins. In support of these observations, previous studies show polyphenolic flavonoids found in GSE offered a significant protection to liver cells via modulating apoptotic regulatory genes such as Bcl-2, c-myc and p53 [47] . Taken together, these data suggest that the UPS might play a crucial role in regulating the stability and localization of p53 and serve as a key regulator of cell death and survival. Hence, part of the anti-apoptotic property of GSE might be mediated via its ability to regulate the UPS for the maintenance of Bcl-2 family proteins, possibly by down-regulating the p53 levels.
We also explored the role of cIAP family proteins that function as RING-finger E3 ligases and specifically target and ubiquitinate caspases and other associated pro-apoptotic proteins [48]. cIAP1 recognizes and ubiquitinates caspase-8 and a few other upstream activators [49] . In this manner, IAP family members inhibit either the activation of various caspases through UPS-mediated direct elimination of caspases or proteins that cause caspase activation. In the present study, RT-PCR and immunoblot analyses revealed that both mRNA and protein levels of cIAP1 in the aged rat heart were remarkably lower than that of the young hearts (Figure 3). These observations are consistent with the recent work that demonstrates cIAP1 is down regulated during aging, suggesting impairment of survival signaling [50] . Therefore, the loss of cIAP1 in the aging heart could be contributing to the increased levels of active caspase (cleaved caspase-3). In support of this idea, a previous study demonstrates that proteasomal inhibition, as observed in the aging heart, can activate caspase-induced cell death [51] . Importantly, our studies indicate that oral supplementation of GSE reversed this trend by decreasing both the active caspase-3 level and the associated cell death.
In summary, the build-up of deleterious proteins due to the functional loss of the UPS can cause myocardial cell loss, a major contributing factor to the development of heart failure. The present work show an age-associated increase in many cell death markers, such as TUNEL and DNA fragmentation which are accompanied by increased levels of p53 in the nucleus, Bax/Bcl-2 ratio, and active caspase-3 and decreased cIAP1 production. Importantly, administration of GSE shows antiapoptotic effects by minimizing these aging associated changes in the heart. Therefore, GSE has therapeutic benefits in reversing many of the aging related deleterious changes and improves ventricular function.
5. ACKNOWLEDGEMENTS
This authors wish to thank Dr. Dhan Kuppuswamy for his helpful suggestions in designing this work.
REFERENCES
- Tamilselvan, J., Jayaraman, G., Sivarajan, K., et al. (2007) Age-dependent upregulation of p53 and cytochrome c release and susceptibility to apoptosis in skeletal muscle fiber of aged rats: Role of carnitine and lipoic acid. Free Radical Biology & Medicine, 43, 1656-1669. doi:10.1016/j.freeradbiomed.2007.08.028
- Hershko, A. and Ciechanover, A. (1998) The ubiquitin system. Annual Review of Biochemistry, 67, 425-479. doi:10.1146/annurev.biochem.67.1.425
- Glickman, M.H. and Ciechanover, A. (2002) The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiological Reviews, 82, 373-428.
- Friguet, B., Bulteau, A.L., Chondrogianni N., et al. (2000) Protein degradation by the proteasome and its implications in aging. Annals of the New York Academy of Sciences, 908, 143-154. doi:10.1111/j.1749-6632.2000.tb06643.x
- Tsukamoto, O., Minamino, T., Okada, K., et al. (2006) Depression of proteasome activities during the progression of cardiac dysfunction in pressure-overloaded heart of mice. Biochemical and Biophysical Research Communications, 340, 1125-1133. doi:10.1016/j.bbrc.2005.12.120
- Birks, E.J., Latif, N., Enesa, K., et al. (2008) Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovascular Research, 79, 472-480. doi:10.1093/cvr/cvn083
- Gray, D.A., Tsirigotis, M. and Woulfe, J. (2003) Ubiquitin, proteasomes, and the aging brain. Science Aging Knowledge Environment, 2003, RE6. doi:10.1126/sageke.2003.34.re6
- Cuervo, A.M. (2008) Autophagy and aging: Keeping that old broom working. Trends in Genetics, 24, 604-612. doi:10.1016/j.tig.2008.10.002
- Vellai, T. (2009) Autophagy genes and ageing. Cell Death & Differentiation, 16, 94-102. doi:10.1038/cdd.2008.126
- Zolk, O., Schenke, C. and Sarikas, A. (2006) The ubiquitin-proteasome system: Focus on the heart. Cardiovascular Research, 70, 410-421. doi:10.1016/j.cardiores.2005.12.021
- Powell, S.R. (2006) The ubiquitin proteasome system in cardiac physiology and pathology. American Journal of Physiology—Heart and Circulatory, 29, H1-H19. doi:10.1152/ajpheart.00062.2006
- Predmore, J.M., Wang, P., Davis, F., et al. (2010) Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation, 121, 997-1004. doi:10.1161/CIRCULATIONAHA.109.904557
- Balasubramanian, S., Mani, S., Shiraishi, H., et al. (2006) Enhanced ubiquitination of cytoskeletal proteins in pressure overloaded myocardium is accompanied by changes in specific E3 ligases. Journal of Molecular and Cellular Cardiology, 41, 669-679.
