Journal of Water Resource and Protection
Vol.6 No.1(2014), Article ID:41978,7 pages DOI:10.4236/jwarp.2014.61001
Seasonal Shifts in the Bacterioplankton Assemblages of High Altitude Middle Atlas Lakes
1Biotechnology Laboratory, School of Science and Engineering, Al Akhawayn University, Ifrane, Morocco
2Department of Biology and the Center for Sustainability and the Environment, Albion College, Albion, USA
Email: *oolapade@albion.edu
Received November 5, 2013; revised December 6, 2013; accepted January 2, 2014
ABSTRACT
Compositional changes of the bacterioplankton assemblages in four high altitude middle Atlas lakes were monitored over a complete seasonal cycle using combinations of culture-dependent and molecular approaches. Viable bacterial numbers varied between seasons, with the lowest numbers recorded in the winter and the highest in the summer in all four lakes. Also, bacterial occurrences were found to be strongly correlated with water temperature in all the four sites, i.e., Lake Aoua (0.88), Ifreh (0.59), Hechlef (0.77) and Affourgagh (0.79) during the study period. Standard microbiological characterization of bacteria isolates from the lakes revealed majority (68%) to be Gram positive. Seasonal variations in the microbial assemblages among the lakes were also validated by restriction fragment length polymorphism (RFLP). Shifts in microbial assemblages, especially of the fecal indicator bacteria appeared to also be influenced by differences in the morphometric and watershed characteristics among the lakes. This study further reveals the need to employ combinations of methodological approaches, including taxonomic, physiological and molecular methods to adequately delineate and fully understand the ecology of microbial assemblages in extreme environments.
Keywords:Bacterial Diversity; High Altitude Lakes; Seasonal Shifts
1. Introduction
Increasing recognition regarding the ecological importance of and contributions by autochthonous microbial assemblages in aquatic systems has significantly heightened the need to further examine and elucidate their community compositions, structures and dynamics [e.g. 1, 6,18,19,27]. Several studies have utilized combinations of molecular approaches in the past to reveal the numerical dominance of bacterial members belonging to the subclasses (especially, α- and β-) of the Proteobacteria as well as the Cytophaga-Flavbacteria cluster in various freshwater environments [2,1]">11]. In particular, bacterial occurrences and compositional changes in microbial assemblages within extreme environments such as in high altitudes have attracted much attention in order to better understand the impacts of seasonal conditions on the occurrences and diversity of indigenous bacterial populations [1,9,1]">1]">21]. Generally, some of these studies have revealed high compositional changes in the microbial community structures between seasons [e.g.,1]">1]">21], while others have documented such shifts between different habitats (i.e. snow, slush and the water body) in freshwater systems in extreme environments [e.g.,1]. However, most of these past studies only based their reports on the elucidation of microbial assemblages within individual lakes, and to the best of our knowledge, very limited information currently exists on microbial occurrence and activities across multiple lakes located in high altitude environments [7].
Therefore, in this study, combinations of culture-dependent and molecular approaches were utilized to examine the autochthonous microbial assemblages in four different lakes (Lakes Aoua, Ifreh, Hechlef (or Iffer) and Affourgagh) located within the Atlas Mountains in Morocco. The Middle-Atlas region of Morocco is known for its high altitude and diverse freshwater bodies in the mountainous areas with great temperature variations, including a typical hot and dry summer period, winter freezing and even snow in the Atlas Mountains [22]. The four lakes were selected because of differences in their morphometric, limnological and catchment area characteristics [15]. In particular, Lakes Aoua and Ifreh have relatively large basins (by surface area) as compared to Lakes Hechlef and Affourgagh (see Figure 1). The main objective of the study was therefore to examine the autochthonous bacterial assemblages in the four selected lakes in response to extreme changes in their environmental conditions as well as potential anthropogenic influences using combinations of culture-dependent and molecular approaches.
2. Materials and Methods
2.1. Description of Study Sites and Sample Collection
Triplicate water and sediment samples were aseptically collected with sterile falcon tubes from the littoral zones of four meromictic high altitude lakes in Morocco. The four lakes are situated in the same geographical region (Lake Aoua—33˚39'9"N, 5˚1'59"W and Lakes Ifreh, Hechlef and Affourgagh—33˚33'33"N, 55'48"W) around the Fes-Boulemane region of Morocco at altitudes around 1500 m above sea level [15].
