Journal of Biomedical Science and Engineering
Vol. 5 No. 4 (2012) , Article ID: 18508 , 9 pages DOI:10.4236/jbise.2012.54020
A new hybrid particle swarm optimization for mutlimodal brain image registration
![]()
Department of Electrical Engineering, Iran University of Science and Technology, Tehran, Iran
Email: fateme.ayat@gmail.com, bshokouhi@iust.ac.ir, ayatollahi@iust.ac.ir
Received 2 September 2011; revised 7 November 2011; accepted 31 January 2012
Keywords: Image Registration; Mutual Information; Particle Swarm Optimization; Multimodal Images
ABSTRACT
Image registration is an important issue in medical analysis. In this process the spatial transformation that aligns the reference image and the floating image is estimated by optimizing a similarity metric. Mutual information (MI), a popular similarity metric, is a reliable criterion for medical image registration. In this paper, we present an improved method for multimodal image registration based on maximization of a new form of normalized MI incorporating particle swarm optimization, PSO, as a searching strategy. Also a new hybrid PSO algorithm is applied to approach more precise and robust results with better performance.
1. INTRODUCTION
Medical image registration plays an increasingly important role in many clinical applications, including the detection and diagnosis of diseases, planning of therapy, guidance of interventions, and the follow-up and monitoring of patients [1]. Image registration is the process of overlaying two or more images taken from same scene at different times, from different viewpoints or by the variety of sensors. It geometrically aligns the images by finding a proper transformation which maps any point of one image to corresponding point of another image.
Medical images which achieved by different sensors (modalities), basically can be grouped in two categories: first, anatomical images such as computed tomography (CT), magnetic resonance (MR) and ultrasound (US) that show body organs in their total structure; second functional images such as positron emission tomography (PET) and single photon emission computed tomography (SPECT) that show soft tissues and their internal activities. The aim of multimodal medical image registration is combining data of different modalities to obtain more complete and detailed information about the patient.
The first step in registering two images is selecting some common properties of images and then matching them [2]. Image registration techniques can be generally classified into two categories: feature-based and intensity-based methods. Feature-based methods require the extraction of features (points, edges, surfaces, shapes) in both images and finding the correspondence between the features. In the earlier literature the features could be markers placed on the human body [3,4] or distinctive anatomical points and other structures visible in the images being registered [5,6]. Manual extraction of features is very time consuming and also depends on the skill of the operator. To overcome these difficulties accurate automatic feature detection is performed by image segmentation. In all feature-based methods, the accuracy of registration is depends on the accuracy of feature detection [7]. On the other hand, intensity-based methods operate directly on the image intensity values (color or grey level), without prior data reduction by the user or segmentation. So their accuracy is not affected by segmentation errors. The intensity-based approaches generally optimize a similarity measure function of the images being registered. This similarity metric can be based on intensity difference, cross correlation and mutual information [7,8]. By adjusting the parameters of an appropriate spatial transformation model, different values of similarity metric are obtained and its maximum value is related to proper values of the transformation parameters. In Registration process, optimization is a searching strategy to find the best transformation parameters. The robustness, accuracy and efficiency of intensity-based Registration method mainly depend on the similarity measure and optimization algorithm.
Over the last few years, mutual information (MI) has become one of the most popular and widely studied similarity criterions for intensity-based registration [9]. MI was first introduced as a measure for medical images by two independent groups: Collignon and Maes [10,11] and Viola and Well [12,13]. Unlike the measure based on intensity difference and cross correlation , MI does not assume a linear intensity relationship between the images under evaluation. Therefore it is suitable highly nonmonotonic function with many local maxima that make it difficult to register images to have a smoother curve with fewer fluctuations, a new form of normalized mutual information (NMI) is proposed [14].
For the optimization of similarity measure, local methods or global methods can be used. Local methods such as steepest descent gradient, Powell’s direction set, conjugate gradient, Levenberg-Marquardt [15] usually trap in local optimum and obtain mis-registration results, so good initial values are necessary. Examples for global optimization method are simulated annealing (SA) [16], genetic algorithm (GA) [17] and particle swarm optimization (PSO) [18]. Though GA is a powerful method for global optimization, it takes a longer computational time and lacks the fine tuning capability. Instead, PSO is a more effective and extremely simple algorithm in comparison with GA and other global optimization algorithms. PSO is a stochastic population based evolutionary computer algorithm [19].
On the other hand, the conventional GA and PSO cannot find the global optimum well. So a new approach named hybrid particle swarm optimization, HPSO, have been proposed which incorporates two concepts (subpopulation and crossover) of GA into the conventional PSO [20]. We present a new hybrid PSO that is more accurate and less time consuming.
In this paper we present brain image registration with affine transformation by maximization a modified logarithmic NMI, MNMI, by using proposed HPSO as an optimization algorithm. In Section 2 registration method and in Section 3 PSO and HPSO will be explained. The experimental results and the conclusion will be presented in Sections 4 and 5 respectively.
2. IMAGE REGISTRATION METHOD
The required steps in image registration is shown in Figure 1. In this process the floating image should be matched to the reference image by applying the T transformation which maps the coordinates of the floating image to reference image. Regarding Eq.1 the best transformation is that gives the maximum similarity metric, MNMI. For this purpose the optimization algorithm searches the parameters of the transformation in the given space to find the best values for the maximum similarity metric.
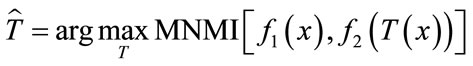 (1)
(1)
In this equation 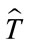 is the best transformation, x is the coordinates of the image and T shows the transformation and its parameters for simplicity.
is the best transformation, x is the coordinates of the image and T shows the transformation and its parameters for simplicity.
2.1. Transformation Model
The image registration algorithm can be classified into two categories of rigid and non-rigid registration. Rigid transformation involves the translation and rotation parameters, whereas non-rigid contains these parameter as well as any other changes.