- Johnston, R.K., Balasubramanian, S., Kasiganesan, H., et al. (2009) Beta3 integrin-mediated ubiquitination activates survival signaling during myocardial hypertrophy. FASEB Journal, 23, 2759-2771.
- Shafiee, M., Carbonneau, M.A., Urban, N., et al. (2003) Grape and grape seed extract capacities at protecting LDL against oxidation generated by Cu2+, AAPH or SIN- 1 and at decreasing superoxide THP-1 cell production. A comparison to other extracts or compounds. Free Radical Research, 37, 573-584. doi:10.1080/1071576031000083152
- Palma, M., Taylor, L.T., Varela, R.M., et al. (1999) Fractional extraction of compounds from grape seeds by supercritical fluid extraction and analysis for antimicrobial and agrochemical activities. Journal of Agricultural and Food Chemistry, 47, 5044-5048. doi:10.1021/jf990019p
- Scalbert, A., Manach, C. and Morand, C. (2005) Dietary polyphenols and the prevention of diseases. Critical Reviews in Food Science and Nutrition, 45, 287-306. doi:10.1080/1040869059096
- Sun, A.Y., Simonyi, A. and Sun G.Y. (2002) The “French Paradox” and beyond: Neuroprotective effects of polyphenols(1,2). Free Radical Biology & Medicine, 15, 314- 318. doi:10.1016/S0891-5849(01)00803-6
- Siemann, E. and Creasy, L. (1992) Concentration of the phytoalexin resveratrol in wine. American Journal of Enology and Viticulture, 43, 49-52.
- Cassidy, A. (2006) Factors affecting the bioavailability of soy isoflavones in humans. Journal of AOAC International, 89, 1182-1188.
- Sato, M., Bagchi, D., Tosaki, A., et al. (2001) Grape seed proanthocyanidin reduces cardiomyocyte apoptosis by inhibiting ischemia/reperfusioninduced activation of JNK-1 and C-JUN. Free Radical Biology & Medicine, 31, 729- 737. doi:10.1016/S0891-5849(01)00626-8
- Du., Y., Guo, H.F. and Lou, H.X. (2007) Grape seed polyphenols protect cardiac cells from apoptosis via induction of endogenous antioxidant enzymes. Journal of Agricultural and Food Chemistry, 55, 1695-1701.
- Fusco, D., Colloca, G., Lo Monaco, M.R., et al. (2007) Effects of antioxidant supplementation on the aging process. Journal of Clinical Interventions in Aging, 2, 377- 387
- Bagchi, D., Sen, C.K., Ray, S.D., et al. (2003) Molecular mechanisms of cardioprotection by a novel grape seed proanthocyanidin extract. Mutation Research, 523-524, 87-97. doi:10.1016/S0027-5107(02)00324-X
- Balu, M., Sangeetha, P., Murali, G., et al. (2006) Modulatory role of grape seed extract on age-related oxidative DNA damage in central nervous system of rats. Brain Research Bulletin, 68, 469-473. doi:10.1016/j.brainresbull.2005.10.007
- Kuppuswamy, D., Kerr, C., Narishige, T., et al. (1997) Association of tyrosine-phosphorylated c-Src with the cytoskeleton of hypertrophying myocardium. Journal of Biological Chemistry, 272, 4500-4508. doi:10.1074/jbc.272.7.4500
- Hayter, J.R., Doherty, M.K., Whitehead, C., et al. (2005) The subunit structure and dynamics of the 20S proteasome in chicken skeletal muscle. Molecular & Cellular Proteomics, 4, 1370-1381. doi:10.1074/mcp.M400138-MCP200
- Sellins, K.S. and Cohen, J.J. (1987) Gene induction by gamma-irradiation leads to DNA fragmentation in lymphocytes. Journal of Immunology, 139, 3199-3206
- Fortun, J., Li, J., Go, J., et al. (2005) Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. Journal of Neurochemistry, 92, 1531-1541. doi:10.1111/j.1471-4159.2004.02987.x
- Leri, A., Liu, Y., Malhotra, A., et al. (1998) Pacing-induced heart failure in dogs enhances the expression of p53 and p53-dependent genes in ventricular myocytes. Circulation, 97, 194-203. doi:10.1161/01.CIR.97.2.194
- Gross, A., McDonnell, J.M. and Korsmeyer, S.J. (1999) BCL-2 family members and the mitochondria in apoptosis. Genes Development, 13, 1899-1911. doi:10.1101/gad.13.15.1899
- Davies, K.J. (2000) Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB Life, 50, 279-289. doi:10.1080/713803728