2.2. Bacterial Enumeration, Isolate Culturing and Microbiological Characterization
Total viable bacterial counts were carried out on Nutrient Agar (NA) plates using standard techniques according to
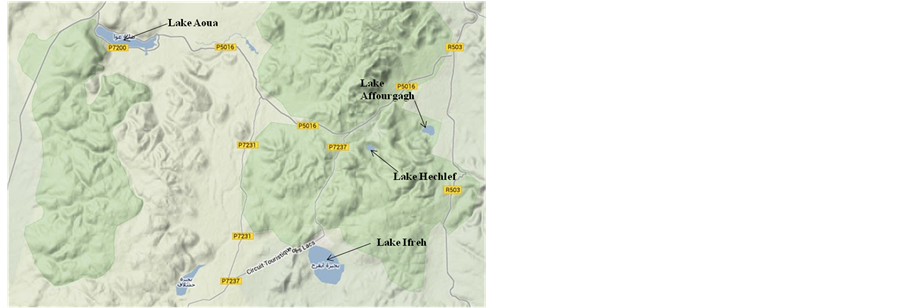
Figure 1. Location map of the four studied lakes in the FesBoulemane and Middle-Atlas region of Morocco.
Leboffe and Pierce [13]. Briefly, 100 µl from the sampled freshwater were plated onto NA agar plates and incubated for 24 h at 37˚C. From sediments, about 1 g was suspended in 100ml of LB broth for 1 h and 100 µL from the suspension were plated onto NA agar plates for 24 h at 37˚C. Distinct bacterial cells were later isolated according to their various morphological characteristics. A series of morphological, physiological and biochemical tests to determine the identities of the isolates were carried out, including Gram staining, growth on selective media e.g., Eosine Methylene Blue (EMB), Mannitol Salt (MSA) and Simmons Citrate media according to standard microbiological methods [13]. Additionally, Carbohydrate utilization test was performed on some of the isolates using M9 minimal medium with 1 mM CaCl2 and 1 mM MgSO4, with several sugars as the only sources of carbon and energy (Table 1).
2.3. Nucleic Acid Extraction and Purification
Genomic DNA was extracted from bacterial isolates using a rapid method. Briefly, from an overnight culture in LB broth biomass harvested from 3 ml was washed twice with 500 µl of solution A containing 100 mM Tris-Cl, 10 mM EDTA, 0.2 mM NaCl, pH 8.0. After the washing step, bacteria were suspended in 500 µl of solution A, SDS was added in a final concentration of 1% (w/v), and incubated for 10 min at 55˚C. Ammonium acetate was then added to a the lysate to a final concentration of 300 mM and the mixture was centrifuged at 15.000 g for 15 min at 4˚C. Supernatants were removed and DNA precipitation was done by adding two volumes of absolute ethanol, mixed well and centrifuged at 15000 g for 15 min at 4˚C. The obtained pellets were washed twice with cold 70% ethanol, dried under fan at 37˚C and suspended in 50 µl Milli-Q water. This extraction method allowed us to extract total genomic DNA from isolates and later analyzed by gel electrophoresis.
2.4. PCR Amplification
The almost complete 16S rDNA genes from the isolated bacteria (longer than 1400 nucleotides) were amplified using the universal primer pair fD1 (5'-AGAGTTTGATCCTGGCTCAG), and rp1 5'-ACGGTTACCTTGTTACGACTT, [24]. The primer’s set PT-2/PT-3 with the respective sequences; 5'-GCGAAAACTGTGGAATTGGG-3' and 5'-TGATGCTCCATAACTTCCTG-3', [3,4] were used to amplify a fragment of 252 bp from the uidA gene encoding β-D-glucuronidase from coliform bacteria. Reaction mixtures for PCR contained 1× buffer, 200 µM of each deoxynucleotide triphosphate, 1.5 mM MgCl2, 0.1 mM of each primer, and 2.5U of Taq DNA polymerase (Promega) in a final volume of 50 µl. the PCRs were performed in a UnoCycler Gradient Thermal Cycler
Table 1 . Carbohydrate utilization tests on bacterial isolates from Lakes Aoua (LAW) and Ifrah (LIS).