The affine transformation is a non-rigid transformation which maps straight lines to straight lines and preserves the parallelism between lines. It estimates rotation, scaling, shear and translation parameter that can be shown as R, S, H and T matrices respectively as below
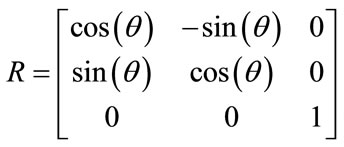 (2)
(2)
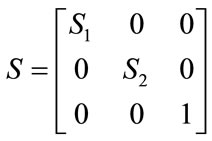 (3)
(3)
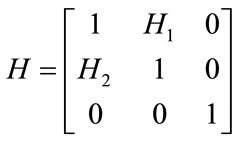 (4)
(4)
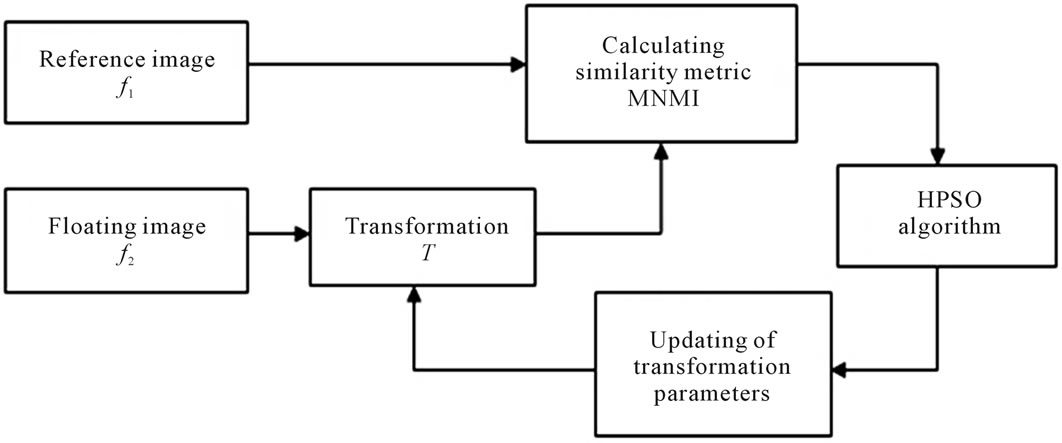
Figure 1. Block diagram of image registration method.
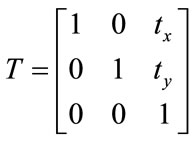 . (5)
. (5)
The affine transformation matrix, A, is
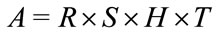 . (6)
. (6)
The two dimensional affine transformation which is used in this paper contains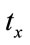 ,
, 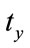 ,
, 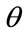 and s that are representing translations along x and y axes, rotation and scaling. Eq.7 shows the mapping of image coordinates based on these parameters.
and s that are representing translations along x and y axes, rotation and scaling. Eq.7 shows the mapping of image coordinates based on these parameters.
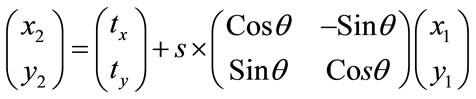 (7)
(7)
2.2. Mutual Information as a Similarity Metric
Mutual information is a reliable and most used method based on the gray levels to measure the similarity metric between two images. It is a concept of information theory that measures the statistic correlations between two data, which is based on the Shannon entropy [9].
Shannon entropy weights the information per outcome by the probability of that outcome occurring. The Shannon entropy can also be computed for an image, in which case we focus on the distribution of the gray values of the image. If each pixel in an image be viewed as random events, the information an image contains can be measured by Shannon entropy. Shannon’s entropy can be viewed as a measure of uncertainty or how much information an image contains.
For an image the probability of pixels with gray level x is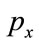 , the Shannon entropy of an image can be defined as
, the Shannon entropy of an image can be defined as
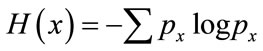 . (8)
. (8)
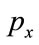 implies occurring probability of gray level x. The Shannon entropy is also a measure of dispersion of a probability distribution.
implies occurring probability of gray level x. The Shannon entropy is also a measure of dispersion of a probability distribution.
In the calculation of MI the joint entropy is used. It is shown as H(A,B) in Eq.9.
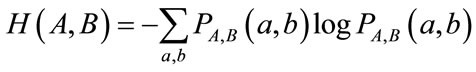 (9)
(9)
In the above equation 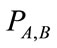 is the joint probability distribution function of the pixels values a and b in the images A and B. The joint probability distribution of the two images is estimated by calculating a joint histogram of the gray values. It is a two dimensional plot showing the combinations of gray values in each of the two images for all corresponding points. The joint probability distribution of the gray values of the images is achieved by dividing each entry in the histogram by the total number of entries. The mutual information of the two images A and B is defined as following equation.
is the joint probability distribution function of the pixels values a and b in the images A and B. The joint probability distribution of the two images is estimated by calculating a joint histogram of the gray values. It is a two dimensional plot showing the combinations of gray values in each of the two images for all corresponding points. The joint probability distribution of the gray values of the images is achieved by dividing each entry in the histogram by the total number of entries. The mutual information of the two images A and B is defined as following equation.
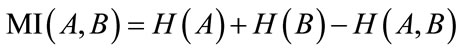 (10)
(10)
In this equation H(A) and H(B) are entropies of the images A and B which are obtained from probability distribution function, Eq.11 and Eq.12.
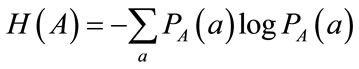 (11)
(11)
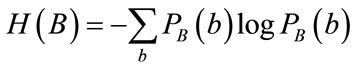 (12)
(12)
In the complete image registration the joint entropy has the lowest value and the MI becomes maximum [9].
Although MI is a good metric, but it is sensitive to overlap regions of the images, so that by decreasing these regions, the samples will be decreased which lessen the power of statistical probability function estimation. Also the MI can be increased with more dismatching of the images. The normalized mutual information, NMI, metric has been proposed to overcome this problem. It has less sensitivity to overlap changes [21]. NMI is as Eq.13 .
