Natural Science
Vol.07 No.11(2015), Article ID:61153,19 pages
10.4236/ns.2015.711051
Thymidine Glycol: The Effect on DNA Structure and DNA Binding by Site-Specific Proteins
Elena A. Kubareva1, Fan Yang2, Alexandra Yu. Ryazanova1, Nina G. Dolinnaya2, Andrei V. Golovin3, Nikolai V. Molochkov4, Elena A. Romanova1, Elizaveta A. Karpova1, Timofei S. Zatsepin2, Tatiana S. Oretskaya1,2*
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, Russia
2Chemistry Department, Lomonosov Moscow State University, Moscow, Russia
3Faculty of Bioengineering and Bioinformatics, Lomonosov Moscow State University, Moscow, Russia
4Institute of Theoretical and Experimental Biophysics, Russian Academy of Science, Moscow Region, Pushchino, Russia

Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 7 October 2015; accepted 13 November 2015; published 16 November 2015

ABSTRACT
Thymidine glycol (5,6-dihydroxy-5,6-dihydrothymidine, Tg) is a major type of oxidative damage in DNA. During chemical oligonucleotide synthesis, Tg residue was incorporated in the different positions of 17 b.p. DNA duplexes, which differ in one base pair in the internal part. According to UV- melting curves, Tg destabilizes the double helix in a sequence independent manner. In contrast, the localized alterations in duplex structure were shown by CD spectroscopy to depend on the type of base pairs flanking the Tg lesion. Molecular dynamics simulations demonstrate that Tg is partially out of the double helix. For the first time, Tg impact on several site-specific DNA-binding proteins is studied, namely p50 and p65 subunits of nuclear factor kappa-B (NF-κB) and DNA methyltransferase SsoII (M.SsoII). Our results show that p50/p50 and p65/p65 homodimers of NF-κB can tolerate a single Tg residue in the binding site quite well. Nevertheless the homodimers have different affinities to the oxidized κB site depending on the Tg position. M.SsoII can act as a transcription repressor when bound to the regulatory site. M.SsoII demonstrates decreased affinity and lowered methylation efficiency when its methylation site contains Tg in the central position. Single Tg in one half of the regulatory site decreases M.SsoII affinity to the oxidized DNA, whereas Tg presence in both half-sites prevents M.SsoII binding to such ligand.
Keywords:
Thymidine Glycol, Oxidative Damage, Molecular Dynamics Simulations, Nuclear Factor Kappa-B, DNA Methyltransferase SsoII

1. Introduction
It is widely established that it is essential to understand the main features of biopolymer interactions in order to develop methods for efficient monitoring the biological activities, as well as the reasons of their changes. Thymidine glycol (5,6-dihydroxy-5,6-dihydrothymidine, Tg) is a major type of pyrimidine residue oxidative damage in DNA. It is formed as a result of thymine base oxidation by genotoxic factors such as ultraviolet or ionizing radiation and chemical oxidants [1] -[3] . This can be also a result of aerobic metabolism [4] . Tg can be formed as well by hydrolytic deamination of 5-methylcytidine followed by thymidine oxidation or vice versa by oxidation of 5-methylcytidine followed by hydrolytic deamination of the unstable 5-methylcytidine glycol [5] . About 400 Tg residues are formed per human cell per day [6] . In medical studies, Tg can be used as a genetic biomarker of oxidative DNA damage along with other biomarkers such as 7,8-dihydro-8-oxo-2'-deoxyguanosine (8-oxoG) [7] - [9] . There are four diastereomers of Tg because it contains two chiral carbon atoms, but Tg exists in solution as either the 5R cis-trans pair (5R,6S; 5R,6R) or the 5S cis-trans pair (5S,6R; 5S,6S) due to epimerization at the C6 position [10] .
In contrast to the canonical DNA bases, thymine glycol is non-planar due to the loss of aromaticity. Biochemical studies have shown that Tg is a weakly mutagenic lesion because it generally pairs with adenine base [11] . However, it presents a strong block to high-fidelity DNA polymerases in vitro and can be lethal in vivo in the absence of translesion DNA synthesis or recombination [3] [12] . According to recent data, the structural and biological consequences of Tg lesion are sequence-dependent [13] . Replacement of thymidine by Tg in a DNA double helix leads to changes in DNA structure and thermodynamic properties. It can also have a certain impact on DNA recognition by DNA binding proteins. Up to date, many researches are dedicated to the Tg influence on the activity of various DNA polymerases and to the Tg removal from DNA by repair enzymes (reviewed in [14] ), but there is a lack of information about Tg impact onto proteins which recognize specific DNA sites. The only research addressing this question is our paper [15] where effect of Tg on the rate of DNA hydrolysis by several restriction endonucleases (MvaI, Bst2UI, MspR9I, Bme1390I, Psp6I, BstSCI, EcoRII, and SsoII) has been studied.
In the present research, we report for the first time the data about interaction of three different site-specific DNA-binding proteins with Tg-containing DNA duplexes. The first two proteins are p50 and p65 subunits of nuclear factor kappa-B (NF-κB), an eukaryotic transcription factor which plays a role in innate and adaptive immune responses, inflammation, cell proliferation, differentiation, and apoptosis [16] - [21] .
The NF-κB family of transcription factors consists of 5 proteins, namely p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), c-Rel, and RelB. NF-κB homo- and heterodimers have been described; different cell types have different repertoires of these dimers [22] - [24] . NF-κB dimers bind to regulatory elements of the genes that contain its recognition sites (κB sites). NF-κB dimers differ in affinity to various promoters; however, the affinity does not correlate with their capability to activate transcription. During gene expression, dimer exchange can be observed [25] . NF-κB translocates into the nucleus in response to several stimuli, including oxidative stress [26] . Yet T can be converted into Tg under the oxidative stress conditions [6] . The sequence of the NF-κB recognition site has many variants [27] , i.e. the protein can tolerate several nucleotide substitutions. On the other hand, conversion of T into Tg can change the overall parameters of the DNA double helix which could appear more harmful for the site recognition than a nucleotide substitution. Several works indicated the importance of the κB site conformation for the proper recognition by NF-κB [28] - [31] . Thus, NF-κB interaction with an oxidized recognition site is an important question and is addressed in the present work. Another common oxidative modification of DNA is 8-oxoG. Its impact on the NF-κB affinity to its target DNA has been determined earlier [32] . Surprisingly, there are no studies about Tg influence on the NF-κB affinity to its recognition site.
