Journal of Cancer Therapy
Vol. 4 No. 5 (2013) , Article ID: 33768 , 6 pages DOI:10.4236/jct.2013.45104
REGγ Mediated Regulation of p21Waf/Cip1, p16INK4a and p14ARF/p19ARF in Vivo
![]()
1The Shanghai Key Laboratory of Regulatory Biology, Institute of Biomedical Sciences, East China Normal University, Shanghai, China; 2Department of Orthopaedic Oncology, Changzheng Hospital, The Second Military Medical University, Shanghai, China; 3Department of Pathology, The Second Chengdu Municipal Hospital, Chengdu, China; 4Department of Molecular Molecular and Cellular Biology, Dan L. Duncan Cancer Center, Baylor College of Medicine, Houston, USA.
Email: #jianruxiao83@163.com, #dangyongyan@gmail.com, #xiaotaol@bcm.edu
Copyright © 2013 Lei Li et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received May 2nd, 2013; revised June 1st, 2013; accepted June 10th, 2013
Keywords: REGγ; p21Waf/Cip1; p16INK4a; p14ARF; p19ARF; Cancer
ABSTRACT
p21Waf/Cip1, p16INK4a and p14ARF (p19ARF in mice) have been demonstrated to be degraded by REGγ-proteasome pathway in an ATPand ubiquitin-independent manner in vitro. However, the in vivo roles of REGγ mediated-degradation of p21Waf/Cip1, p16INK4a and p14ARF remain unclear. In this study, we showed enhanced expression of p21Waf/Cip1, p16INK4a and p19ARF in multiple tissues from REGg–/– mice compared to REGg+/+ mice. Furthermore, we examined the expression of p21Waf/Cip1, p16INK4a and p14ARF in different cancer tissues and observed that the REGγ protein levels were highly expressed in different human cancers while the level of p21Waf/Cip1, p16INK4a and p14ARF appears to be inversely correlated. These results demonstrate that REGγ may exert its function in physiological and pathological conditions through degradation of p21Waf/Cip1, p16INK4a and p14ARF in vivo.
1. Introduction
The INK4a/ARF locus encodes two proteins, p16INK4a and p14ARF (equivalent to p19ARF in mice) [1,2]. Both proteins demonstrate tumor suppressor activity and play an important role in cell proliferation, cell progression and cell growth arrest. Mice lacking either p19ARF or p16INK4a are more prone to tumorigenesis compared to wild-type littermates [3,4]. P16INK4a is a well characterized tumor suppressor and was shown to inhibit the kinase activities of the cyclin D-dependent kinases CDK4 and CDK6 [5,6]. P16INK4a inactivation has been implicated in the deregulation of the cell cycle in the malignant melanoma [7,8].
P14ARF and p19ARF proteins are present in normal cells at low levels but accumulate in response to oncogene activation [9-12]. High p14ARF and 19ARF protein levels promote cell cycle arrest, senescence, or apoptosis by binding directly to the p53-degrading ubiquitin ligase Mdm2 and protecting p53 from Mdm2-mediated degradation [12-14]. The biological properties of p16INK4a, p14ARF and p19ARF have been studied extensively, but the regulation of these proteins remains unknown.
REGγ (also known as PA28g or PSME3) is a member of the 11S proteasomes which binds and activates the 20S proteasome to promote ATPand ubiquitin-independent protein degradation. Recent observations show that REGg could enhance proteasomal degradation of some proteins, such as steroid receptor coactivator-3, p21Waf/Cip1 and smurf1 [15-17] and REGg was also involved in p16INK4a and p19ARF degradation in vitro [16]. Furthermore, there is some evidence suggesting that REGg is involved in cancer progression and REGg has been reported to be overexpressed in colorectal cancer, thyroid cancer, liver cancer and lung cancer [18-22]. However, there is no evidence about the degradation of p21Waf/Cip1, p16INK4a, p14ARFand p19ARF by REGg in vivo.
In this study, we demonstrate that REGγ mediated the degradation of p21Waf/Cip1, p16INK4a and p14ARF/p19ARF in multiple mouse tissues and in human cancer tissues. In addition, the mRNA levels of p21Waf/Cip1, p16INK4a and p19ARF were not affected by REGg in mouse tissues. Our results provide the first evidence that REGγ plays an important role in the degradation of p21Waf/Cip1, p16INK4a and p14ARF/p19ARF in vivo.