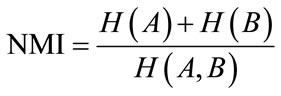 (13)
(13)
By every change in the parameters values in each step, a new transformation is applied to the floating image. So its entropy is changed. As the result the MI measure is not a uniform function and has many fluctuations. To have a smoother curve a logarithmic normalized mutual information, LMNI, has been used [14] as below.
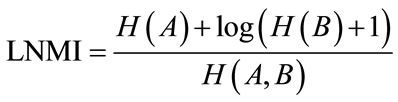 (14)
(14)
In this paper we propose a modified normalized mutual information, MNMI, which is more efficient and has smoother curve than LNMI, and is as following expression.
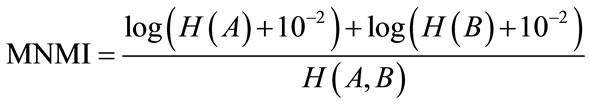 (15)
(15)
As we see in the LNMI equation, the entropy of the floating image H(B) is less effective than entropy of the reference image H(A), so it has less role in estimation of the transformation parameter. However as the Eq.15 shows the effects of both images are similar in MNMI.
3. HYBRID PARTICLE SWARM OPTIMIZATION
3.1. Particle Swarm Optimization
PSO has been used as a searching strategy for finding the transformations parameters. It is a populated searching method based on the stochastic technique that is inspired by social behavior of bird flocking and fish schooling [19].
In PSO, a population of individuals is evolved by cooperation and competition among the individuals themselves through iterations. Each individual, named particle, of the population, called swarm, represents a potential solution to a problem. Each particle changes its position in search space and updates its velocity according to its own movement experience and neighbors’ movement experience, aiming at a better position for itself. All of particles have fitness values which are evaluated by the fitness function to be optimized.
PSO is initialized with a number of random particles as a group. The ith particle of the group is defined by a velocity vector 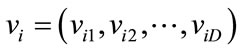 and a position vector
and a position vector 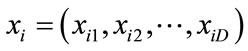 in a D dimensional space. In each iteration, the best position that gives the most fitness for each particle, pbest, and for all of the particles, gbest, are achieved. According to these values, particles update their position and velocity by the Eq.16 and Eq.17.
in a D dimensional space. In each iteration, the best position that gives the most fitness for each particle, pbest, and for all of the particles, gbest, are achieved. According to these values, particles update their position and velocity by the Eq.16 and Eq.17.
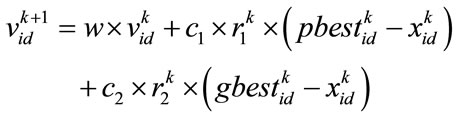 (16)
(16)
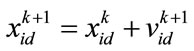 (17)
(17)
In these equations k is the iteration number, d = 1, 2, ···,D, i = 1, 2, ···, N and N is the size of the population. 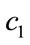 and
and 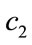 are acceleration coefficient and usually have constant value of 2.
are acceleration coefficient and usually have constant value of 2. 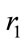 and
and 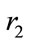 are random number between 0 and 1. w is inertia coefficient which will vary according to the Eq.18.
are random number between 0 and 1. w is inertia coefficient which will vary according to the Eq.18.
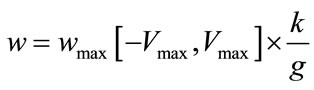 (18)
(18)
 ,
, 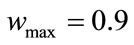 and g is the maximum number of iteration. The velocity of particles should be in the
and g is the maximum number of iteration. The velocity of particles should be in the 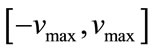 range to be ensured that particle does not exit from allowed searching space. The process will be stopped when it reaches to a predetermined number of iterations or a minimum error.
range to be ensured that particle does not exit from allowed searching space. The process will be stopped when it reaches to a predetermined number of iterations or a minimum error.
PSO is done in following steps:
1) PSO is initialized with N number of random particles in searching space.
2) The fitness function is calculated for each particle in initial population. And the pbest and gbest is determined.
3) The velocity and position vectors of particles is updated according to Eq.16 and Eq.17 .
4) The fitness function is evaluated again.
5) If 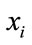 is better than
is better than 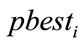 then
then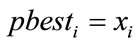 .
.
6) If 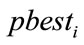 is better than
is better than 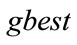 then
then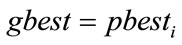 .
.
7) If the stopping condition is satisfied the algorithm will be terminated, else repeat from step 3.
3.2. Hybrid Particle Swarm Optimization
In this paper, we propose a new hybrid particle swarm optimization (HPSO), which incorporates two concepts of genetic algorithms, subpopulation and crossover, into the PSO. In this algorithm, the particles will be grouped in the M number of subpopulations, Each of them has its own global best particle,  , for m = 1, ···, M.
, for m = 1, ···, M.
Here, the best four subpopulations with most  is determined and the particle related to the
is determined and the particle related to the  in each of these four subpopulations will be a candidate to be a parent for crossover. These candidates will be ranked 1 to 4 according to the
in each of these four subpopulations will be a candidate to be a parent for crossover. These candidates will be ranked 1 to 4 according to the  values, in which 1 is related to highest value. The four parents then will be chosen among the candidates with the probability
values, in which 1 is related to highest value. The four parents then will be chosen among the candidates with the probability 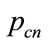 allocated to each candidate as Eq.19 .
allocated to each candidate as Eq.19 .
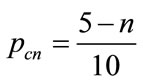 (19)
(19)
where n is the ranking number.
Each pair of parents 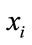 and
and 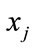 (in the case of
(in the case of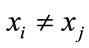 ) generate two children
) generate two children 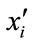 and
and 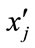 by arithmetic crossover shown as below.
by arithmetic crossover shown as below.
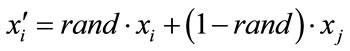 (20)
(20)
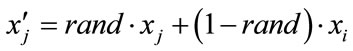 (21)
(21)
where rand is a uniformly distributed random number among 0 to 1. The velocities are given by
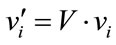 (22)
(22)
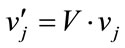 (23)
(23)
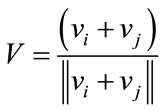 . (24)
. (24)
The children then will be replaced the worth particles of their parents’ subpopulations. If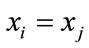 , a candidate will be randomly chosen with equal probability and will substitute one of the parents.
