Open Journal of Statistics
Vol.05 No.06(2015), Article ID:60526,17 pages
10.4236/ojs.2015.56059
Statistical Inference in Generalized Linear Mixed Models by Joint Modelling Mean and Covariance of Non-Normal Random Effects
Yin Chen1, Yu Fei2, Jianxin Pan3*
1School of Insurance, University of International Business and Economics, Beijing, China
2School of Statistics and Mathematics, Yunnan University of Finance and Economics, Kunming, China
3School of Mathematics, University of Manchester, Manchester, UK
Email: chenyin@uibe.edu.cn, feiyukm@aliyun.com, *jianxin.pan@manchester.ac.uk
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/



Received 8 September 2015; accepted 20 October 2015; published 23 October 2015
ABSTRACT
Generalized linear mixed models (GLMMs) are typically constructed by incorporating random effects into the linear predictor. The random effects are usually assumed to be normally distributed with mean zero and variance-covariance identity matrix. In this paper, we propose to release random effects to non-normal distributions and discuss how to model the mean and covariance structures in GLMMs simultaneously. Parameter estimation is solved by using Quasi-Monte Carlo (QMC) method through iterative Newton-Raphson (NR) algorithm very well in terms of accuracy and stabilization, which is demonstrated by real binary salamander mating data analysis and simulation studies.
Keywords:
Generalized Linear Mixed Models, Multivariate t Distribution, Multivariate Mixture Normal Distribution, Quasi-Monte Carlo, Newton-Raphson, Joint Modelling of Mean and Covariance

1. Introduction
Generalized linear mixed models (GLMMs) are very helpful and widely used for analyzing discrete data and data from exponential family distributions. Statistical inference of GLMMs is challenging due to the incorporation of random effects, especially for example, the marginal likelihood has the form of analytical intractable high dimensional integration. The existing and popular statistical methods include: 1) analytical Laplace approximation: the uncorrelated penalized quasi-likelihood (PQL) by Breslow and Clayton (1993) [1] and correlated PQL by Lin and Breslow (1996) [2] ; hierarchical generalized linear models (HGLM) procedure by Lee and Nelder (2001) [3] ; 2) numerical technique: Bayesian approach with sampling by Karim and Zeger (1992) [4] ; MCEM algorithm by Booth and Hobert (1999) [5] ; Gauss-Hermite quadrature (GHQ) by Pan and Thompson (2003) [6] ; Quasi-Monte Carlo (QMC) and Randomized QMC method by Pan and Thompson (2007) [7] and Aleid (2007) [8] .
One common idea in above literatures is random effects in GLMMs, which represent latent effects between individuals, follow normal distribution with mean zero and identity covariance matrix. However, these assumptions are not always valid in practice because individual has its own special natures and may not be normal distributed. To address this issue, multivariate t distribution and multivariate mixture normal distribution will be assumed for random effects in GLMMs. Quasi-Monte Carlo (QMC) method through iterative Newton- Raphson (NR) algorithm solves the high-dimensional integration problem in the marginal likelihood under the non-normal assumptions.
Review of GLMMs and the marginal Quasi-likelihood will be proposed in Section 2. QMC approximation, the simplest Good Point set (square root sequence) and analytical form of maximum likelihood estimates (MLEs) in GLMMs will be discussed in Section 3. The idea of modified cholesky decomposition and joint modelling of mean and covariance structure will be explained in Section 4. In Section 5, the salamander data will be analyzed as an example under the assumption of multivariate t distribution of random effects, then simulation study with same design protocol as salamander data followed in Section 6. Discussions on the related issues and further studies are given in Section 7.
2. Generalized Linear Mixed Models
The generalized linear mixed models (GLMMs) are typically constructed by incorporating random effects into the linear predictor of a conditionally independent exponential family model (McCulloch, 2003, [9] ).
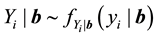 ;
;
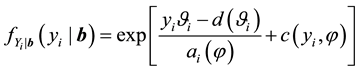 ;
;
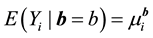 ;
;
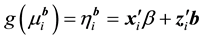 ;
;
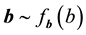 ;
;
where 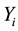 denotes the observed response,
denotes the observed response, 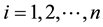 ,
, 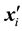 is the
is the 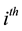 row of design matrix;
row of design matrix; 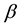 is
is 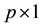 fixed effect parameter;
fixed effect parameter; 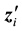 is the
is the 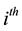 row of the second design matrix;
row of the second design matrix;  is
is 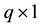 random effect.
random effect.
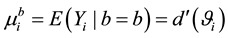 and
and