2. Materials and Methods
2.1. Animals
The REGg–/– mice with C57BL/6 genetic background were acquired from Dr. John J. Monaco and bred in the Animal Core. To generate the mice required in this study, we kept REGg+/– mice intercrosses between males and females. Genotyping of REGg+/+ and REGg–/– mice was carried out by PCR analysis of genomic DNA as described.
The REGg+/+ and REG–/– mice were born at Mendelian frequency, grew normally and used in the subsequent experiments.
2.2. Antibody and Cell Culture
Antibodies were purchased from Invitrogen (REGg/PA 28g), BD Pharmingen (p21Cip/WAF1), Sigma (β-actin, p14), Santa Cruz (p16, p19). The HeLa stable cell lines were generated by retroviral shRNA vectors specific for REGg or a control vector from OriGene (Rockville, MD). These two cell lines were cultured in DMEM supplemented with 10% fetal bovine serum (Gibco).
2.3. RNA Analyses
Tissues were homogenized in 1 ml RNAisoTM Plus (TAKARA). Total RNA was extracted and 2 μg RNA was reversely transcribed into cDNA with M-MLV reverse transcriptase (Invitrogen) following the manufacturer’s instruction. The mouse gene-specific primers are as follows:
REGg sense primer, 5-TCCTCACCAATAGCCACG-3;
REGg antisense primer, 5-CTCGATCAGCAGCCGAAT-3;
p16 sense primer, 5-GCTGCAGACAGACTGGCCA-3;
p16 antisense primer, 5-GTCCTCGCAGTTCGAATCTG-3;
p19 sense primer, 5-CGCAGGTTCTTGGTCACTGT-3;
p19 antisense primer, 5-TGTTCACGAAAGCCAGAGCG-3;
p21 sense primer, 5-CCTGGTGATGTCCGACCTG-3;
p21 antisense primer, 5-CCATGAGCGCATCGCAATC-3.
2.4. Western Blot Analysis
Tissues were homogenized in liquid nitrogen and lysed using RIPA buffer (1.0 mM EDTA 150 mM 1% Triton X-100 50 mM Tris-HCl (pH 7.4) NaCl 0.1% SDS 1% sodium deoxyholate) for 30 min. Then the homogenized tissues were centrifuged at 12,000 g for 15 min and the supernatants were collected. Equivalent amount of protein from each sample was analyzed by using primary antibodies specific for p16INK4a, p21Waf/Cip1, p19ARF and REGg overnight at 4˚C. After incubation with fluorescent labeled secondary antibody, specific signals for proteins were visualized by LI-COR Odyssey Infrared Imaging System.
2.5. Immunohistochemistry (IHC) and H&E Staining
Tissues were fixed with 4% paraformaldehyde for 48 hours. Then the samples were dehydrated through a graded series of ethanol, embedded in paraffin and sectioned at 4 μm.
Immunohistochemistry (IHC) and H&E staining were performed as described [23]. Antibodies against p16INK4a, p21Waf/Cip1, p19ARF and REGg were used at 1:500, 1:600, 1:600, and 1:400 dilutions, respectively.
2.6. Data Collection and Statistical Analysis The statistical data was got by GraphPad Prism 5.0 software. The results were expressed as the mean ± s.d. Statistical analysis was performed using the two tailed, paired Student’s t test. A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. The p21Waf/Cip1, p16INK4a, and p19ARF Expression is Higher in REG(–/– Mouse Tissues
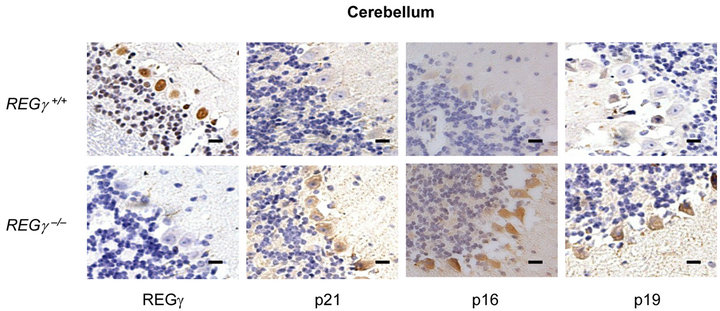
Numerous REGγ targets proteins such as p21Waf/Cip1, p16INK4a, and p14ARF/p19ARF (p19ARF in the mouse) of REGγ-proteasome, have been identified in vitro [16, 17]. However, the molecular details and in vivo biological significance of REGγ, p21Waf/Cip1, p16INK4a and p19ARF interplay remain elusive. We investigated cell specific expression patterns of REGγ, p21Waf/Cip1, p16INK4a and p19ARF by IHC analysis in cerebellum (Figure 1(a)), kidney (Figure 1(b)), stomach (Figure 1(c)) and liver (Figure 1(d)) from REGg+/+ and REGg–/– littermate mice. The staining of REGγ was negative in the tissues from REGg–/– mice. Intriguingly, there was an increased expression of p21Waf/Cip1, p16INK4a and p19ARF in REGg depleted tissues, suggesting that REGg may regulate the expression p21Waf/Cip1, p16INK4a and p19ARF in vivo.