, a candidate will be randomly chosen with equal probability and will substitute one of the parents.
The above procedure of HPSO is added after every evaluating of fitness function in conventional PSO algorithm.
4. EXPERIMENTAL RESULTS
In this section, we used several experiments to show the better performance of MNMI metric than LNMI and NMI metrics, and the advantage of using proposed HPSO algorithm over GA, conventional PSO and HPSO proposed by Chen [22].
4.1. Comparing MNMI with LNMI and NMI
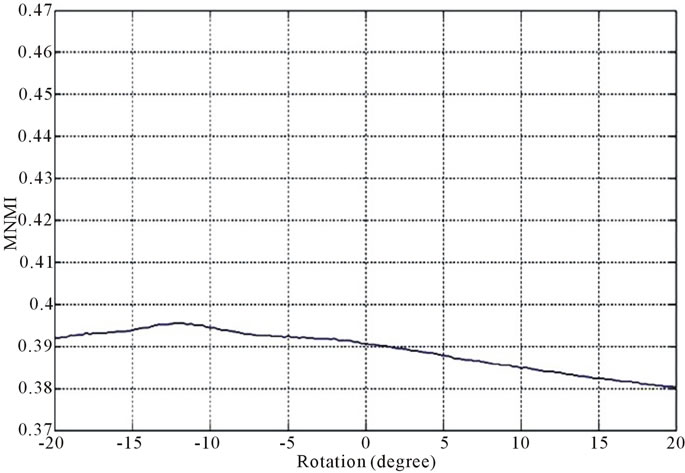
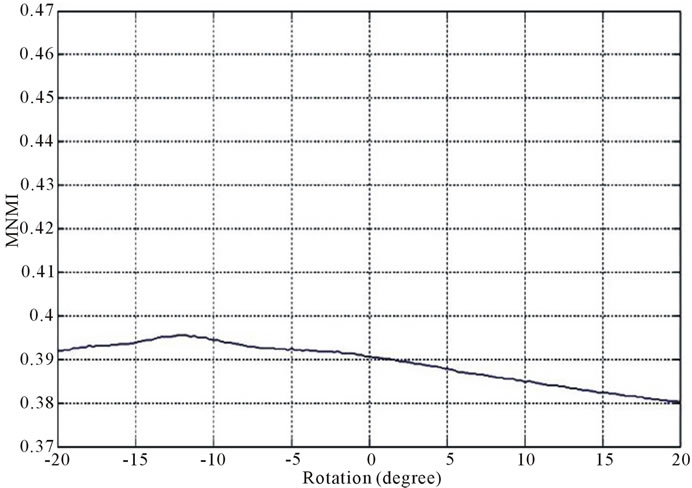
In this experiment, we have used a pair of MR, CT images as data set. The MR image is the reference image, and CT image which is rotated 12˚, is considered as floating image. The registration of these two images is performed with three similarity metrics MNMI, LNMI and NMI individually by a comprehensive search (without optimizing algorithm) in the range of [−20˚, 20˚].
Figure 2 and Figure 3 show the similarity metrics
 (a)
(a)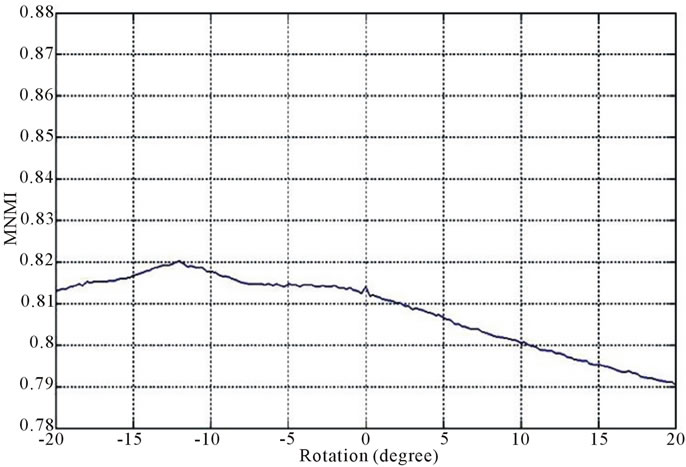 (b)
(b)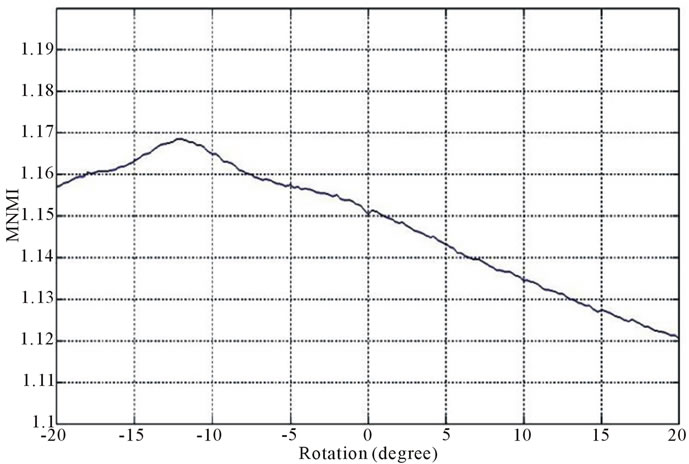 (c)
(c)
Figure 2. Diagrams of similarity metrics, (a) MNMI; (b) LMNI, (c) NMI for different rotation in the range of [−20˚, 20˚].
values versus rotation. The maximum value of the similarity metrics occurs at −12˚. As these figures demonstrate, our MNMI measure has very smoother curve with less fluctuations. It causes that optimization algorithms do not trap in local optima.
4.2. HPSO as Optimization Algorithm
Here, the results of performing HPSO for image registration based on maximization of MNMI is presented . The algorithm has been applied to a pair of CT images of the brain as monomodal; and a pair of “MR-T2, MR-PD” and “MR-T2, CT” as multimodal.