A, GLUC+; B, FRUC+; C, SORBITOL-; D, ARABITOL+; E, MYOINOSITOL+; F RIBOSE YE+; G, DALCITOL+; H, MESO-ERYTHRITOL YE+; I, RHAMNOSE+; J, SORBOSE+; K, SUCCINIC ACID +; L, RAFFISOC+; M, CITRIC ACID+; N, GLUCONIC ACID+; O, MYOINOSITOL+; P, MALTOSE+; Q, GALACTOSE+; R, MELIBIOSE+; S, RHAMNOSE-; T, SORBOSE-; U, SUCCINIC ACID-; V, RUFFISOC-; W, CITRIC ACID-; X, GLUCONIC ACID-; Y, MYOINOSITOL-.
(VWR-Belgium) using an initial denaturation step at 95˚C (5 m), followed by 28 cycles of denaturation at 95˚C (1 m), annealing at 55˚C (40 s) and extension at 72˚C (1 m 30 s), a final extension step at 72˚C (5 m) and subsequent cooling to 4˚C to complete the reaction. The PCR products related to each sample (2 µL) was verified by gel electrophoresis in 1% agarose (Applichem) in 0.5× TAE buffer. PCR products were purified using a DNA Gel extraction Kit (VWR-Belgium).
2.5. Screening of PCR Products by Restriction Fragment Length Polymorphism (RFLP)
The purified PCR product (3 µg) from each isolate was digested for 1h at 37˚C with 2 U Alu I (New England Biolabs). The restriction fragments were separated by gel electrophoresis in 1.5% (w/v) agarose (Applichem) in 0.5× TAE buffer. The gel was stained with ethidium bromide (0.5 mg/ml) and visualized under UV excitation (Genosmart, VWR). Restriction profiles were compared using Genosoft software from (VWR-Belgium), and isolates were classified according to distinct restriction fragment length polymorphism (RFLP) patterns.
2.6. Statistical Analyses
Relationships between bacterial populations and water chemistry characteristics in each of the lakes were examined using Linear Regression and Pearson Correlation analyses (SPSS for Windows, version 15, SPSS Inc., Chicago, IL).
3. Results
There was clear seasonality in both water chemistry, particularly temperature (Figure 2) and in the occurrences of total bacterial populations (Figure 3) among the four lakes examined during the study period. Specifically, temperature values were as low as 3˚C in December to as high as 30˚C in August through September. Viable bacterial counts followed the same seasonal trend as recorded for temperature among the four lakes, ranging averagely from 1.375 × 106 CFU/mL in to about 6.714 × 106 CFU/mL (Figure 2). There were strong correlations between bacterial densities and temperature and in all the four sites i.e., Lake Aoua (0.88), Ifreh (0.59), Hechlef (0.77) and Affourgagh (0.79).
A total of 44 bacterial isolates cultured from samples collected the four lakes were characterized based on combinations of their morphological, physiological and biochemical attributes. Twenty eight (64%) of these isolates appeared rod-shaped, while 16 (36%) of them were spherical or cocci-shaped (Table 2). Gram staining revealed that 30 (68%) of the cells possessed the Gram positive cell wall, while the remaining 14 (28%) stained gram negatively. Overall, the distribution of the isolates
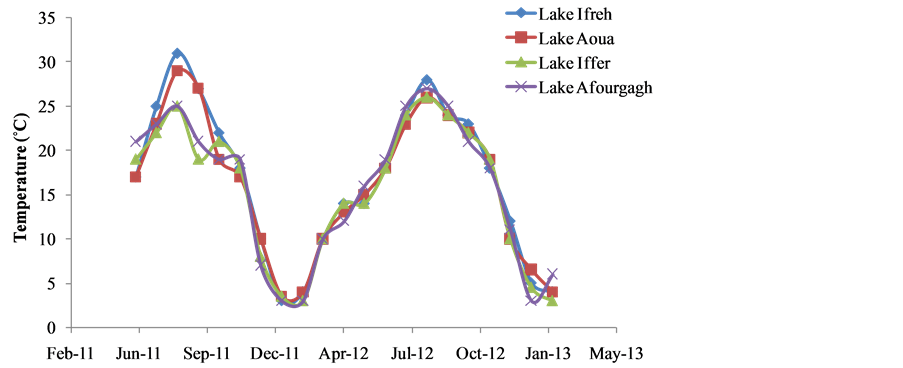
Figure 2. Seasonal trends in water temperature values recorded in the four lakes during the study period.