 (a)
(a)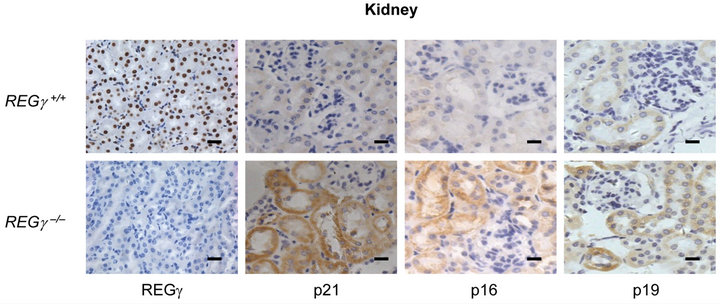 (b)
(b)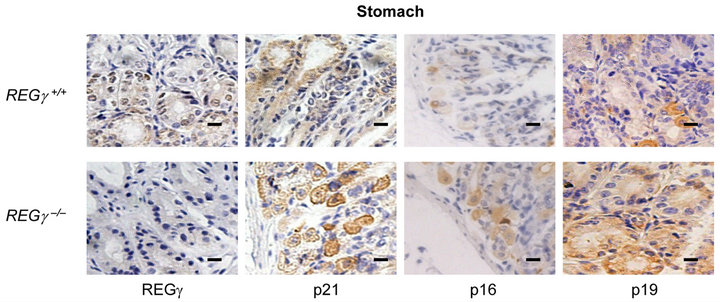 (c)
(c)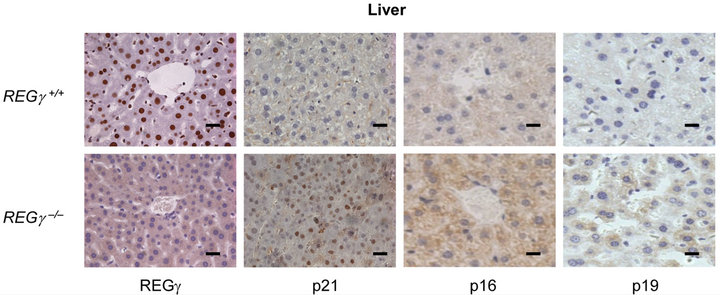 (d)
(d)
Figure 1. The expression of p21Waf/Cip1, p16INK4a and p19ARF is higher in REGg–/– mouse tissues. IHC analysis of REGg, p21Waf/Cip1 p16INK4a and p19ARF in REGg+/+ and REGg–/– cerebellum (a); kidney (b); stomach (c) and liver (d) mouse tissues. The result revealed increased p21Waf/Cip1 p16INK4a and p19ARF expression in REGg–/– tissues. Scale bar, 100 μm.
3.2. Comparative Analysis of REG( Protein and mRNA Expression
To verify the REGγ expression observed by IHC, p21Waf/Cip1, p16INK4a and p19ARF protein and mRNA levels were examined using extracts from different mouse tissues. Firstly, the relative protein levels were normalized to β-actin and the results showed that p21Waf/Cip1, p16INK4a and p19ARF protein levels were higher in REGg deficient cerebellum (Figure 2(a)), stomach (Figure 2(b)) and liver (Figure 2(c)). The comparison of relative mRNA and protein levels in different tissues may provide intact information about tissue-specific roles for a gene. Next, we consistently observed a higher mRNA expression of p16INK4a and p19ARF in multiple tissues, indicating that REGg regulated p16INK4a and p19ARF in protein level (Figure 2(d)). To further demonstrate that REGg is involved in p21Waf/Cip1, p16INK4a and p19ARF degradation, we generated stable HeLa cell line constitutively expressing either a control non-specific shRNA (shN) or a REGg-specific shRNA (shR). We monitored the p21Waf/Cip1, p16INK4a and p14ARF protein levels in Hela stable cells. As expected, the p21Waf/Cip1, p16INK4a and p14ARF protein levels were higher in Hela-ShR cells compared with Hela-shN cells (Figure 2(e)). The same phenomenon was also found in Hela cells described previously [16].