known overdispersion scalar, 

In this definition we see the usual ingredients of a generalized linear model. First, the distribution of 













with the mean of 



is a 





with mean






In GLMMs, statistical inferences of 







where


determinates the conditional log quasi-likelihood of 



It is extremely challenging to obtain the maximum likelihood estimates (MLEs), 
3. Quasi-Monte Carlo Integration and Estimation
3.1. Quasi-Monte Carlo Integration
Quasi-Monte Carlo (QMC) sequences are a deterministic alternative to Monte Carlo (MC) sequences (Niederreiter, 1992, [11] ). For the q-dimensional unit cube domain

where sample points 
If the integrand f has finite variation, then the error of the quasi-Monte Carlo quadrature can be bounded using the Koksma-Hlawka inequality

where 




where A is an arbitrary q-dimensional subcube parallel to the coordinate axes and originating at the centre, 

For carefully selected sample points, the discrepancy and consequently the error of low-discrepancy sequ-
ences (Niederreiter, 1992, [11] ) can be in the order of
the probabilistic error bound of Monte Carlo methods (Fang and Wang, 1994, [12] ). Moreover, quasi-Monte Carlo methods guarantee this accuracy in a deterministic way, unlike Monte Carlo methods where the error bound is also probabilistic.
In the QMC approach, there exist precise construction algorithms for generating the required points. These algorithms can be divided into two families, the Lattice rule and Digital-net. Only the good point set, which is belong to lattice rule (Fang and Wang, 1994, [12] ) will be used and discussed in following sections.
3.2. The Good Point (GP) Set
A good point


points of the set

y. In practice, the following forms of the good point 

where


3.3. MLE in GLMMs by QMC Estimation
When QMC method applied to marginal quasi-likelihood function (4)

where 



as the Cholesky decomposition of 
Let










where


The weight 



The score Equations (7) and (8) in general have no analytical solutions for 


where 


lihood.
Calculation of the Hessian matrix is difficult and its computation is intensive. When the difference between 



the MLEs, 

4. Modified Cholesky Decomposition and Covariance Modelling for Random Effects
Modified Cholesky decomposition of




where

It offers a simple unconstrained and statistically meaningful reparametrization of the covariance matrix. So 



where 












The linear joint models of the autoregressive coefficients (ACs) and the logarithms of innovation variances (IVs)

where 





5. Salamander Data Analysis
The famous salamander interbreeding data set was published by McCullagh and Nelder (1989), which came from three repeated same protocol design experiments. The salamanders originally lived in two geographically isolated populations, Roughbutt (RB) and Whiteside (WS). Each experiment involved in two closed groups and each group involved 5 females and 5 males from each population. In a fixed period, each female mated with 6 males, 3 from each population. So 60 correlated binary observations were created in each closed group, and totally, 360 binary observations (1 for successful interbreeding, 0 for failed interbreeding) were in three ex- periments. The primary two objectives of experiment are to study whether the successful mating rate is sig- nificant between populations and if heterogeneity between individuals in the mating probability exists. This binary data set is challenging because it is block crossed, correlated, balanced (as each female mated with a total of six males and vice verse) and incomplete (as each female mated with just three out of a possible five males, and vice verse).
In a closed group, 5 females and 5 males from each population involved, so total 20 salamander were indexed by i or j. Denoting by 

where the conditional probability 





Here the random effect is not assumed as normal distribution but multivariate t distribution with degree of freedom