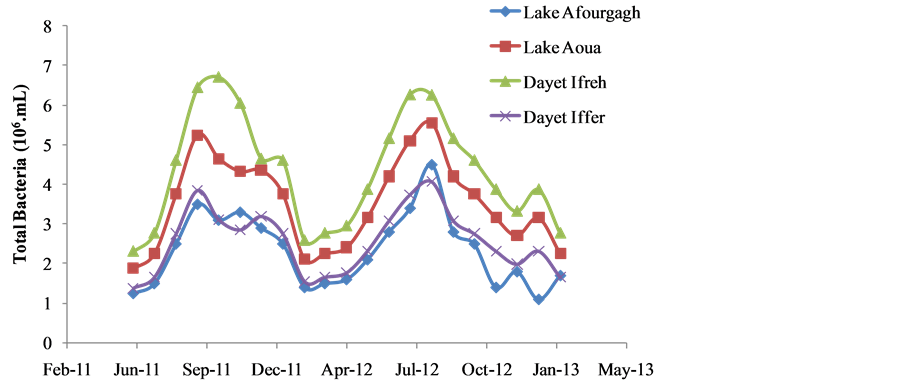
Figure 3. Seasonal trends in total bacterial numbers enumerated from water samples collected from the four lakes during the study period.
Table 2. Characteristics of bacteria isolated from the four lakes.
LAW: Lake Aoua, LIS: Lake Ifreh, LIF: Lake Hechleff (Iffer) and LAF: Lake Afourgagh. Percentage values are in parentheses.
by sampling source, revealed that the Gram positive taxa numerically dominated in three of the four lakes examined, by accounting for between 14% and 27% of total isolate numbers, except in Lake Ifreh, where only 3 (7%) were characterized as Gram Positive. Results of the Carbohydrate Utilization test revealed diverse preferences and affinity for majority of the organic carbon sources by almost all the isolates examined (Table 1). While there was 100% utilization of glucose, fructose, ribose, galactose and melibiose by all the ioslates, however in contrast, meso-erythritol, rhamnose and citric acid were not utilized by any of the bacterial isolates examined.
The results from the restriction fragment length polymorphism {RFLP} profiles generated with Alu I restriction enzyme after the amplification of total genomic DNA from the different lakes with primers fd1 and rp1 also clearly revealed seasonal changes in bacterial community composition in the study sites (Figure 4). The RFLP profiles showed that there were more patterns during the summer compared to the winter in all the sites, with very close similarities in the banding patterns found in samples collected from Lakes Aoua, Hechlef and Affourgagh as compared to those in Lake Ifreh. Also, PCR detection of fecal indicator bacteria (FIB) from the four lakes performed using the set of primers PT2/PT3 targeting a fragment from the uidA gene coding for β-galacturonidase showed higher amplification of the gene in samples from Lake Ifreh compared to the other three lakes (i.e. Aoua, Hechlef and Afourgagh), with relatively low and/or undetectable levels of amplification (Figure 5).
4. Discussion
The application of combinations of culture-dependent

Figure 4. Comparative Restriction Fragment Length Polymorphism {RFLP} generated from the four lakes (Lane 1: Lake Aoua; 2: Lake Iffer; 3: Lake Afourgagh and 4: Lake Ifreh) during the study period.

Figure 5. PCR detection and gel electrophoresis of Fecal Indicator Bacteria (FIB) in water samples collected from the lakes using primer sets PT2 and PT3 that targets the uidA gene. (Lane 1: Lake Ifreh; 2: Lake Aoua; 3: Lake Afourgagh and 4: Lake Iffer).