3.3. Highly Expressed REG( and Reduced Expression of p21Waf/Cip1, p16INK4a and p14ARF are Observed in Human Cancers
It is known that REGg is high expressed in lungs, liver, thyroid and colon cancer [20,24]. However, the biological links between REGg and its correlated genes such as p21Waf/Cip1, p16INK4a and p14ARF in cancers remain unknown. Immunohistochemistry (IHC) analysis of REGg regulated proteins revealed that the REGg level was high expressed in human cancers such as kidney (Figure 3(a)), lungs (Figure 3(b)) and brain (Figure 3(c)) while the protein level of p21Waf/Cip1, p16INK4a and p14ARF was decreased. However, the protein level of p21Waf/Cip1, p16INK4a and p14ARF was increased when REGg level was lower in stomach cancer (Figure 3(d)). These results suggest that REGγ mediated degradation of p21Waf/Cip1, p16INK4a and p14ARF is one of the important mechanisms by which REGγ mediates its physiological function in different cancers.
4. Discussion
p21Waf/Cip1, p19ARF and p16INK4a are important tumor-suppressor proteins. p21Waf/Cip1 is encoded by CDKN1A gene, which can bind to and inhibit the activity of cyclin-CDK2 or -CDK4 complexes, and thus functioning as a regulator of cell cycle progression at G1. p16INK4a inhibits Cdk4 and Cdk6 activity leading to activating RBp19ARF stabilizes p53 protein levels by binding to Mdm2 and promoting its degradation [25]. Mice lacking p21Waf/Cip1, p16INK4a or p19ARF are prone to form tumors induced by carcinogens. Currently, several studies showed that REGγ can promote the degradation of p21Waf/Cip1 and p53 in vitro. However, the detailed link between REGγ and the cyclin-dependent kinase inhibitors is still unclear. This study systematically demonstrated that REGγ can promote the degradation of