In the present work, the Tg residue (the 5R,6S isomer) has been incorporated site-specifically into three 17-bp DNA duplexes which contained different variants of the NF-κB recognition site. The Tg lesion is located in different positions of the site in order to find out the effect of the Tg・A pair microenvironment. The affinities of р50/р50 and р65/р65 homodimers to these oxidized recognition sites are evaluated.
The third protein studied in this work is DNA methyltransferase SsoII (M.SsoII), a prokaryotic bifunctional protein that acts as a methyltransferase and as a transcription factor.
The protein consists of two domains: a larger one (residues 72 - 379) possesses the DNA methyltransferase activity, while a smaller one (residues 1 - 71) regulates transcription of its own gene [33] - [35] . These two functions are provided by M.SsoII recognition of two different specific sites in DNA. Each domain binds to its own DNA sequence (methylation or regulatory site). In the present work the effect of Tg on M.SsoII functioning is evaluated. A set of Tg-containing DNA duplexes is used for this purpose. We demonstrate for the first time that DNA methylation is decreased when the methylation site contains Tg. The Tg presence in the regulatory site significantly impairs M.SsoII binding to it. Thus, M.SsoII could not act as a transcription factor under conditions of oxidative stress, in contrast to NF-κB.
The effect of a single Tg lesion on melting behavior and structural alterations of oligonucleotide duplexes is determined by UV spectroscopy, circular dichroism spectroscopy, and molecular dynamics simulation.
2. Materials and Methods
2.1. Protein Expression and Purification
Plasmid pGEX-1 encoded a recombinant protein in which glutathione-S-transferase (GST) was fused to the N-terminus of the NF-κB р50 subunit; this construction did not allow the cleavage of the GST. This plasmid was kindly provided by Prof. A. Israёl (Institut Pasteur, France). The fusion with GST allows one-step purification of the target protein from E. coli cell culture by affine chromatography [36] [37] . To improve the purity and the stability of the protein we modified the standard protocol. Competent E. coli HB-101 cells (Promega, USA) were transformed with the plasmid pGEX-1. The cell culture was grown in LB medium with 100 μg/ml ampicillin at 37˚C up to an A500 value of 0.6 - 0.8. Protein expression was induced by adding isopropyl-β-D-1-thioga- lactopyranoside (IPTG) up to 1 mM and the cell culture was kept at 37˚C for 3 - 4 h. The cells were harvested by centrifugation at 10,000 rpm for 20 min (J-6 centrifuge with JA-10 rotor, Beckman, USA). The cell pellets were resuspended in 25 - 30 ml buffer A (20 mM Tris-HCl, рН 8.0, 25 mM NaCl, 0.1 mM EDTA) with 0.2 mg/ml lysozyme and 1 mM phenylmethanesulfonyl fluoride (PMSF). The cells were lysed using French press. The lysate was clarified by centrifugation at 15,000 rpm for 20 min (J-6 centrifuge with JA-14 rotor, Beckman, USA); the supernatant was transferred to the tubes and centrifuged again at 35,000 rpm for 2 h using L5-75 centrifuge with Ti-60 rotor (Beckman, USA). The resulting supernatant was loaded onto a 50-ml column containing an anion exchange resin Toyopearl DEAE-650S (Tosoh Bioscience, Japan) pre-equilibrated with buffer A. The target protein was eluted with a linear NaCl gradient (0.025 - 1 M for 100 min) in buffer A at 1 ml/min. The elution was monitored by absorbance at 280 nm using Uvicord S densitometer (LKB, Sweden). The presence of р50-GST in the collected fractions was confirmed by SDS-polyacrilamide gel (PAG) electrophoresis. The fractions which contained the target protein were dialysed against PBS buffer (10 mM Na2HPO4, 1.8 mM KH2PO4, pH 8.0, 140 mM NaCl, 2.7 mM KCl) for 10 - 12 h and then loaded onto a 2-ml column containing Glutathione Sepharose 4B (GE Healthcare, USA) pre-equilibrated with PBS. The column was washed with 4 column volumes of PBS at 1 ml/min. The sorbent was eluted with 3 - 5 column volumes of 10 mM reduced glutathione in 50 mM Tris-HCl buffer (pH 8.0). The elution was monitored using Uvicord S; the purity of р50-GST in the collected fractions was analysed by SDS- PAGE.
Prof. N.D. Perkins (Newcastle University, Newcastle Upon Tyne, UK) kindly provided us with plasmid pGEX-KT RelA which was constructed in Dr. Gary Nabel’s laboratory at the University of Michigan (USA). This plasmid encoded a recombinant protein in which GST was fused to the N-terminus of NF-κВ р65 subunit fragment (amino acid residues 1 - 306); this construction did not allow the cleavage of the GST. Competent E. coli BL21 cells (Novagen, Germany) were transformed with the plasmid. Protein expression and cell lysis were performed the same way as for the р50-GST purification (see above). Then 0.5 ml 50% (v/v) Glutathione Sepharose 4B (GE Healthcare, USA) was added to the supernatant and incubated for 30 min at 4˚C. The mixture was centrifuged for 1 min at 15,000 g. The precipitate was washed 4 times with 2 ml PBS. The protein was eluted with 0.5 ml 10 mM reduced glutathione in 50 mM Tris-HCl (pH 8.0). The purity of р65-GST in the collected fractions was analysed by SDS-PAGE. The proteins р50-GST and p65-GST were dialysed against buffer A, aliquotted, frozen in liquid nitrogen and stored at −70˚C.
Plasmids pQESso9 (encoding restriction endonuclease SsoII (R.SsoII)) and pQMSsoII (encoding M.SsoII) were kindly provided by Prof. A.S. Karyagina (Gamaleya Research Institute of Epidemiology and Microbiology, Russia). Both recombinant proteins contained N-terminal His6-tags which allowed protein purification by affine chromatography on Ni-NTA agarose. Protein expression and purification was performed according to standard protocols [38] [39] .