In the experiment, first image is the reference image, and by applying the translations of 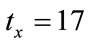 and
and 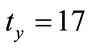 pixels, the rotation of
pixels, the rotation of 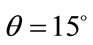 and the scaling of
and the scaling of 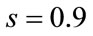 to the second image, the floating image is obtained. The used HPSO algorithm has the population of 40 particles, the number of 8 subpopulations and 40 iterations. The desired parameters for proper registration is achieving the minus value of
to the second image, the floating image is obtained. The used HPSO algorithm has the population of 40 particles, the number of 8 subpopulations and 40 iterations. The desired parameters for proper registration is achieving the minus value of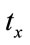 ,
, 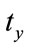 ,
, 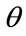 and
and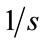 .
.
To evaluate the performance of our algorithm, it has been compared to a GA and a PSO with similar population and iterations; and also the HPSO introduced in [22] with only two best subpopulations for crossover.
The proposed algorithm is implemented in MATLAB and evaluated using real patient brain images from the Whole Brain Atlas, WBA, [23].
Figure 4, Figure 5 and Figure 6 show the results of performing our HPSO algorithm for “CT, CT”, “MR-T2, MR-PD” and “MR-T2, CT” image registration, respectively. Table 1, Table 2 and Table 3 are related to above figures and present the average and standard deviation (STD) of 10 times running for each of mentioned algorithms.
 (a)
(a)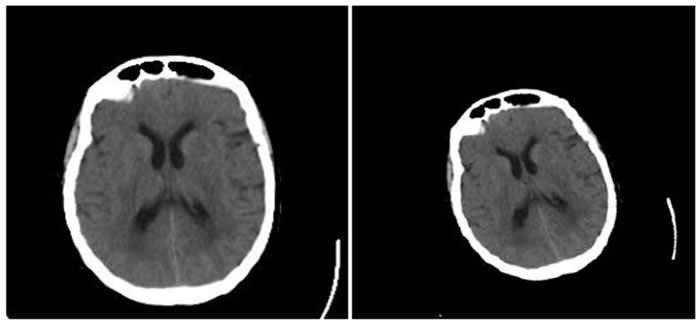 (b)
(b)
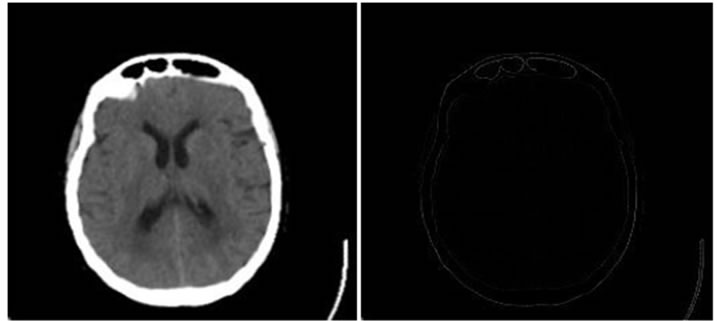
Figure 4. Results of performing HPSO algorithm for CT, CT image registration, (a) Reference image; (b) floating image; (c) registered image; (d) difference image.
 (a)
(a)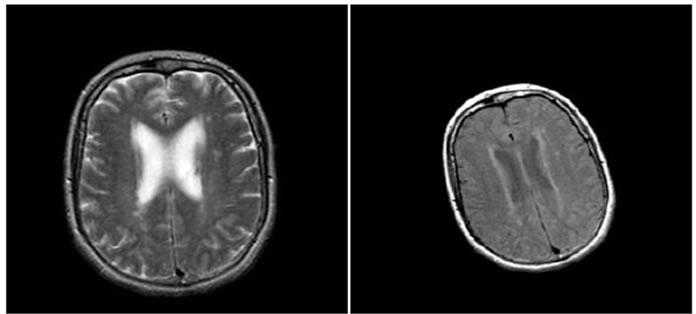 (b)
(b)
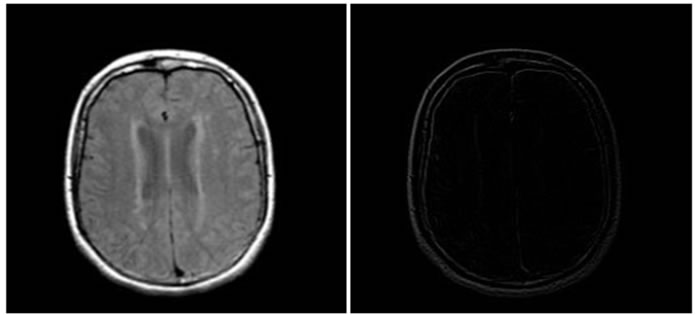
Figure 5. Results of performing HPSO algorithm for MR-T2, MR-PD image registration, (a) Reference image; (b) floating image; (c) registered image; (d) difference image.
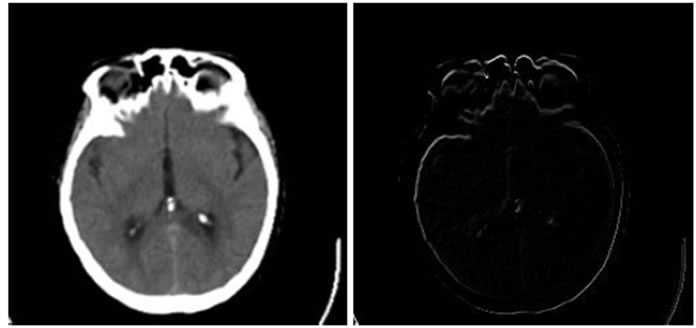 (a)
(a)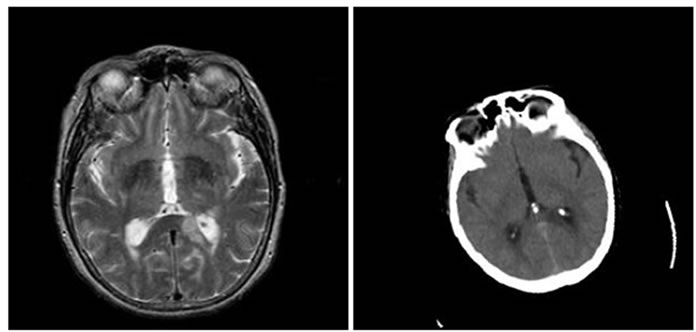 (b)
(b)
Figure 6. Results of performing HPSO algorithm for MR-T2, CT image registration, (a) Reference image; (b) floating image; (c) registered image; (d) difference image.