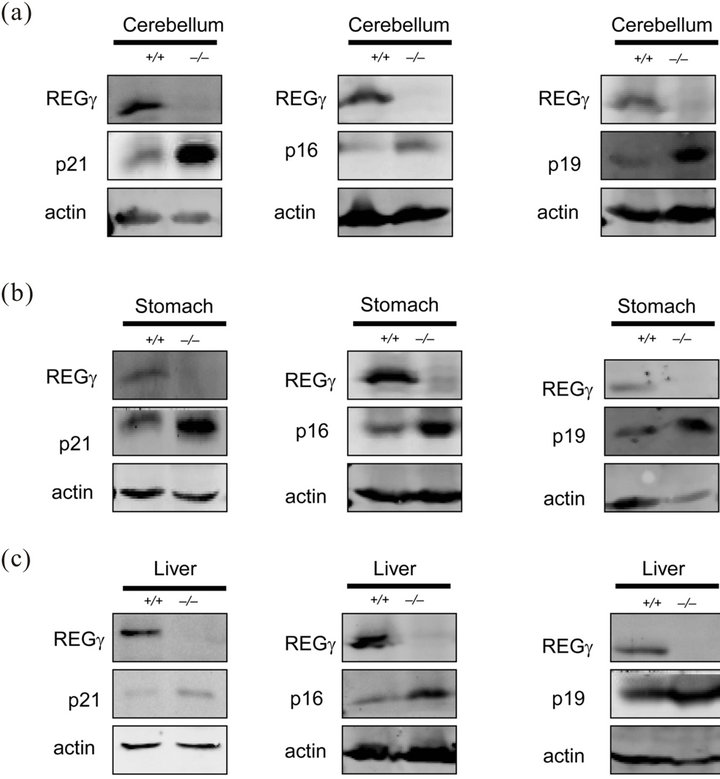
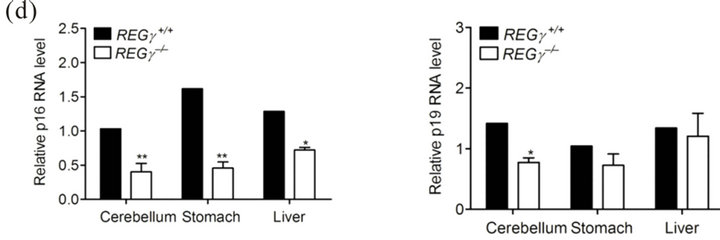

Figure 2. Comparative analysis of REGg protein and mRNA expression. (a) The protein level of REGg, p21Waf/Cip1, p16INK4a and p19ARF in REGg+/+ and REGg–/– mouse cerebellum tissue. Overexpression of p21Waf/Cip1 p16INK4a and p19ARF is observed in REGg–/– cerebellum tissue; (b) The protein expression of REGg, p21Waf/Cip1, p16INK4a and p19ARF in REGg+/+ and REGg–/– mouse stomach tissue. The expression of p21Waf/Cip1, p16INK4a and p19ARF is higher in REGg–/– stomach tissue; (c) The REGg, p21Waf/Cip1, p16INK4a and p19ARF expression of in REGg+/+ and REGg–/– mouse liver tissue. The protein level of p21Waf/Cip1, p16INK4a and p19ARF is higher in REGg–/– liver tissue; (d) The mRNA levels of p16INK4a and p19ARF in mouse tissues were tested in REGg+/+ (n = 3) and REGg–/– (n = 3) mouse tissues. The mRNA levels of p16INK4a and p19ARF is higher in REGg+/+ mouse tissues compared with REGg–/– mouse tissues. (*p < 0.05; **p < 0.01); (e) Expression of REGg, p21Waf/Cip1, p16INK4a and p14ARF in Hela-shN and Hela-shR cell lines.
p21Waf/Cip1, p16INK4a and p19ARF in vivo.
REGγ-deficient mice [23,26] have been demonstrated growth retardation and cell-specific mitotic defects, in-
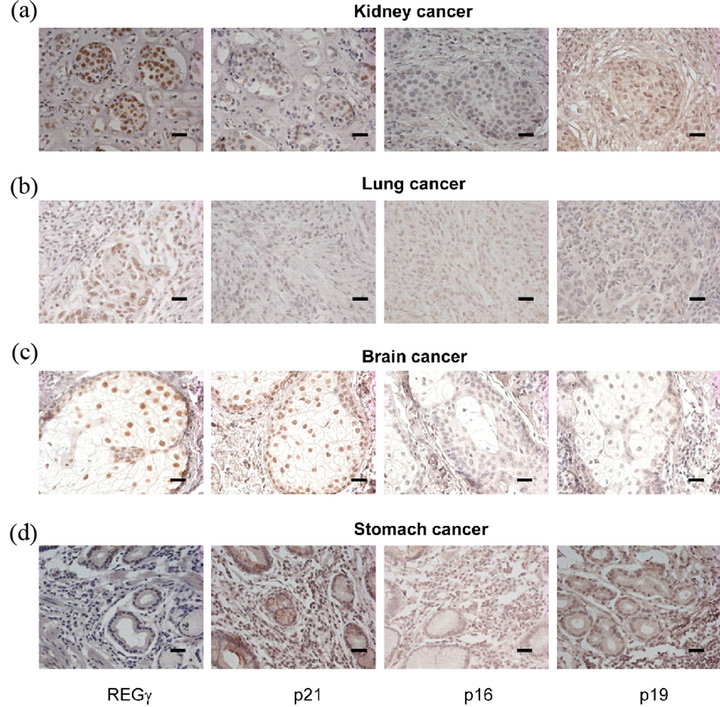
Figure 3. Highly expressed REGg and decreased REGg, p21Waf/Cip1, p16INK4a and p14ARF protein level are observed in human cancers. IHC analysis of REGg, p21Waf/Cip1, p16INK4a and p14ARF in human kidney cancers (a); lung cancers (b); brain cancers (c) and stomach cancers (d). The result revealed that the REGg level is high expressed in different human cancers while the protein level of p21Waf/Cip1, p16INK4a and p14ARF was decreased. Scale bar, 100 μm.