2.2. Oligodeoxyribonucleotides and DNA Duplexes
Non-modified oligodeoxyribonucleotides and oligodeoxyribonucleotides containing a single Tg residue were assembled by the standard phosphoramidite method using the automatic ABI 394 DNA/RNA synthesizer (Applied Biosystems, USA). The coupling time for 5R,6S Tg phosphoramidite (Glen Research, USA) was prolonged up to 5 min. Mild cleavage and deprotection with ammonium hydroxide at room temperature for 2 h was used to obtain the required oligonucleotide products containing the Tg residue still protected by the tert-butyl- dimethylsilyl (TBDMS) group. To remove the TBDMS groups, triethylamine trihydrofluoride was applied at 40˚C overnight. The mixtures were desalted on NAP-20 columns (GE Healthcare, USA). The deprotected oligonucleotides were purified by electrophoresis in 20% PAG with 7 M urea. Their purity was confirmed by reverse-phase HPLC (ion-pair mode) (Waters, USA). Mass spectra of the Tg-containing oligonucleotides were recorded using Reflex IV MALDI-TOF mass spectrometer (Bruker, Germany). A mixture of 2',6'-dihydroxya- cetophenone (40 mg/ml in methanol) and ammonium hydrogencitrate (80 mg/ml in water) in 1:1 ratio (v/v) was used as a matrix. The results of the MALDI-TOF analyses coincide with the calculated values for the modified oligonucleotides (data not shown). Oligonucleotide concentrations were determined spectrophotometrically using the extinction coefficients from the nearest-neighbor data [40] .
Oligonucleotides were radioactively labeled by mixing with (γ-32P) ATP and 10 units T4 polynucleotide kinase (Thermo Fisher Scientific, USA) in 10 μl of buffer В (50 mM Tris-HCl, pH 7.6, 10 mM MgCl2, 5 mM dithiothreitol, 0.1 mM spermidine, 0.1 mM EDTA) and incubating for 30 min at 37˚C. The labeled oligonucleotides were purified by gel-filtration using G-25 Illustra MicroSpinTM columns (GE Healthcare, USA). The radioactivity was determined by the Cherenkov method using Tracor Analytic Delta 300 counter (ThermoQuest/ CE Instruments, USA); the experimental error did not exceed 2%. DNA duplexes were prepared by mixing equimolar amounts of complementary oligonucleotides in water, adding 32Р-labeled oligonucleotide (if necessary), heating the mixture up to 90˚C and cooling down slowly to room temperature (annealing).
2.3. Temperature Dependence of the Duplex Stability
The duplexes (Table 1, Table 2) were prepared in 2 - 4 μM concentrations by mixing the complementary DNA strands in 1:1 molar ratio in buffer С (10 mM Tris-HCl, pH 7.6, 10 mM MgCl2). The samples were then heated up to 80˚C - 90˚C and slowly cooled down to the room temperature. There were no differences in the annealing protocol for the Tg-containing and the unmodified DNA duplexes. The dependence of sample absorbance (A260) vs. temperature was measured in a 500-μl quartz microcuvette (Hellma, Germany) with a 10-mm path length using the U-2900 UV/Visible Spectrophotometer (Hitachi, Japan) equipped with a thermoelectric temperature controller. The thermally-induced unfolding transitions were monitored between 35˚C and 85˚C at 260 nm with the heating rate of 0.5˚C/min. The Tm value for each duplex was determined as the temperature corresponding to the maximum on its differential melting curve (dA/dT vs. T). The hyperchromic effect (h260) accompanying the duplex melting was calculated as (A80 − A35/A80) × 100%, where A80 and A35 were the absorbance values at 80˚C and 35˚C, respectively. The mixtures of the oligonucleotides after the melting procedure were desalted on NAP-5 columns (GE Healthcare, USA) and analyzed by reverse-phase HPLC (ion-pair mode) to confirm the absence of the oligonucleotide degradation products.
2.4. Circular Dichroism (CD) Measurements
DNA duplex solutions for the CD measurements were prepared the same way as for the melting experiments. The CD spectra of the samples dissolved to ~2 µM concentration in buffer С were measured in a quartz cuvette
Table 1. Tg impact on thermal stability of DNA duplexes and NF-κB affinity to the duplexes. The three pairs of duplexes (I and II, III and IV, V and VI) differ in the κB site sequence and in position of Tg lesion.
*The NF-κB binding sites are in bold and the superscripts indicate the nucleotide positions in the sites. Tg (or the corresponding T) is underlined. **The hyperchromic effect accompanying the double helix melting; relatively low h260 value is explained by the presence of single-stranded prominent (4- and 2-nucleotides) termini flanking the duplex core, which absorb the UV light at 260 nm, but do not contribute to secondary structure unfolding.
Table 2. Effect of Tg-modification on the affinity of M.SsoII protein to the DNA duplexes containing its recognition sites.
*The methylation site (5 bp) and the regulatory site (15 bp) are in bold. The Tg (or the corresponding T in control duplexes) is underlined. **These data are already published in [15] . ***The methylation rate is measured under conditions when the strand with Tg (or the corresponding T) was 32P-labeled. Methylation rate for the duplex VIII is taken as 1.
with a 10-mm path length at 22˚C using the Chirascan CD spectrometer (Applied Photophysics Ltd., UK) equipped with a thermoelectric temperature controller. The cell compartment was continuously purged with dry nitrogen. The measurements were performed from 330 to 220 nm, with the signal averaging time of 2 s and the scan speed of 30 nm/min. The spectrum of the solution without DNA (buffer С) was taken as a baseline and subtracted from the spectra of the samples. The CD spectra were averaged over 3 scans. The CD intensity was expressed as mean oligonucleotide dichroism, Δε (cm−1・M−1). The spectra were smoothed in Origin 8.0 (demonstration version) using the Savitzky-Golay filter.
2.5. Molecular Dynamics Simulations
The structure of DNA duplex II containing the cis-5R,6S Tg lesion was analyzed by molecular dynamics (MD) simulations using the GROMACS package with parmbsc0 force field [41] . The RESP method [42] was used to derive partial charges of Tg residue isomers. The initial model was based on the known data for thymidine. Geometry optimization and molecular electrostatic potential refinement were performed using quantum mechanics approach with the RHF theory level and using 6-31G basis set (http://dx.doi.org/10.1063/1.1674902, DOI: 10.1021/cr00074a002) in GAMESS [43] . These optimized conformations were used for calculation of electrostatic potential (ESP) for all the Tg isomers with RHF 6-31+G (d, p) basis. The derived partial charges for the 5R,6S Tg lesion are indicated in Figure 1(a).