Table 1. Average results of 10 times running of optimization algorithms for MR-T2, CT image registration.
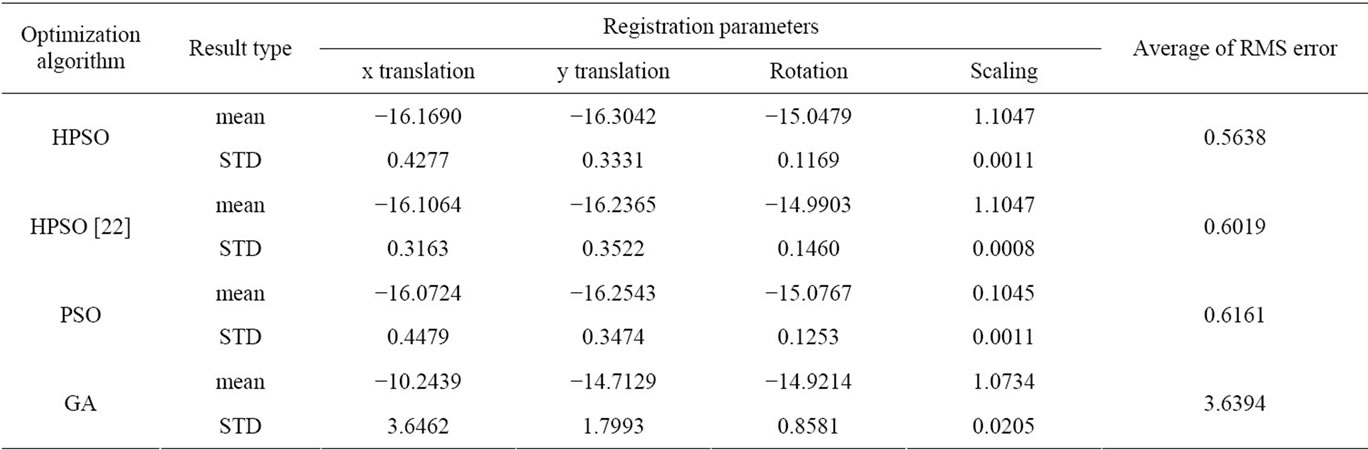
Table 2. Average results of 10 times running of optimization algorithms for MR-T2, MR-PD image registration.
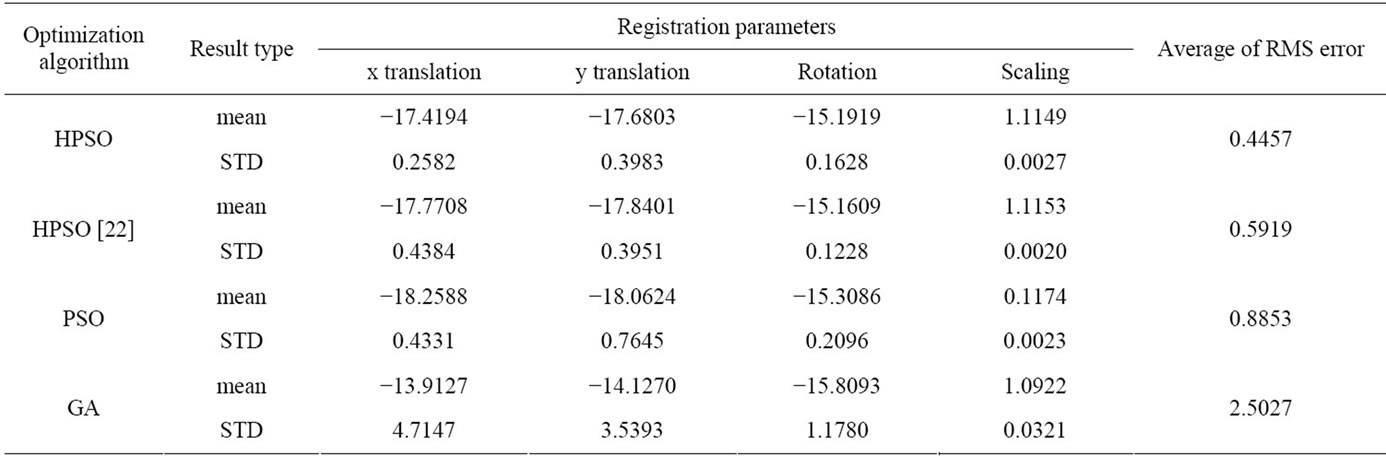
Table 3. Average results of 10 times running of optimization algorithms for MR-T2, CT image registration.
According to Table 1, although all algorithms have good performance in above monomodal image registration, but the results of our algorithm show less dispersion i.e. it is more accurate.
The result values in Table 2 show that the average of RMS error of our algorithm is less than the others.
In the last experiment, as it is obvious in Table 3, all parameters of averaging, STD and RMS error of proposed algorithm are better than the other algorithms.
Figure 7 demonstrates the MNMI value in the end of each iteration for the algorithms. This proves that our HPSO approach the final result in less iteration and consequently takes less time.
At the end, we can conclude that our proposed algorithm has better performance in nearly all aspects especially in multimodal image registration.