dicating a role of REGγ in regulation of cell cycle and cell proliferation. It is well known that p16INK4a, p19ARF, smARF and p21Waf/Cip1 are important CDK inhibitors. Therefore, we used the REGγ-knockout mice to detect the effects of REGγ on p16INK4a/p19ARF/p21Waf/Cip1 in several normal tissues. As expected, our IHC and blot results showed that the levels of p16INK4a/p19ARF/ p21Waf/Cip1 were markedly increased in cerebellum, stomach, kidney and liver when REGγ was deleted. Our result demonstrated that REGγ can promote the degradation of CDK inhibitors in normal tissues. These results indicated that REGγ plays an important role in regulating the dynamic balance of body such as organizational self-renewal and tissue remodeling. These results also provide the first time to study the effects of REGγ on CDK inhibitors in vivo. Recent studies demonstrate that REGγ can promote proteasome-dependent degradation of cyclin-dependent kinase inhibitors p21Waf/Cip1, p16INK4a and p19ARF, in a ubiquitinand ATP-independent manner [16,17]. However, their conclusion is mainly limited to in vitro studies. Thus, our finding is consistent with previous suggestion that REGγ is involved in the regulation of cell cycle and cell proliferation.
Besides as the CDK inhibitors, p16INK4a/p19ARF/ p21Waf/Cip1 are well known as the tumor suppressor. The first genetic evidence supporting a tumor suppressor activity for p21Waf/Cip1 came from the discovery that CDKN1A deficient mice developed spontaneous tumours. Many human cancers such as colorectal, cervical, head and neck, and small-cell lung cancers are associated with reduced p21Waf/Cip1 expression. The frequent mutations and deletions of p16INK4a and p14ARF in many types of human cancers cancer cell lines suggested an important role for p16INK4a and p14ARF in carcinogenesis. Thus, the effects of REGγ on p16INK4a/p19ARF/p21Waf/Cip1 were investigated in several human cancer tissues. In cancer tissues of stomach, lung, kidney and brain, we found that REGγ has different expression patterns. REGγ’s expression was higher in the kidney and lung cancer compared with stomach cancer. These results indicated that REGγ showed its specificity for participating in carcinogensis. Consistently, previous studies revealed that REGγ can degrade not only tumor suppressor but also tumor activator, which suggests its dual function in cancer. Thus, we assumed that the mechanism of REGγ involved in cancer is complicated. Interestingly, we found the levels of p16INK4a/p19ARF/p21Waf/Cip1 are firmly regulated by REGγ both in normal tissues and in cancer tissues. When REGγ levels are high in the kidney and lung cancer, the expressions of p16INK4a/p19ARF/p21Waf/Cip1 are low. On the contrary, the high expression of p16INK4a/p19ARF/ p21Waf/Cip1 was observed in the stomach cancer which showed low expression of REGγ. Our results demonstrated that REGγ can promote the degradation of p16INK4a/p19ARF/p21Waf/Cip1 not only in normal tissues but also in cancer tissues. REGγ has been reported to be involved in some types of cancer [22,27]. Therefore, we assumed that REGγ maybe play its role in the formation and development of cancer by altering the levels of CDK inhibitors under some conditions.
Recently, REGγ was shown to facilitate the turnover of tumor suppressor p53 by promoting MDM2-mediated p53 ubiquitination [28]. P53 is known as “guardian of the genome” because it shuts down cell division in response to DNA damage. Previous research had linked the function of p14ARF to p53. It is also reported that p53 mediates the DNA damage-induced checkpoint through transactivation of p21Waf/Cip1. Thus, the relations between REGγ and p53 in vivo are also required to be systemically studied in the future. The members in the CDK inhibitor family also included p15 (INK4B), p18 (INK4C) in INK4 subfamily and p27 (KIP1), p57 (KIP2) in the KIP subfamily. It is still unclear if REGγ can promote the degradation of p15, p18, p27 and p57. Furthermore, the effects of REGγ on p16INK4a/p19ARF/p21Waf/Cip1 in the other tissues of mice or in the other types of cancer are needed to be clarified.
In summary, this study is the first to systematically study the effects of REGγ on p16INK4a/p19ARF/p21Waf/Cip1 in vivo. We found that REGγ can promote the degradation of CDK inhibitor both in normal tissue of mice and in human caner tissues. The data suggest that inhibition of REGγ may be of potential benefit in preventing and treating some types of human cancer.
5. Acknowledgements
This work was supported by National Institutes of Health (1R01CA131914, P30 CA125123), Norman Hackerman Advanced Research Program (1082318401; PN00 4949- 0012-2009), and supported in part by the Pilot/Feasibility Program of the Diabetes & Endocrinology Research Center (P30-DK079638) at Baylor College of Medicine. This manuscript was also funded in part by the National Natural Science Foundation of China (81261120555, 81071657, 81272943, 31100946, 31200878); the Science and Technology Commission of Shanghai Municipality (11DZ2260300, 10JC1404200, 11ZR1410000, 12ZR14 09300); the National Basic Research Program (2009 CB918402, 2011CB504200); and the Fundamental Research Funds for the Central Universities (78210101 and 78210139).