Note that each experiment includes two closed groups and involves 40 salamanders so that the log-likelihood for each experiment involves a 40-dimensional integrals. The dimensionality for each experiment can be further reduced to the sum of two 20-dimensional integrals due to the block design of two closed groups, see Karim and Zeger (1992) [4] and Shun (1997) [15] for more details about the design. When the three experiments data are pooled as in model (13), the log-likelihood 


Modelling 




where 








where







The logarithms of innovation variances (IVs) model is
where 






where



In the logarithms of innovation variances (IVs) model, originally there were 4 covariates including the item 





The salamander data is now analyzed by using QMC approach to calculate the MLEs of the fixed effects and variance components involved in the model. Specifically, we implement the simplest QMC point, square root sequences to estimate the parameters for the modelling of the pooled data. A set of 20-dimensional points on the unit cube 
through using transformation, 

variate t distribution, 

mate the integrated log-likelihood, then we use the Newton-Raphson algorithm (10) to maximize the appro- ximated log-likelihood function.
The covariance modelling for










the parameter is reduced from 

computation easier. Note that, the higher the dimension q, the more significant this advantage. Third, the modified Cholesky decomposition confirms that unconstrained parameters lead to strict condition that is the 
In the calculations, the convergence accuracy is set to be
numerical results from Tables 1-4 show that for each of the degree of freedom, QMC method make the maximum log-likelihood similar when increasing the number of QMC points from 10,000 to 100,000 which indicates any number of points between these could prove reasonable estimate of the parameters. In addition, the parameter estimates using these sequences become stable quickly. Bearing in mind that the integral space is
Table 1. MLEs of the parameters by covariance modelling for the pooled salamander data using square root sequence points when varying K, the number of square root sequence points (standard errors in parentheses) (random effect-multivariate
Table 2. MLEs of the parameters by covariance modelling for the pooled salamander data using square root sequence points when varying K, the number of square root sequence points (standard errors in parentheses) (random effect-multivariate
Table 3. MLEs of the parameters by covariance modelling for the pooled salamander data using square root sequence points when varying K, the number of square root sequence points (standard errors in parentheses) (random effect-multivariate
Table 4. MLEs of the parameters by covariance modelling for the pooled salamander data using square root sequence points when varying K, the number of square root sequence points (standard errors in parentheses) (random effect-multivariate
20-dimensional unit cube so that even if the number of the QMC points is chosen as
For mean model, the estimators for fixed effects, 









A criteria easy to be executed to decide which covariate is significant is if the estimator is greater than twice of standard error. So the four fixed effect parameters of 










Another point is why the random effect is not assumed as multivariate mixture normal distribution for the salamander data because Newton-Raphson algorithm does not achieve convergency until the mixture normal distribution degenerates to normal distribution.
6. Simulation
We conducted a simulation study to assess the efficiency of the QMC estimation in the GLMMs, in particular, to assess the performance of the GP set point when the distribution of random effect is multivariate t distribution (1) and multivariate mixture normal distribution (2). Based on the logistic model in (13), we simulate the sala- mander pooling data, which have 360 correlated binary observations. In the simulation study, we run 100 simulations. The same protocol design as the salamander mating experiment is adopted for the simulated data and the log-likelihood function for each simulated data set thus involves six 20-dimensional integrals that are analytically intractable.
When using the QMC approximation, we generate 


The Newton-Raphson algorithm (10) is used to maximize the approximated log-likelihood function. The true values are chosen to be (1.20, −2.80, −1.00, 3.60) for the fixed effects, 


It is clear that estimation by modified Cholesky decomposition with QMC approach is able to produce reasonably good results in GLMMs when the random effect is not normal distribution. When the random effects followed by multivariate t distribution (Table 5), the estimates of regression fixed effects and variance com- ponents are those on average of 100 simulation, which has little bias. That means the method of modified
Table 5. The average of the 100 parameter estimates in the simulation study, where the QMC approximation uses 