- Bonoli, M., Bendini, A., Cerretani, L., et al. (2004) Qualitative and semiquantitative analysis of phenolic compounds in extra virgin olive oils as a function of the ripening degree of olive fruits by different analytical techniques. Journal of Agricultural and Food Chemistry, 52, 7026-7032. doi:10.1021/jf048868m
- Kwak, M.K., Wakabayashi, N., Greenlaw, J.L., et al. (2003) Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Molecular and Cellular Biology, 23, 8786-8794. doi:10.1128/MCB.23.23.8786-8794.2003
- Kwak, M.K., Cho, J.M., Huang, B., et al. (2007) Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free Radical Biology & Medicine, 43, 809-817. doi:10.1016/j.freeradbiomed.2007.05.029
- Kostin, S., Pool, L., Elsasser, A., et al. (2003) Myocytes die by multiple mechanisms in failing human hearts. Circulation Research, 92, 715-724. doi:10.1161/01.RES.0000067471.95890.5C
- Saraste, A., Pulkki, K., Kallajoki, M., et al. (1999) Cardiomyocyte apoptosis and progression of heart failure to transplantation. European Journal of Clinical Investigation, 29, 380-386. doi:10.1046/j.1365-2362.1999.00481.x
- Freude, B., Masters, T.N., Robicsek, F., et al. (2000) Apoptosis is initiated by myocardial ischemia and executed during reperfusion. Journal of Molecular and Cellular Cardiology, 32, 197-208. doi:10.1006/jmcc.1999.1066
- Maki, C.G. and Howley, P.M. (1997) Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Molecular and Cellular Biology, 17, 355-363.
- Latonen, L., Kurki, S., Pitkanen, K., et al. (2003) P53 and MDM2 are regulated by PI-3-kinases on multiple levels under stress induced by UV radiation and proteasome dysfunction. Cellular Signalling, 15, 95-102. doi:10.1016/S0898-6568(02)00044-X
- Mihara, M., Erster, S., Zaika, A., et al. (2003) P53 has a direct apoptogenic role at the mitochondria. Molecular Cell, 11, 577-590. doi:10.1016/S1097-2765(03)00050-9
- Chipuk, J.E., Kuwana, T., Bouchier-Hayes, L., et al. (2004) Direct activation of bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science, 303, 1010-1014. doi:10.1126/science.1092734
- Leu, J.I., Dumont, P., Hafey, M., et al. (2004) Mitochondrial p53 activates bak and causes disruption of a Bak-Mcl1 complex. Nature Cell Biology, 6, 443-450.
- Danial, N.N. and Korsmeyer, S.J. (2004) Cell death: Critical control points. Cell, 116, 205-219 doi:10.1016/S0092-8674(04)00046-7
- Desagher, S. and Martinou, J.C. (2000) Mitochondria as the central control point of apoptosis. Trends in Cell Biology, 10, 369-377. doi:10.1016/S0962-8924(00)01803-1
- Siu, P.M., Pistilli, E.E., Murlasits, Z., et al. (2006) Hindlimb unloading increases muscle content of cytosolic but not nuclear Id2 and p53 proteins in young adult and aged rats. Journal of Applied Physiology, 100, 907-916. doi:10.1152/japplphysiol.01012.2005
- Joshi, S.S., Kuszynski, C.A. and Bagchi, D. (2001) The cellular and molecular basis of health benefits of grape seed proanthocyanidin extract. Current Pharmaceutical Biotechnology, 2, 187-200. doi:10.2174/1389201013378725
- Salvesen, G.S. and Duckett, C.S. (2002) IAP proteins: Blocking the road to death’s door. Nature Reviews Molecular Cell Biology, 3, 401-410. doi:10.1038/nrm830
- Bertrand, M.J., Milutinovic, S., Dickson, K.M., et al. (2008) cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Molecular Cell, 30, 689-700. doi:10.1016/j.molcel.2008.05.014
- Kisiswa, L., Albon, J., Morgan, J.E., et al. (2010) Cellular inhibitor of apoptosis (cIAP1) is down-regulated during retinal ganglion cell (RGC) maturation. Experimental Eye Research, 91, 39-747. doi:10.1016/j.exer.2010.08.024
- Henderson, C.J., Aleo, E., Fontanini, A., et al. (2005) Caspase activation and apoptosis in response to proteasome inhibitors. Cell Death and Differentiation, 12, 1240-1254. doi:10.1038/sj.cdd.4401729