The superposition of 3D structures of thymine glycol isomers to thymine was done in PyMol with fitting of atom pairs (N1, N3, C5).
The structure of the unmodified DNA duplex I was generated using X3DNA program package (doi:10.1038/ nprot.2008.104) and then the duplex II with the 5R,6S Tg lesion was build in PyMol (The PyMOL Molecular Graphics System, Version 1.2.1 Schrödinger, LLC). For the equilibrated structure calculation, the force field parmbsc0 was used, which is specially optimized for proteins and nucleic acids simulations [41] . The Tg torsion angles O3'-P-O5'-C5' and O5'-C5'-C4'-C3' were corrected in this force field. The system charge of the average structure from the final round of the simulated annealing calculations was neutralized by adding Na+ ions and placed in a truncated octahedral TIP4P water box. The solvated system was optimized and subjected to MD simulation with trajectory length of 40 ns and integration time step of 2 fs. Then the trajectories were analyzed with GROMACS 4.5 package [41] . In order to avoid the structural perturbations due to the terminal regions, we simulated only the central part of the duplexes I and II. Analysis of atom fluctuations was done with utility in GROMACS package where DNA coordinates during trajectory were superimposed to the starting structure and atom fluctuations were averaged through the whole trajectory.
2.6. DNA-Protein Complex Formation and Electrophoretic Mobility Shift Assay
Complex formation between NF-κВ subunits and 32Р-labeled DNA duplexes I - VI (Table 1) was carried in 10 μl of buffer D (7.5 mM HEPES, pH 8.0, 34 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT, 10% glycerol) containing 25 ng/μl poly(dI・dC) for 30 min at 25˚C. The concentrations of DNA duplexes varied from 20 to 200 nM. The protein concentration varied from 1.0 to 4.2 μM (for p50-GST) and from 3.2 to 6.5 μM (for p65-GST).
Complexes formation between M.SsoII and 32Р-labeled DNA duplexes VIII - XIII (Table 2), were carried in 20 μl of buffer E (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM dithiothreitol) containing 50 ng/μl poly(dI・dC) and 0.5 mM S-adenosyl-L-homocysteine (AdoHcy) for 20 min at 37˚C. The concentrations of DNA duplexes varied from 20 to 100 nM. The M.SsoII concentration was 0.92, 0.92, 0.80, 1.2, 1.6, and >2.4 μM when mixed with the duplexes VIII, IX, X, XI, XII, and XIII, correspondingly. Since M.SsoII is characterized by high non-specific DNA binding, not only poly (dI·dC) but also AdoHcy are necessary for the specific complex formation [44] [45] .
The DNA-protein complexes were analyzed by electrophoretic mobility shift assay (EMSA) in 7% non-de- naturing polyacrylamide gel (PAG). The radioactive bands were visualized using Molecular Dynamics PhosphorImager SI (Molecular Dynamics, USA). The yield of DNA-protein complex was determined as ratio of radioactivity of the band corresponding to DNA-protein complex to the whole radioactivity of the bands corres-
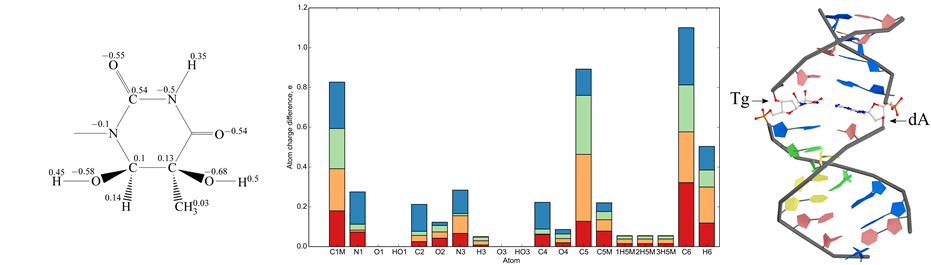
Figure 1. Characteristics of Tg isomers. (a) Formula and derived partial charges for the cis-5R,6S isomer of N-1-methyl-5, 6-dihydroxy-5,6-dihydrothymine; (b) Atom charge difference of N-1-methyl-5,6-dihydroxy-5,6-dihydrothymine isomers as compared with N-1-methylthymine in stacked bar diagram: cis-5R,6S - red bar, cis-5S,6R - orange bar, trans-5R,6R - green bar and trans-5S,6S - blue bar; (c) Position of cis-5R,6S Tg residue in the central part of the duplex II.
ponding to free DNA and DNA-protein complex. The Kd values were obtained using the classical Scatchard method [46] . Each experiment was performed no less than three times; the average values of Kd and the standard deviation (SD) were determined.
2.7. Determination of DNA-Protein Complexes Stoichiometry
To establish the stoichiometry of р50-GST or р65-GST complexes with DNA duplexes, a set of molecular mass markers for nondenaturing electrophoresis (Sigma, USA) was used during EMSA. This set comprised α-lactal- bumin (14,200 Da), carbonic anhydrase (29,000 Da), ovalbumin (45,000 Da), bovine serum albumin (BSA; monomer of 66,000 Da and dimer of 132,000 Da), and urease (trimer of 272,000 Da and hexamer of 545,000 Da). The marker proteins were dissolved in 10 mM Na-phosphate (pH 7.1) with 50 mM NaCl. Experimental procedures and data processing were performed according the manufacturer’s recommendations. EMSA was carried out in a set of PAG containing 4%, 4.5%, 5%, 5.5%, 6%, 7%, 8%, 9%, and 10% polyacrylamide. The relative mobility (Rf) was determined for every protein and every DNA-protein complex as a ratio of the protein or the complex migration distance to the tracking dye (bromophenol blue) migration distance. A plot was made of (100 × log (Rf × 100)) value dependence on the gel percentage and the slope of the obtained line was calculated. Then a calibrating graph was made for the marker proteins, namely a dependence of the negative slope on the molecular mass (in logarithmic coordinates). Using this graph, the molecular mass of the DNA-protein complexes was estimated.