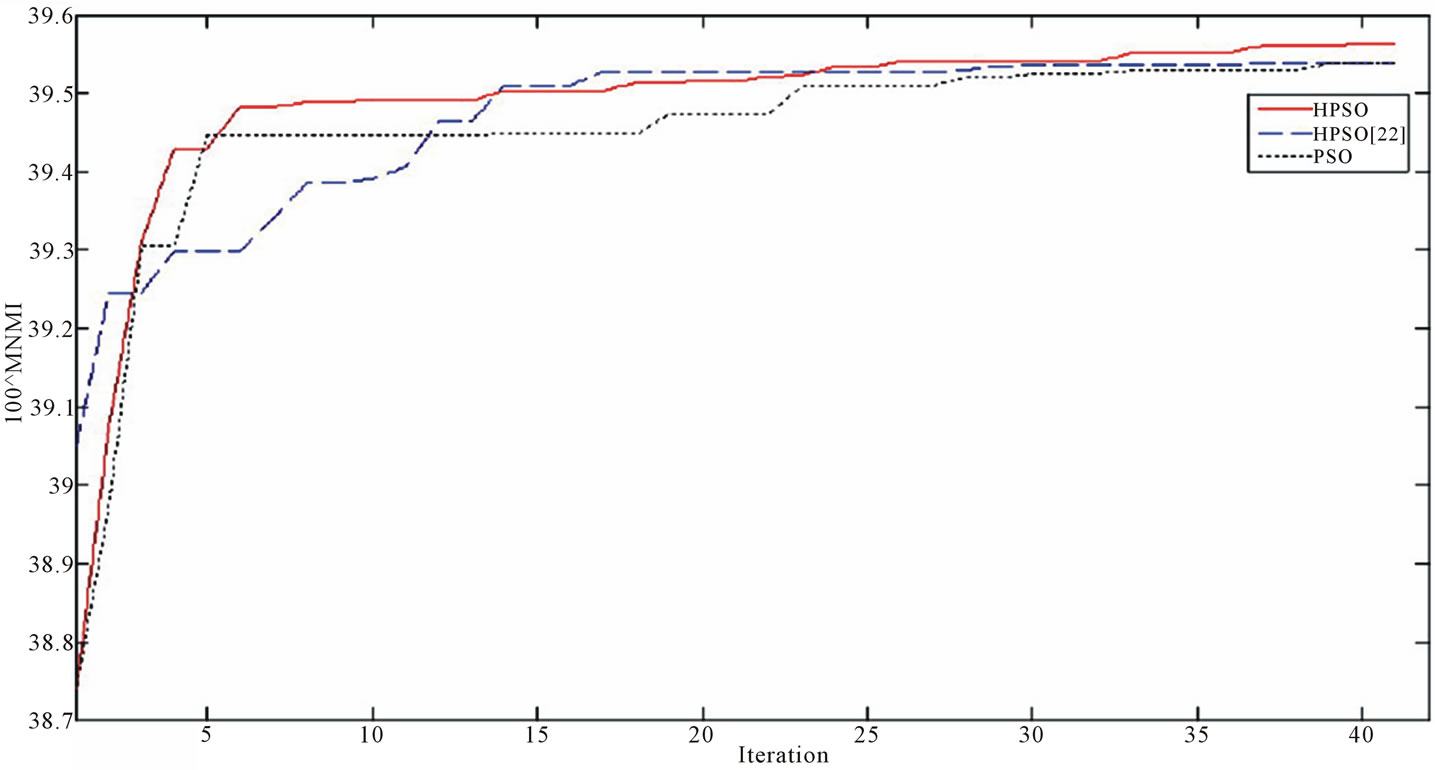
Figure 7. the MNMI value in the end of each iteration for the optimization algorithms.
5. CONCLUSION
In this paper we proposed a modified normalized mutual information, MNMI, as a similarity metric for image registration with a smoother curve which prevents trapping of optimization algorithm in local optima. In addition, a new hybrid PSO which incorporates subpopulation and crossover of GA into the conventional PSO was proposed. As the results of experiments, this optimization algorithm has more accurate and less time consuming performance and so it is very suitable for image registration especially in multimodal cases.
The drawback of this method is that it is not very accurate in the presence of large shear distortion between images.
![]()
![]()
REFERENCES
- Rueckert, D. and Aljabar, P. (2010) Nonrigid registration of medical images: Theory, methods, and applications. IEEE Signal Processing Magazine, 27, 113-119. doi:10.1109/MSP.2010.936850
- Brown, L.G. (1992) A survey of image registration techniques. ACM Computing Surveys (CSUR), 24, 326-376. doi:10.1145/146370.146374
- Wang, M.Y., Fitzpatrick, J.M. and Maurer, C.R. (1995) Design of fiducials for accurate registration of CT and MR volume images. In: Loew, Ed., Medical Imaging, 96- 108.
- Fuchs, M., Wischmann, H., Neumann, A., Weese, J., Zylka, W., Sabczynski, J., Kuhn, M.H., Buzug, T.M., Schmitz, G., and Gieles, P.M.C. (1996) Accuracy analysis for image-guided neurosurgery using fiducial skin markers, 3D CT imaging, and an optical localizer system. In: Lemke, H.U., Vannier, M.W., Inamura, K. and Farman, A.G., Eds., Computer Assisted Radiology, Elsevier, Amsterdam, 770-775.
- Rubinstein, R., Karger, H., Pietrzyk, U., Siegal, T., Gomori, J.M. and Chisin, R. (1996) Use of 201 thallium brain SPECT, image registration, and semi-quantitative analysis in the follow-up of brain tumors. European Journal of Radiology, 21, 188-195. doi:10.1016/0720-048X(95)00726-7
- Evans, A.C., Collins, D.L., Neelin, P. and Marrett, T.S. (1996) Correlative analysis of three-dimensional brain images. In: Taylor, R.H., Lavall’ee, S., Burdea, G.C. and M¨osges, R., Eds., Computer-Integrated Surgery, Technology and Clinical Applications, Chapter 6, MIT Press, Cambridge, 99-114.