(viable bacterial count) and culture-independent molecular (i.e. PCR detection and RFLP) methods utilized in this study, both established the seasonal shifts in the bacterioplankton assemblages within the four high altitude lakes. Temperature changes between the very warm summer (starting from late May through early October), probably due to thermal mixing of the water body and cold winter (November through December) months appear to be the major drivers of autochthonous microbial assemblage shifts in all the study sites. The trend observed in this study strongly validates the drastic temperature fluctuations that have been observed in water bodies located in Atlas Mountains, that are typically characterized by humid summer months (up to 30˚C in August) to winter freezing (around 3˚C) and even snow around December [22]. Pernthaler et al. [21] also observed a similar seasonal shift in the bacterial abundance of a high mountain lake. Results they obtained from the utilization of both fluorescently labeled oligonucleotide probes (FISH) and image-analyzed microscopy revealed seasonal variation in bacterial assemblages in both the summer and beneath winter ice cover in Lake Gossenköllessee, a small oligotrophic high-mountain lake located within the Central Alps in Australia. It is worth noting that the four lakes examined in our study appear to have in common many limnological as well as watershed characteristics with Lake Gossenköllessee, especially regarding their relatively small morphometric basins and geographic locations, with all lakes situated at altitudes far exceeding 1500 m above sea level [15,22].
Another environmental variable that co-varied with water temperature and probably influenced the temporal shifts in microbial communities in the studied lakes could be organic matter availability. The ecological significance of various types of dissolved organic matters, especially those associated with phytoplankton biomass in freshwater environments have been well documented [8,17,23]. In this study, phytoplankton blooms were mostly observed covering Lake Ifreh for most of the summer through the end of fall, as compared to the other three lakes that had moderate algal blooms during the same period. The occurrences of the phytoplankton blooms probably further promoted high bacterial abundance, especially during the summer period, given the strong algal-bacterial co-variation that had been previously reported in various aquatic systems [e.g.,8]. Moreover, the high propensity exhibited by the bacterial isolates for the various carbohydrates examined here is further indicative of the ubiquity and direct influences of such diverse organic carbon sources on the metabolic and physiological processes of the isolates found within the studied lakes.
The numerical dominance of Gram negative bacteria that is phylogenetically affiliated to the subclasses (e.g.α, β, and γ) of the Proteobacteria, and the CytophagaFlavobacteria-Bacteroidetes in freshwater environments have been previously well documented [1,2,6,9,11,17, 21,27]. However, in this study based on the results of the culture-dependent, viable bacterial analysis of the communities revealed the predominance of Gram positive rod-shaped bacteria over the Gram negative cells, in most of the lakes examined. While this result is somewhat unusual and contradicting, previous observations that consistently reveal the numerical dominance of Gram negative populations, relatively high abundances of Gram positives, especially those with high GC contents are however not at all uncommon in high mountain lakes, where they have been revealed to have accounted for up to 63% of total bacterioplankton biomass [e.g.,9,10]. The high dominance of Gram positive, rod-shaped bacteria observed here could be as a direct result of various extreme conditions that includes intensive humidity, high radiation and temperature extremes common to these high altitude locations especially in the summer [22], which ultimately ensure the selection and survival of hardy bacterial cells in the studied lakes. Therefore, it is probably accurate to conclude that the prevalence of Gram positive bacteria in these lakes during the study period is mainly due to their resistance to these extreme conditions of temperature and high exposure to UV light. Additionally, differences in the morphometric and watershed characteristics among the lakes could also have influenced the autochthonous bacterial composition. Particularly, Lake Ifreh with the lowest numbers of Gram positive bacteria also slightly differed in the RFLP profile results as compared to the other three lakes that seemed to share relatively more similar basin shape as well as watershed characteristics.
The high PCR detection of the uidA gene coding for β-galacturonidase and ubiquitously associated with the presence of coliform bacteria populations (i.e., mainly Escherichia, Enterococci and Shigella species) in Lake Ifreh is probably indicative of allochthonous influxes of the cells directly through various domestic activities (human and animal) around this lake as compared to the other three sites. In addition, the high phytoplankton biomass that covered the lake during the summer months could also be harboring the fecal bacterial populations from where they continuously regenerated and then released into the water body [e.g.,16,25,26]. Overall, the clear differences and uniqueness of the bacterioplanton assemblages that existed in Lake Ifreh were further revealed and validated in both the bacterial isolate composition (indicating mostly Gram negatives) and the RFLP patterns that also differed from the other three lakes examined. This further validated the need to employ combinations of methodological approaches, including taxonomic, physiological and molecular methods in order to adequately delineate and understand the ecology of microbial assemblages indigenous to extreme freshwater environments.