Table 6. The average of the 100 parameter estimates in the simulation study, where the QMC approximation uses 



Cholesky decomposition of covariance modelling with QMC approach is quite useful and effective. When the random effects followed by multivariate mixture normal distribution (Table 6), the estimates of regression fixed effects are acceptable although bigger bias, especially for fixed effects. The estimates of variance components are quite accurate. That is, when random effects change to two peaks distribution, the accuracy need to be en- hanced.
7. Conclusion and Discussion
It is quite common in the literatures that random effects are independent identically distributed normal variables in generalized linear models (GLMMs). In longitudinal study, the covariance-variance matrix of random effects for individuals is usually assumed to be homogeneous. However, this assumption may not be valid in practice. In this article, the assumption extends to random effects follow multivariate t and mixture normal distribution. Approximation of marginal quasi-likelihood and parameter estimation in GLMMs is very difficult and challenging because they involve integral on random effects, especially for high-dimensional problems.
In this article, we have used Quasi-Monte Carlo (QMC) sequence approximation to calculate the maximum likelihood estimates and marginal log quasi-likelihood in GLMMs. The key idea of QMC sequences is to generate integration points which are scatted uniformly in the integration domain. The good point, one simple QMC point, is used through this article. Newton-Raphson (NR) algorithm is the iterative method to obtain op- timum. QMC-NR method converges very quickly and performs very well in terms of accuracy and stabilization. In longitudinal studies, the covariance structure plays a crucial role in statistical inferences. The correct modelling of covariance structure improves the efficiencies of the mean parameters and provides much more reliable estimates (Ye and Pan, 2006, [16] ). The joint modelling (JM) for mean and for covariance-variance components for random effects reduces the number of parameters and has a clear statistical meaning. QMC-NR method with joint modelling can be called as QMC-NR-JM method. Although QMC-NR-JM method performs well in stability and produces accurate estimation of the parameters in GLMMs, there is no way to avoid intensive computation in real data analysis and simulation study. Particularly, the binary cross-designed sala- mander dataset, involving six 20-dimensional integrals in the log quasi-likelihood function, has been analyzed and the same protocol design is adopted for simulations.
Note that a practical issue is how to select of the number of integration points? My solution is increasing the number of QMC points gradually until the MLEs become stable. It correspondingly leads to increasing the computational time and efforts. This problem is quite obvious for simulation study, especially when the number of parameters increases. Another worth noting issue is that a range of starting values for the fixed effects and variance components have been tried and we find that the different starting values almost can’t change the results when algorithm reaches convergency.
In the QMC-NR-JM approximation method, the estimators of random effects cannot be calculated at the same time when estimating MLEs for fixed effects and variance components. Pan and Thompson (2007) [7] proposed an iterative method to predict of the random effects, implying that an initial value of b substituted into the equation to yield a new prediction of b. This process is then repeated until the convergence of
Acknowledgements
We thank the editors and the reviewers for their comments. This research is funded by the National Social Science Foundation No. 12CGL077, National Science Foundation granted No. 71201029, No.71303045 and No. 11561071. This support is greatly appreciated.