- Maintz, J.B.A. and Viergever, M.A. (1998) A survey of medical image registration. Medical Image Analysis, 2, 1-37. doi:10.1016/S1361-8415(98)80001-7
- Gholipour, A., Kehtarnavaz, A., Briggs, R., Devous, M. and Gopinath, K. (2007) Brain functional localization: A survey of image registration techniques. IEEE Transactions on Medical Imaging, 26, 427-451. doi:10.1109/TMI.2007.892508
- Pluim, J.P.W., Maintz, J.B.A. and Viergever, M.A. (2003) Mutual information based registration of medical images: A survey. IEEE Transactions on Medical Imaging, 22, 986-1004. doi:10.1109/TMI.2003.815867
- Collignon, A., Maes, F., Delaere, D., Vandermeulen, D., Suetens, P. and Marchal, G. (1995) Automated multimodality medical image registration using information theory. Proceedings of the 14th International Conference of Information Processing and Medical Imaging, 263-274.
- Maes,F. Collignon, A., Vandermeulen, D., Marchal, G. and Suetens, P. (1997) Multimodality image registration by maximization of mutual information. IEEE Transactions on Medical Imaging, 16, 187-198. doi:10.1109/42.563664
- Viola, P. and Wells, W.M. III (1995) Alignment by maximization of mutual information. Proceedings of the 5th International Conference Computer Vision, 20-23 June 1995, 16-23.
- Wells, W.M. III, Viola, P., Atsumi, H., Nakajima, S. and Kikinis, R. (1996) Multi-modal volume registration by maximization of mutual information. Medical Image Analysis, 1, 35-51. doi:10.1016/S1361-8415(01)80004-9
- Nejad, A.G. and Ayatollahi, A. (2010) Genetic algorithm as the main optimizer for medical image registration. Proceedings of the 17th Iranian Conference on Biomedical Engineering (ICBME), 1-3.
- Maes, F., Vandermeulen, D. and Suetens, P. (1999) Comparative evaluation of multi-resolution optimization strategies for multimodality image registration by maximization of mutual information. Medical Image Analysis, 1, 373-386. doi:10.1016/S1361-8415(99)80030-9
- Matsopoulos, G.K., Mouravliansky, N.A., Delibasis, K.K. and Nikita, K.S. (1999) Automatic retinal image registration scheme using global optimization techniques. IEEE Transactions on Information Technology in Biomedicine, 3, 47-60. doi:10.1109/4233.748975
- Rouet, J.M., Jacq, J.J. and Roux, C. (2000) Genetic algorithms for a robust 3-D MR-CT registration. IEEE Transactions on Information Technology in Biomedicine, 4, 126-136. doi:10.1109/4233.845205
- Wachowiak, M.P., Smolikova, R., Zheng, Y.F., Zurada, J.M. and Elmaghraby, A.S. (2004) An approach to multimodal biomedical image registration utilizing particle swarm optimization. IEEE Transactions on Evolutionary Computation, 3, 289-301. doi:10.1109/TEVC.2004.826068
- Eberhart, R. and Kennedy, J. (1995) A new optimizer using particles swarm theory. Proceedings of the 6th International Symposium on Micro Machine and Human Science, 4-6 October 1995, 39-43. doi:10.1109/MHS.1995.494215
- Lovberg, M., Rasmussen, T.K. and Krink, T. (2001) Hybrid particles swarm optimizer with breeding and subpopulations. Proceedings of the 3rd Genetic Evolutionary Computation Conference, San Francisco, 469-476.
- Studholme, C., Hill, D.L.G. and Hawkes, D.J. (1999) An overlapinv ariant entropy measure of 3D medical image alignment. Pattern Recognition, 32, 71-86. doi:10.1016/S0031-3203(98)00091-0
- Chen, Y.W. and Mimori, A. (2009) Hybrid particle swarm optimization for medical image registration. Proceedings of the 5th International Conference on Natural Computation (ICNC), Tianjin, 14-16 August 2009, 26-30.
- Johnson, K. and Becker, J. (2008) The whole brain atlas. http://www.med.harvard.edu/AANLIB/home.html


 (a)
(a)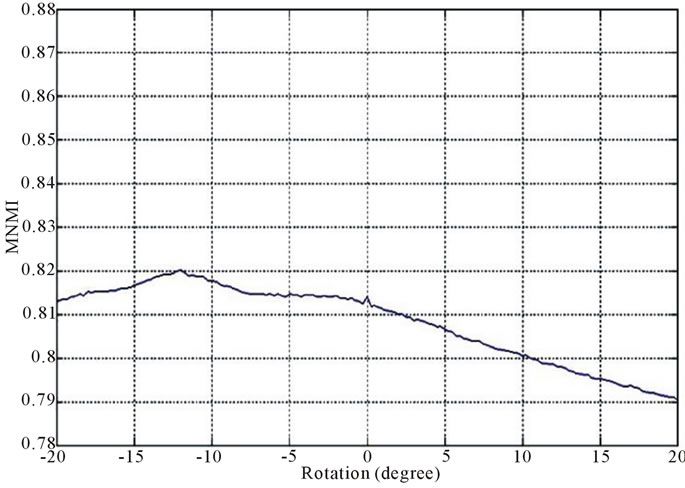 (b)
(b)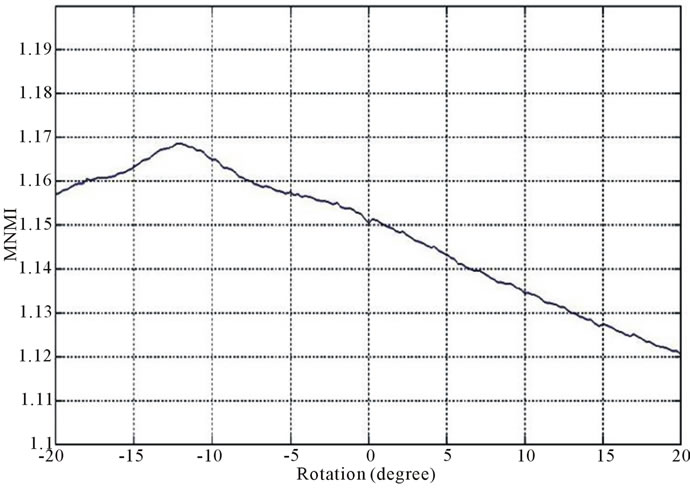 (c)
(c)