REFERENCES
- D. E. Quelle, F. Zindy, R. A. Ashmun, et al., “Alternative Reading Frames of the INK4a Tumor Suppressor Gene Encode Two Unrelated Proteins Capable of Inducing Cell Cycle Arrest,” Cell, Vol. 83, No. 6, 1995, pp. 993-1000. doi:10.1016/0092-8674(95)90214-7
- S. W. Lowe and C. J. Sherr, “Tumor Suppression by INK4a-Arf: Progress and Puzzles,” Current Opinion in Genetics & Development, Vol. 13, No. 1, 2003, pp. 77- 83. doi:10.1016/S0959-437X(02)00013-8
- M. Serrano, H. Lee, L. Chin, et al., “Role of the INK4a Locus in Tumor Suppression and Cell Mortality,” Cell, Vol. 85, No. 1, 1996, pp. 27-37. doi:10.1016/S0092-8674(00)81079-X
- P. Krimpenfort, K. C. Quon, W. J. Mooi, et al., “Loss of p16INK4a Confers Susceptibility to Metastatic Melanoma in Mice,” Nature, Vol. 413, No. 6851, 2001, pp. 83-86. doi:10.1038/35092584
- M. Serrano, G. J. Hannon and D. Beach, “A New Regulatory Motif in Cell-Cycle Control Causing Specific Inhibition of Cyclin D/CDK4,” Nature, Vol. 366, No. 6456, 1993, pp. 704-707. doi:10.1038/366704a0
- C. J. Sherr, “Principles of Tumor Suppression,” Cell, Vol. 116, No. 2, 2004, pp. 235-246. doi:10.1016/S0092-8674(03)01075-4
- E. Sharpless and L. Chin, “The INK4a/ARF Locus and Melanoma,” Oncogene, Vol. 22, No. 20, 2003, pp. 3092- 3098. doi:10.1038/sj.onc.1206461
- R. Ben-Saadon, I. Fajerman, T. Ziv, et al., “The Tumor Suppressor Protein p16INK4a and the Human Papillomavirus Oncoprotein-58 E7 Are Naturally Occurring LysineLess Proteins That Are Degraded by the Ubiquitin System. Direct Evidence for Ubiquitination at the N-Terminal Residue,” The Journal of Biological Chemistry, Vol. 279, No. 40, 2004, pp. 41414-41421. doi:10.1074/jbc.M407201200
- S. Bates, A. C. Phillips, P. A. Clark, et al., “p14ARF Links the Tumour Suppressors RB and p53,” Nature, Vol. 395, No. 6698, 1998, pp. 124-125. doi:10.1038/25867
- I. Palmero, C. Pantoja and M. Serrano, “p19ARF Links the Tumour Suppressor p53 to Ras,” Nature, Vol. 395, No. 6698, 1998, pp. 125-126. doi:10.1038/25870
- A. Radfar, I. Unnikrishnan, H. W. Lee, et al., “p19ARF Induces p53-Dependent Apoptosis during Abelson VirusMediated Pre-B Cell Transformation,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 95, No. 22, 1998, pp. 13194-13199. doi:10.1073/pnas.95.22.13194
- F. Zindy, R. T. Williams, T. A. Baudino, et al., “ARF Tumor Suppressor Promoter Monitors Latent Oncogenic Signals in Vivo,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 100, No. 26, 2003, pp. 15930-15935. doi:10.1073/pnas.2536808100
- R. Honda, and H. Yasuda, “Association of p19ARF with Mdm2 Inhibits Ubiquitin Ligase Activity of Mdm2 for Tumor Suppressor p53,” The EMBO Journal, Vol. 18, No. 1, 1999, pp. 22-27. doi:10.1093/emboj/18.1.22
- C. A. Midgley, J. M. Desterro, M. K. Saville, et al., “An N-Terminal p14 ARF Peptide Blocks Mdm2-Dependent Ubiquitination in Vitro and Can Activate p53 in Vivo,” Oncogene, Vol. 19, No. 19, 2000, pp. 2312-2323.