Acknowledgements
The study was supported mostly by the School of Science and Engineering, Al Akhawayn University (AUI), Morocco and the Middle Eastern North African (MENA) exchange program grant to OAO through the Great Lakes College Alliance (GLCA), Ann Arbor, Michigan, USA.
REFERENCES
- A. Alfreider, J. Pernthaler, R. Amann, B. Sattler, F.-O. Glockner, A. Willie and R. Psenner, “Community Analysis of the Bacterial Assemblages in the Winter Cover and Pelagic Layers of a High Mountain Lake by in Situ Hybridization,” Applied and Environmental Microbiology, Vol. 62, No. 6, 1996, pp. 2138-2144.
- M. Bahr, J. E. Hobbie and M. L. Sognin, “Bacterial Diversity in an Arctic Lake: A Freshwater SAR11 Cluster,” Aquatic Microbial Ecology, Vol. 11, No. 3, 1996, pp. 271- 277. http://dx.doi.org/10.3354/ame011271
- A. K. Bej, R. J. Steffan, J. DiCesare, L. Haff and R. M. Atlas, “Detection of Coliform Bacteria in Water by Polymerase Chain Reaction and Gene Probes,” Applied and Environmental Microbiology, Vol. 56, No. 2, 1990, pp. 307-314.
- T. A. Cebula, W. L. Payne and P. Feng, “Simultaneous Identification of Strains of Escherichia coli Serotype O157:H7 and Their Shiga-Like Toxin Type by Mismatch Amplification Mutation Assay-Multiplex PCR,” Journal of Clinical Microbiology, Vol. 33, No. 1, 1995, pp. 248-250.
- B. B. Cho and F. Azam, “Major Role of Bacteria in Biogeochemical Fluxes in the Ocean’s Interior,” Nature, Vol. 332, 1988, pp. 441-443. http://dx.doi.org/10.1038/332441a0
- M. T. Cottrell, L. A. Waidner, L. Yu and D. L. Kirchman, “Bacterial Diversity of Metagenomic and PCR Libraries from the Delaware River,” Environmental Microbiology, Vol. 7, No. 12, 2005, pp. 1883-1895. http://dx.doi.org/10.1111/j.1462-2920.2005.00762.x
- M. Felip, R. Sattler, R. Psenner and J. Cataln, “Highly Active Microbial Communities in the Ice and Snow Cover of High Mountain Lakes,” Applied and Environmental Microbiology, Vol. 61, 1995, pp. 2394-2401.
- X. Gao, O. A. Olapade, M. W. Kershner and L. G. Leff, “Algal-Bacterial Co-Variation in Freshwater Streams: A Cross-Stream Comparison,” Archiv für Hydrobiologie, Vol. 159, No. 2, 2004. pp. 253-261. http://dx.doi.org/10.1127/0003-9136/2004/0159-0253
- F.-O. Glockner, E. Zaichikovg, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler and R. Amann, “Comparative 16S rRNA Analysis of Lake Bacterioplankton Reveals Globally Distributed Phylogenetic Clusters Including an Abundant Group of Actinobacteria,” Applied and Environmental Microbiology, Vol. 66, No. 11, 2000, pp. 5053- 5065. http://dx.doi.org/10.1128/AEM.66.11.5053-5065.2000
- E. A. Gontang, W. Fenical and P. R. Jensen, “Phylogenetic Diversity of Gram-Positive Bacteria Cultured from Marine Sediments,” Applied and Environmental Microbiology, Vol. 73, No. 10, 2007, pp. 3272-3282. http://dx.doi.org/10.1128/AEM.02811-06
- W. D. Hiorns, B. A. Methe, S. A. Nierzwicki-Bauer and J. P. Zehr, “Bacterial Diversity in Adirondack Mountain Lakes as Revealed by 16S rRNA Gene Sequences,” Applied and Environmental Microbiology, Vol. 63, 1997, pp. 2957-2960.
- M. J. Leboffe and B. E. Pierce, “Microbiology Laboratory Theory & Application,” 2nd Edition, Morton Publishers, 2006, 630 pages.
- R. M. Maier, I. L. Pepper and C. P. Gerba, “Environmental Microbiology,” Academic Press, 2000, 585 pages.