2.8. Detection of DNA Methylation Efficiency by M.SsoII
The 32Р-labeled duplexes VIII and IX (Table 2) were used. The label was in that strand which contained Тg (or the corresponding T in control). 350 nM solution of each duplex in buffer E containing 0.1 mM AdoMet was incubated with 10 nM M.SsoII at 37˚C for 0.5 - 60 min. Then the reaction mixture was kept for 10 min at 65˚C in order to inactivate M.SsoII and cooled down slowly to re-anneal the duplexes. Further, MgCl2 was added up to 1 mM followed by R.SsoII addition up to 240 nM. The reaction mixture was incubated for 1 h at 37˚C. R.SsoII does not hydrolyse the double-stranded sequence 5'-CCNGG-3' when it is methylated [47] . In the control experiment, R.SsoII was added to duplexes VIII and IX without pre-treatment with M.SsoII. The reaction products were analyzed by electrophoresis in 20% PAG with 7 M urea. The degree of DNA methylation by M.SsoII was assessed as the degree of the DNA protection from cleavage by R.SsoII, i.e. as the percentage of the DNA duplex not hydrolysed by R.SsoII. The initial methylation rate was determined graphically as an angular coefficient of the initial linear part of the corresponding kinetic curve. The calculation was carried out not less than for 5 time points. The relative methylation rate was determined as the ratio between the initial methylation rate of the modified DNA duplex and the same value of the corresponding non-modified substrate. Each measurement was performed no less than three times; the SD did not exceed 10%.
3. Results and Discussion
3.1. Synthesis of Tg-Containing Oligonucleotides
The oligonucleotides containing a single Tg lesion were synthesized by the standard phosphoramidite chemistry using the 5R,6S Tg phosphoramidite building block [15] . Since Tg is labile under alkaline conditions, we applied a short cycle of ammonia treatment at room temperature for oligonucleotide deprotection (see Materials and Methods). The duplexes (Table 1, Table 2) were prepared by annealing the complementary DNA strands in equimolar ratio without any differences in the protocol for the Tg-containing and the non-modified DNA duplexes.
3.2. Thermal Stability of DNA Duplexes Containing the Tg Lesion
Each Tg-containing DNA duplex was studied in comparison with a duplex of the same sequence but without oxidative lesion. UV-monitored melting profiles at 260 nm were recorded for the three sets of DNA duplexes, which differ only in one base pair in the internal part and in the position of the Tg residue. The loss of aromaticity in the thymine glycol base resulted in a significant decrease of the duplex stability. The melting temperature (Tm) values of the Tg-containing DNA duplexes were 10˚C - 11˚C lower in comparison with the corresponding duplexes without the Tg lesions (Table 1). These results agree with the data reported in [10] . All the samples demonstrate a single reversible helix-coil transition. However, the melting profiles become broader and hyperchromic effects accompanying the DNA duplex unfolding are distinctly lower as the Tg lesion replaces thymidine in the duplex structure (Table 1). According to our data, the Tm reduction induced by the oxidized thymidine does not depend on the nature of the base pairs surrounding the damaged site. The oligonucleotide mixtures after the melting procedure were analyzed by reverse-phase HPLC (ion-pair mode) in order to determine the integrity of the DNA strands. The analysis proved that a single Tg lesion did not result in oligonucleotide decomposition at elevated temperatures (data not shown). Thus, the increase in optical density at 260 nm of the DNA duplexes was associated only with the double helix melting as mentioned in [15] .
3.3. Effect of the Tg Lesion on DNA Duplex Structure
All the three sets of the DNA duplexes (Table 1) were studied by CD method to estimate the double helix conformational alteration induced by the oxidative damage. For longer DNAs, the local structural disturbances induced by a particular modified nucleotide residue could not be studied by this method, because the CD signal of such a residue is buried in the total signal of the double helix. However, taking into account that the sign and the amplitude of the CD spectrum depend on a geometrical relationship between the coupled electronic transition dipoles on adjacent nucleotides and on the distance between the bases, a sequence-dependent impact of the Tg lesion onto the CD spectra of short 17-bp DNA duplexes would be expected.
At the temperature conventionally used in CD experiments (22˚C), all the complementary oligonucleotides are in duplex form, including the modified ones. The CD spectra of all the DNA duplexes have a conservative shape with the positive Cotton effect, which is typical for the DNA B-form (Figure 2). Nevertheless, the spectral changes induced by Tg are still observed in each set of the DNA duplexes, their character being dependent on the type of the base pairs flanking the Tg lesion. Incorporation of the Tg/A pair (instead of T/A) between G/C and C/G base pairs (duplex II) does not change the spectrum shape, but leads to a reduced intensity of both positive and negative peaks centered at 275 nm and 240 nm, respectively (Figure 2(a)). The CD spectrum of the duplex IV containing Tg flanked by A/T and T/A base pairs undergoes more significant changes in comparison with the undamaged duplex III. They include broadening and increased intensity of the positive peak, a blue shift in the maximum of this peak (from 278 to 272 nm) and in zero point transition (Figure 2(b)). This shift together with decreasing the low wavelength negative peak is indicative of the B to A form partly conformational transition. The spectral changes resulted from the Tg residue flanked by the A/T and G/C base pairs (duplex VI) are comparable to those of the duplex IV, where Tg is positioned between the A/T and T/A base pairs, but they are less intense (Figure 2(c)).
The role of sequence context in the Tg-induced changes in the CD spectra could be explained by difference in stacking and/or steric interactions between the Tg residue and the flanking nucleotide pairs. Thymine glycol is known to remain in the Watson-Crick orientation with respect to the complementary adenine, although it disrupts hydrogen bonding interactions within the Tg/A pair [10] . The C5 methyl group at the 5R Tg is shown to protrude axially from the damaged pyrimidine ring and to hinder stacking with the 5'-adjacent base, especially with purine. In the 3'-direction, stacking between Tg and the adjacent nucleotide pair is disrupted as well [10] . In addition, Tg causes destabilization of the 5'-neigboring nucleotide pair. An NMR structure of a duplex DNA containing Tg in the sequence 5'-ATgA-3' suggests that the Tg lesion is largely extrahelical [48] , while in the

Figure 2. CD spectra of the unmodified DNA duplex (solid line) and the corresponding Tg-containing duplex (dashed line). The duplex pairs are the following: I and II (а), III and IV (b), and V and VI (c).
sequence 5'-GTgG-3', Tg remains either stacked or partially stacked within the double helix [49] . According to our data, the extent of the alterations in the CD spectra induced by Tg depends on the number of the A/T pairs flanking the damaged site. These alterations could be attributed to a different degree of the Tg extrahelicity or/and to the Tg shift to the helix periphery, as in the case of A-form DNA.