Cite this paper
YinChen,YuFei,JianxinPan, (2015) Statistical Inference in Generalized Linear Mixed Models by Joint Modelling Mean and Covariance of Non-Normal Random Effects. Open Journal of Statistics,05,568-584. doi: 10.4236/ojs.2015.56059
References
- 1. Breslow, N.E. and Clayton, D.G. (1993) Approximate Inference in Generalized Linear Mixed Models. Journal of the American Statistical Association, 88, 9-25.
- 2. Lin, X. and Breslow, N.E. (1996) Bias Correction in Generalized Linear Mixed Models with Multiple Components of Dispersion. Journal of the American Statistical Association, 91, 1007-1016.
http://dx.doi.org/10.1080/01621459.1996.10476971 - 3. Lee, Y. and Nelder, J.A. (2001) Hierrarchical Generalized Linear Models: A Synthesis of Generalized Linear Models, Random Effect Models and Structured Dispersions. Biometrika, 88, 987-1006.
http://dx.doi.org/10.1093/biomet/88.4.987 - 4. Karim, M.R. and Zeger, S.L. (1992) Generalized Linear Models with Random Effects; Salamnder Mating Revisited. Biometrics, 48, 681-694. http://dx.doi.org/10.2307/2532317
- 5. Booth, J.G. and Hobert, J.P. (1999) Maximizing Generalized Linear Mixed Model Likelihoods with an Automated Monte Carlo EM Algorithm. Journal of the Royal Statistical Society, 61, 265-285.
http://dx.doi.org/10.1111/1467-9868.00176 - 6. Pan, J. and Thompson, R. (2003) Gauss-Hermite Quadrature Approximation for Estimation in Generalized Linear Mixed Models. Computational Statistics, 18, 57-78.
- 7. Pan, J. and Thompson, R. (2007) Quasi-Monte Carlo Estimation in Generalized Linear Mixed Models. Computational Statistics and Data Analysis, 51, 5765-5775.
http://dx.doi.org/10.1016/j.csda.2006.10.003 - 8. Al-Eleid, E.M.O. (2007) Parameter Estimation in Generalized Linear Mixed Models Using Quasi-Monte Carlo Methods. PhD Dissertation, The University of Manchester, Manchester.
- 9. McCullouch, C.E. (2003) Generalized Linear Mixed Models. NSF-CBMS Regional Conference Series in Probability and Statistics Volume 7. Institution of Mathematical Statistics and the American Statistical Association, San Francisco.
- 10. McCulloch, P. and Nelder, J.A. (1989) Generalized Linear Models. 2nd Edition, Chapman and Hall, London.
http://dx.doi.org/10.1007/978-1-4899-3242-6 - 11. Niederreiter, H. (1992) Random Number Generation and Quasi-Monte Carlo Methods. SIAM, Philadelphia.
http://dx.doi.org/10.1137/1.9781611970081 - 12. Fang, K.T. and Wang, Y. (1994) Number-Theoretic Methods in Statistics. Chapman and Hall, London.
http://dx.doi.org/10.1007/978-1-4899-3095-8 - 13. Pourahmadi, M. (1999) Joint Mean-Covariance Models with Applications to Longitudinal Data: Unconstrained Parameterization. Biometrika, 86, 677-690.
http://dx.doi.org/10.1093/biomet/86.3.677 - 14. Pourahmadi, M. (2000) Maximum Likelihood Estimation of Generalized Linear Models for Multivariate Normal Covariance Matrix. Biometrika, 87, 425-435.
http://dx.doi.org/10.1093/biomet/87.2.425 - 15. Shun, Z. (1997) Another Look at the Salamander Mating Data: A Modified Laplace Approximation Approach. Journal of the American Statistical Association, 92, 341-349.
http://dx.doi.org/10.1080/01621459.1997.10473632 - 16. Ye, H. and Pan, J. (2006) Modelling Covariance Structures in Generalized Estimating Equations for Longitudinal Data. Biometrika, 93, 927-941.
http://dx.doi.org/10.1093/biomet/93.4.927
Appendix A. Second-Order Derivatives of Log-Likelihood Newton-Raphson Algorithm
where 


likelihood.
The second-order derivatives of log likelihood
Appendix B. MLEs for Covariance Modeling
The first and second derivative of 









matrix 

Every element in 



We can put the 














The above relationship is a one-to-one correspondence between i and






are all lower triangular matrices. Denote 

The first derivative can be calculated as

where 

The second derivative can be calculated as


because of




where




NOTES
*Corresponding author.