- M. Malki, I. Marín, A. Essahale, R. Amils and M. Moumni, “Physico-Chemical Parameters and Bacterial Diversity in the Moroccan’s Sebou River,” Journal of Food, Agriculture & Environment, Vol. 6, No. 1, 2008, pp. 172- 176.
- N. C. Morgan, “An Ecological Survey of Standing Waters in North West Africa III: Site Descriptions for Morocco,” Biological Conservation, Vol. 24, No. 3, 1982, pp. 161- 182. http://dx.doi.org/10.1016/0006-3207(82)90055-6
- O. A. Olapade, M. M. Depas, E. T. Jensen and S. L. McLellan, “Microbial Communities on Cladophora Mats along the Coastlines of Lake Michigan,” Applied and Environmental Microbiology, Vol. 72, No. 3, 2006, pp. 1932- 1938. http://dx.doi.org/10.1128/AEM.72.3.1932-1938.2006
- O. A. Olapade and L. G. Leff, “Seasonal Response of Stream Biofilm Communities to Dissolved Organic Matter and Nutrient Enrichments,” Applied and Environmental Microbiology, Vol. 71, No. 5, 2005, pp. 2278-2287. http://dx.doi.org/10.1128/AEM.71.5.2278-2287.2005
- O. A. Olapade, “Molecular Analyses of the Diversity in Marine Bacterioplankton Assemblages along the Coastline of the Northeastern Gulf of Mexico,” Canadian Journal of Microbiology, Vol. 56, No. 10, 2010, pp. 853-863. http://dx.doi.org/10.1139/W10-069
- A. Oren, “Molecular Ecology of Extremely Halophilic Archaea and Bacteria,” FEMS Microbiology Ecology, Vol. 39, No. 1, 2003, pp. 1-7. http://dx.doi.org/10.1111/j.1574-6941.2002.tb00900.x
- P. J. H. Jackman, “Microbial Systematics Based on Electrophoretic Whole-Cell Protein Patterns,” Methods in Microbiology, No. 19, 1987, pp. 209-225.
- J. Pernthaler, F.-O. Glockner, S. Unterholzner, A. Alfreider, R. Psenner and R. Amann, “Seasonal Community and Population Dynamics of Pelagic Bacteria and Archaea in a High Mountain Lake,” Applied and Environmental Microbiology, Vol. 64, 1998, pp. 4299-4306.
- M. Ramdani, N. Elkhiati and R. J. Flower, “Lakes of Africa: North of Sahara. Reference Module in Earth Systems and Environmental Sciences,” Encyclopedia of Island Waters, 2009, pp. 544-554.
- M. Simon, B. C. Cho and F. Azam, “Significance of Bacterial Biomass in Lakes and the Ocean: Comparison to Phytoplankton Biomass and Biogeochemical Implications,” Marine Ecology Progress Series, Vol. 86, 1992, pp. 103- 110. http://dx.doi.org/10.3354/meps086103
- W. G. Weisburg, S. M. Barns, D. A. Pelletier and D. J. Lane, “16S Ribosomal DNA Amplification for Phylogenetic Study,” Journal of Bacteriology, Vol. 173, No. 2, 1991, pp. 697-703.
- R. L. Whitman, D. A. Shivey, H. Pawlik, M. B. Nevers and M. N. Byappanahalli, “Occurrence of Escherichia coli and Enterococci in Cladophora (Chryophyta) in Neashore Water and Beach Sand of Lake Michigan,” Applied and Environmental Microbiology, Vol. 69, No. 8, 2003, pp. 4714-4719. http://dx.doi.org/10.1128/AEM.69.8.4714-4719.2003
- R. L. Whitman, S. E. Byers, D. A. Shivey, D. M. Ferguson and M. Byappanahalli, “Occurrence and Growth Characteristics of Escherichia coli and Enterococci within the Accumulated Fluid of the Northern Pitcher Plant (Sarracenia purpurea L.),” Canadian Journal of Microbiology, Vol. 51, No. 12, 2005, pp. 1027-1037. http://dx.doi.org/10.1139/w05-091
- W. Zwister, N. Selje and M. Simon, “Seasonal Patterns of the Bacterioplankton Community Composition in a Large Mesotrophic Lake,” Aquatic Microbial Ecology, Vol. 31, No. 3, 2003, pp. 211-225. http://dx.doi.org/10.3354/ame031211
NOTES
*Corresponding author.