Thus, the CD spectroscopy data confirm that the Tg-containing DNA double helixes adopt the expected B-type structure in solution and the oxidative damages do not provoke global structural disturbances. The 5R,6S Tg lesion induces significant localized conformational changes which are sequence-dependent.
3.4. Molecular Dynamics Simulation of Tg-Containing B-DNA Duplex
Molecular dynamics simulations were performed for the DNA duplex II containing the cis-5R,6S Tg. As an accurate estimation of free energy difference in simulation requires substantial increase of computer resources, we focused on the Tg influence on duplex structure and its behavior during temperature increase. Since Tg is not included in the list of canonical nucleosides, the structures for the cis-5R,6S, cis-5S,6R, trans-5R,6R, and trans- 5S,6S isomers of Tg were created on the basis of thymidine structure.
Charges of heterocyclic base were determined for every Tg isomer (see e.g. Figure 1(a)). Sugar-phosphate backbone was replaced with a СН3 group. Such substitution permitted us to decrease the calculation time. The comparison of atom charges of different thymine glycol isomers with thymine revealed that the smallest standard deviation value belongs to cis-5R,6S isomer (0.079) while the largest one is associated with cis-5S,6R isomer (0.097). Thus, cis-5R,6S isomer of thymine glycol used in our work is most similar to thymine in terms of atom charge distribution (Figure 1(b)).
Superposition of 3D structure of each of thymine glycole isomers to thymine demonstrated similarity of modified and non-modified bases. The minimal difference is observed for cis-5S,6R isomer of thymine. The unmodified duplex I and the duplex II containing the cis-5R,6S Tg were subjected to MD simulations in explicit solvent using Parmbsc0 force field [41] .
The Tg residue was found to be shifted towards the DNA major groove compared to the original T residue (Figure 1(c)). Despite this shift and the loss of aromaticity, one hydrogen bond remains between Tg and A, namely between H3 of Tg and N1 of А. (The bond between О4 of Tg and hydrogen of 6-NH2 group of А is disrupted). Thus, the Tg position in the duplex II can be considered partially extrahelical (Figure 1(c)), as it is observed by NMR in [48] . The Tg also destabilizes its 5'-neighboring G/C pair [49] .
Since MD simulations allow observing system dynamics only in narrow time windows, it is difficult to simulate system behavior near equilibrium. We focused on the local perturbations around Tg to evaluate the initial changes in the modified duplex. Obviously, Tg presence influences stacking interactions and hydrophobicity of the whole molecule. Hydrophobic effect depends on temperature and we focused on conformational fluctuations during simulation with temperature change. Tg results in an increase of atom fluctuations in the duplex II in comparison to the unmodified duplex I, mainly on the A/T-rich 5'-side from the Tg residue (Figure 3). The root-mean-square deviation of the DNA backbone heavy atoms is calculated to estimate the structure mobility of the duplexes I and II. Several superimposed atom fluctuation curves for the Tg-containing duplex II in the temperature range 300 - 365 K together with those for the duplex I at 300 K are presented in Figure 3. Observed atom fluctuations increase at higher temperature supports the mechanical duplex instability near Tg. For all the MD trajectories, the Tg lesion exhibits an increased hydrophobic surface relative to thymidine, thus supporting the weakened base stacking between the cis-5R,6S Tg and the neighboring nucleotide pairs, and partly extrahelical conformation of the thymine glycol base (Figure 1(c)).
3.5. NF-κB Interaction with the Tg-Containing DNA Duplexes
Transcription factors from the NF-κB family have a region of high sequence similarity at the N-terminus which is called Rel-homology region (RHR). The NF-κB subunits associate in a combinatorial manner forming homo- or heterodimers. Their target sites in DNA differ in length from 9 to 11 bp and are referred to as κB DNA. The consensus sequence for the 10-bp κB site is the following: 5'-GGGRNWYYCC-3'/3'-GGRRWNYCCC-5' (where R = A or G, N = any nucleotide, W = A or T, and Y = C or T). Since the sequence is degenerated, many variants can be found in gene promoters [27] [50] . Several rules for the target recognition by different NF-κB dimers were deduced from the X-ray structures [23] . First, p50 and p52 recognize the 5'-GGGRN-3'/3'-NYCCC-5' half-site, while p65 binds to the 5'-YYCC-3'/3'-GGRR-5' half-site. One of the specificity determinants is the 5'-terminal
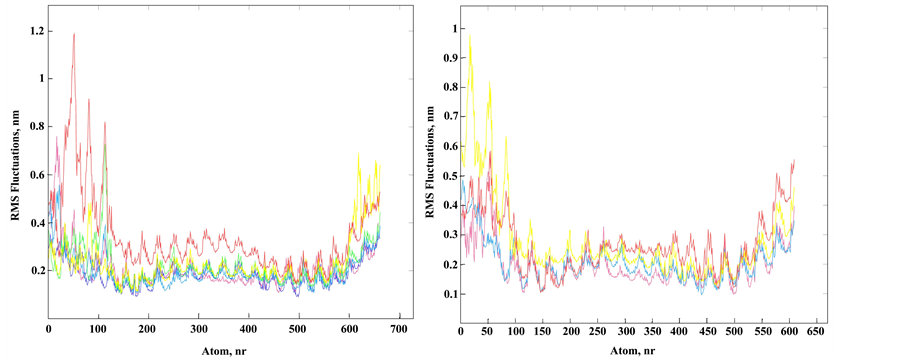
Figure 3. Structural fluctuation in MD of the DNA duplexes I and II. (a) Superimposed atom fluctuations for the Tg strand of the Tg-containing duplex II and appropriate strand of unmodified duplex I. Fluctuations for duplex I at 300 K (magenta) and fluctuations for duplex II in spectrum colors from blue to red according to temperatures 300 K (blue), 320 K, 336 K, 350 K and 356 K (red); (b) The same plots for the strand complementary to Tg strand (unmodified strand) of duplex II (spectrum colors) and appropriate strand of duplex I (magenta).