- X. Li, D. M. Lonard, S. Y. Jung, et al., “The SRC-3/AIB1 Coactivator Is Degraded in a Ubiquitinand ATP-Independent Manner by the REGγ Proteasome,” Cell, Vol. 124, No. 2, 2006, pp. 381-392. doi:10.1016/j.cell.2005.11.037
- X. Chen, L. F. Barton, Y. Chi, et al., “Ubiquitin-Independent Degradation of Cell-Cycle Inhibitors by the REGγ Proteasome,” Molecular Cell, Vol. 26, No. 6, 2007, pp. 843-852. doi:10.1016/j.molcel.2007.05.022
- X. Li, L. Amazit, W. Long, et al., “Ubiquitinand ATPIndependent Proteolytic Turnover of p21 by the REGgamma-Proteasome Pathway,” Molecular Cell, Vol. 26, No. 6, 2007, pp. 831-842. doi:10.1016/j.molcel.2007.05.028
- T. Okamura, S. Taniguchi, T. Ohkura, et al., “Abnormally High Expression of Proteasome Activator-Gamma in Thyroid Neoplasm,” The Journal of Clinical Endocrinology & Metabolism, Vol. 88, No. 3, 2003, pp. 1374- 1383.
- M. Roessler, W. Rollinger, L. Mantovani-Endl, et al., “Identification of PSME3 as a Novel Serum Tumor Marker for Colorectal Cancer by Combining Two-Dimensional Polyacrylamide Gel Electrophoresis with a Strictly Mass Spectrometry-Based Approach for Data Analysis,” Molecular & Cellular Proteomics, Vol. 5, No. 11, 2006, pp. 2092-2101. doi:10.1074/mcp.M600118-MCP200
- Y. Hong, K. S. Ho, K. W. Eu, et al., “A Susceptibility Gene Set for Early Onset Colorectal Cancer That Integrates Diverse Signaling Pathways: Implication for Tumorigenesis,” Clinical Cancer Research, Vol. 13, No. 4, 2007, pp. 1107-1114. doi:10.1158/1078-0432.CCR-06-1633
- H. L. Jia, Q. H. Ye, L. X. Qin, et al., “Gene Expression Profiling Reveals Potential Biomarkers of Human Hepatocellular Carcinoma,” Clinical Cancer Research, Vol. 13, No. 4, 2007, pp. 1133-1139. doi:10.1158/1078-0432.CCR-06-1025
- J. He, L. Cui, Y. Zeng, et al., “REGγ Is Associated with Multiple Oncogenic Pathways in Human Cancers,” BMC Cancer, Vol. 12, 2012, p. 75. doi:10.1186/1471-2407-12-75
- G. Yu, Y. Zhao, J. He, et al., “Comparative Analysis of REGγ Expression in Mouse and Human Tissues,” Journal of Molecular Cell Biology, Vol. 2, No. 4, 2010, pp. 192- 198. doi:10.1093/jmcb/mjq009
- G. Salvatore, T. C. Nappi, P. Salerno, et al., “A Cell Proliferation and Chromosomal Instability Signature in Anaplastic Thyroid Carcinoma,” Cancer Research, Vol. 67, No. 21, 2007, pp. 10148-10158. doi:10.1158/0008-5472.CAN-07-1887
- Y. Zhang, Y. Xiong and W. G. Yarbrough, “ARF Promotes MDM2 Degradation and Stabilizes p53: ARFINK4a Locus Deletion Impairs Both the Rb and p53 Tumor Suppression Pathways,” Cell, Vol. 92, No. 6, 1998, pp. 725-734. doi:10.1016/S0092-8674(00)81401-4
- L. F. Barton, H. A. Runnels, T. D. Schell, et al., “Immune Defects in 28-kDa Proteasome Activator Gamma-Deficient Mice,” The Journal of Immunology, Vol. 172, No. 6, 2004, pp. 3948-3954.
- I. Mao, J. Liu, X. Li, et al., “REGγ, a Proteasome Activator and beyond?” Cellular and Molecular Life Sciences, Vol. 65, No. 24, 2008, pp. 3971-3980. doi:10.1007/s00018-008-8291-z
- Z. Zhang and R. Zhang, “Proteasome Activator PA28 Gamma Regulates p53 by Enhancing Its MDM2-Mediated Degradation,” The EMBO Journal, Vol. 27, No. 6, 2008, pp. 852-864. doi:10.1038/emboj.2008.25
NOTES
*These authors contributed equally to this work.
#Corresponding authors.