G/C pair in the 5-bp half-site. It is contacted by a side chain of His residue which is specific for p50 and p52 subunits and is replaced by Ala in p65 and c-Rel subunits [31] . Moreover, the 5'-G/C pair presence negatively affects p65 and c-Rel binding by altering the geometry of the adjacent GG/CC block. Second, p65 and c-Rel homo- and heterodimers prefer the κB sites which contain five or six A/T pairs in the center. Thus, structural and biochemical works allowed to divide κB sites in two classes: class I κB DNA comprises 10-bp and 11-bp κB sites which bind to p50 and p52 subunits, while class II κB DNA has a length of 9 bp and is recognized specifically by p65 and c-Rel subunits [31] [51] .
In the present work, two types of human NF-κB transcription factors are studied, р50 and р65. Each subunit is a fusion protein and carries N-terminal glutathione-S-transferase (GST) domain. GST allows purification of the target protein from E. coli cells via affine chromatography [36] . The GST-p50 and GST-p65 fusions have been successfully used in a number of in vitro studies (for example, [37] [52] - [57] ). The recombinant р50 contains the full-length sequence of the wild-type p50 while the recombinant p65 contains only residues 1 - 306 of the wild-type p65 (i.e. the RHR). The recombinant proteins have molecular masses of 70.1 kDa (р50-GST) and 61.8 kDa (p65-GST).
Three DNA duplexes (Table 1) were used to assess Tg impact on the κВ site recognition by NF-κB. Tg (or T in the control duplexes) was placed into one of the degenerate positions in the κВ site. The DNA duplex I contains κB site from immunoglobulin κ (Ig-κB) light chains enhancer. Each one of the duplexes III and V carries one substitution in the site: T/A instead of G/C in the position 16 and T/A instead of A/T in the position 15, respectively. In each case, the resulting sequence is compatible with the consensus for 10-bp κВ site.
Complex formation between р50 or р65 and each one of the DNA duplexes I - VI has been analyzed using EMSA (see Figure 4 as an example).
Unexpectedly, two different complexes (C1 and C2 in Figure 5) are detected in the case of both modified and parent duplexes. Under the experimental conditions, almost no complexes are observed between р50 or р65 and a non-specific duplex 5'-TCAGCACCCAGGGTGCC-3'/3'-AGTCGTGGGTCCCACGG-5' (VII), i.e. the yield is less than 1% (Figure 4). Thus, one can conclude that complexes C1 and C2 are the specific ones. The stoichiometry of these DNA-protein complexes is determined using Ferguson plots [58] . Briefly, molecular mass of each particle is calculated based on its electrophoretic mobility in polyacrylamide gels of different percentage. A commercial set of proteins is used to obtain the calibration curve. Despite a large experimental error, the method accuracy is suitable for discrimination of different protein oligomeric states. The molecular masses obtained point unambiguously to large complexes containing the protein dimers and tetramers (Table 3).
NF-κB is functionally active as a dimer which interacts with one κB site in double-stranded DNA, as is testified
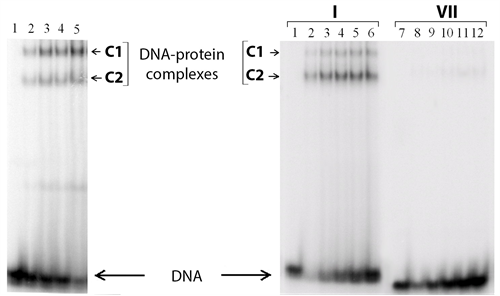
Figure 4. Analysis of р50-GST and р65-GST interaction with DNA duplexes. The reaction mixtures were incubated at 25˚C for 30 min and analysed by non-denaturing PAG electrophoresis followed by radioautography. (а) Complex formation between p65-GST and 32P-labeled DNA duplex I (40 nM; contains the NF-κВ recognition site) in the presence of 25 μg/ml poly(dI・dC). Protein concentrations in the mixtures were 1.6, 3.2, 4.8, and 8.4 µM per monomer (lanes 2 - 5, respectively). Lane 1 is a control mixture which contains only DNA without protein; (b) Complex formation between p50-GST (1.9 µM per monomer) and 32P-labeled DNA duplexes I (with the κВ site) or VII (without the κВ site) in the absence of poly (dI・dC). The concentrations of duplexes I and VII were 40, 80, 120, 160, and 200 nM (lanes 2 - 6 and 8 - 12, respectively). Lanes 1 and 7 are control mixtures which contain only DNA.
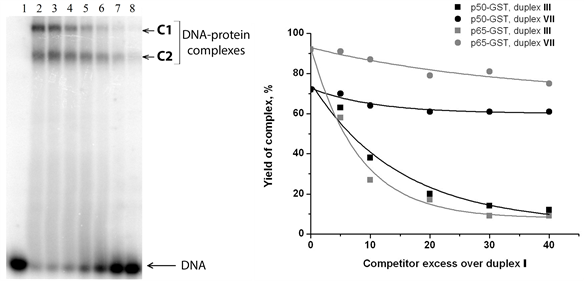
Figure 5. Analysis of р50-GST and р65-GST interaction with DNA duplexes containing the NF-κВ recognition site in the presence of competitor DNA. (а) Complex formation between p50-GST (2.1 µM per monomer) and 32P-labeled DNA duplex V (100 nM; contains the κВ site) in the presence of poly(dI・dC) at different concentrations: 3.13, 6.25, 12.5, 25, 50, or 100 μg/ml (lanes 2 - 8, respectively). Lane 1 is a control mixture containing only DNA without protein. The reaction mixtures were incubated at 25˚C for 30 min and analysed by non-denaturing PAG electrophoresis followed by radioautography; (b) The specificity of complex formation between 32P-labeled duplex I (50 nM) and р50-GST (3.8 µM per monomer) or p65-GST (0.3 µM per monomer) was tested by competition assay. Unlabeled duplex III (with the κВ site) was used as a specific competitor, while unlabeled duplex VII (without the κВ site) served as a non-specific competitor. The yield of the 32P-labeled DNA-protein complexes decreases depending on the competitor excess over the 32P-labeled duplex I. Average values of 3 experiments are shown; the standard deviation did not exceed 5%.
Table 3. Stoichiometry of complexes formed between DNA duplex III (6.3 kDa) and р50-GST (70.1 kDa) or р65-GST (61.8 kDa).
by various X-ray structures [59] - [61] . However, homotetramers can be formed by full-length p50 [62] [63] . The C1/C2 complex ratio remains constant at the DNA concentration ranged from 0 to 200 nM (data not shown) which corroborates the assumption that oligomerization does not depend on the DNA-binding domain, i.e. it is provided by GST. Indeed, GST exits as dimer in crystal structures [64] [65] and does not interact with DNA [37] . Complex of р50-GST with the 32P-labeled duplex I was titrated with non-labeled duplexes III or VII (Figure 5(a)). Increasing concentrations of duplex III disrupted the complexes between р50-GST and duplex I, both C1 and C2 forms. This fact indicates that DNA-binding centre of p50-GST retains its specificity when this protein is recruited into the large complex (C1) by the GST domain. The duplex VII had only minor effect, which can be explained by a weak affinity to any non-specific DNA typical for every DNA-binding protein. The same results were obtained for the complex between р65-GST and 32P-labeled duplex I titrated with non-labeled duplexes III and VII (Figure 5(b)). Taken together, these results suggest partial tetramerization of p50-GST and p65- GST provided by the GST domain.
The dissociation constants for the DNA-protein complexes were determined using the Scatchard plot [46] . This method implies a one-step reaction where only one complex is formed. In our case, two complexes are present in every reaction mixture with the constant C1/C2 ratio over the whole range of DNA protein concentration ratios testifying no connection between the oligomerization and the DNA-binding center. Therefore, we take the sum of the two complexes in each sample and use the classical Scatchard method.
The Kd values of р50-GST complexes with the non-modified duplexes I, III, and V differ among themselves up to 1.4-fold (Table 1). Thus, all the one-nucleotide substitutions do not change dramatically р50 interaction with the κB site. The Kd values of р50-GST complexes with the oxidized duplexes II, IV, and VI depend on the Tg position inside the κB site. Surprisingly, Т oxidation into Tg slightly improves р50-GST binding to the duplexes II and VI where the Tg/A pair is located between two G/C pairs or between G/C and A/T pairs. A similar improvement was observed for р50 homodimer binding to the NF-κB promoter element where G residue preceding the κB site was replaced by 8-oxoG [32] .
On the other hand, the presence of Tg/A pair between two А/Т pairs reduces р50-GST affinity to the DNA (duplex IV) ~2.7-fold. As mentioned above (see also [48] ) oxidation of thymidine residue (especially flanked by А/Т pairs from the both sides) results in Tg extrahelical localization and in the changed conformation of the 5'- phosphate group of the Tg nucleotide. A generalized scheme of interaction between р50/р50 dimer and its target DNA [66] suggests that maximal number of the DNA protein contacts should be broken when Tg is incorporated in the position 16 of the κB site, as found here.
In contrast to р50-GST, р65-GST demonstrates different affinity to the non-modified duplexes I, III, and V (difference in Kd up to 4.5-fold). The most effective binding is observed for the duplex III which contains five A/T pairs at the center, in agreement with the rules described in [31] . The Тg presence in the κB site impairs р65-GST binding to such DNA 3-4-fold (Table 1). The weaker is р65-GST interaction with the non-modified duplex, the weaker is its interaction with the oxidized duplex. In the crystal structure of р65 homodimer with the 9-bp κB-33 site (5'-G2A3A4A5T6T7T8C9C10-3'/3'-C19T18T17T16A15A14A13G12G11-5', positions numbered corresponding to the general consensus of the κB site; PDB entries: 1 ram, 2 ram), the methyl groups of T residues in positions 16 and 17 form van der Waals contact with the side chain of Tyr36 [61] . Most likely, Tg presence in the positions 16 or 17 impairs this contact leading to the decrease of р65-GST affinity to such DNA.
Until now the relationship between efficiency of binding of NF-κB to DNA and its transcriptional activity has not yet been revealed. For example, it is still unclear if stable binding of NF-κB to DNA is obligatory for its function, or if weaker interactions compatible with transitory contacts are more preferable [67] . However, а fundamental feature of NF-κB-mediated transcriptional response to extracellular signals is its high selectivity that provides activation of only one set of targeted genes. So, we can suppose that lack of proper DNA recognition by this protein could lead to some of biological effects because NF-κB stays at the intersection of different metabolic pathways and physiological processes [68] .
Thymidine glycol, as many other products of oxidation process is repaired in a cell readily and does not lead to a permanent change in DNA sequence. There are several types of repair systems which recognize and eliminate Tg, thus restoring the original canonical structure of DNA [14] . However, in case one Tg residue in a κB site remains unrepaired, it could influence functioning of NF-κB transcription factors. We have shown that the observed effect depends on the dimer composition and on the Tg position in the κB site. p50-GST is relatively tolerant to thymidine oxidation in the case of duplexes II and VI. А minor impact on p50 homodimer binding affinity was also observed in the case of another type of oxidative modification when G1 or G3 in the κB site was substituted by 8-oxoG [32] . The difference in Kd values of the p50-GST complexes with the oxidized duplex IV and the corresponding unmodified duplex III does not reach 3-fold. In a similar way, the affinity of p50 dimer to a duplex with oxidized G2 is decreased 4-fold [32] .
The changes in binding to Tg-containing DNA are more significant in the case of p65-GST (Table 1). The affinity of p65-GST decreases at interaction with all the three oxidized duplexes (II, IV, and VI) up to 4.5-fold. Is this difference essential or not? On one hand, the differences in NF-κB affinity to the κB sites based on variations in their oxidation status are not so big and are comparable with the ones derived from the natural diversity of the κB site sequences. For example, р65-GST affinity to the unmodified variants of the κB site (duplexes I and III) also differs up to 4.5-fold (Table 1). This fact demonstrates that a protein activated during oxidative stress should be resistant to the oxidative damages in DNA up to a certain degree.
Оn the other hand, we are sure that at least 4-fold decrease of p65 affinity to its oxidized site should be essential for biological processes. It is known that NF-κB dimers, when they bind to the κB sites in promoter/enhancer areas of their target genes, regulate transcription by involving coactivators and corepressors in this process. Importantly, transcription activation domain (TAD) is needed for positive regulation of gene expression. The only p65, c-Rel и RelB from NF-κB family have this domain. p50 or p52 homodimers which are not able to involve coactivator complexes, can occupy the κB sites in non-stimulated cells [68] . Some other nuclear proteins also could be bound to the κB sites and might be strong competitors for p65 homodimer binding to its target sequence. As a result, p65/p65 might not activate transcription under such conditions, because the oxidized sites may be occupied by other proteins and simply may not be accessible for p65 homodimer. For example, based on our data κB site with Tg modification decreases affinity of p65 to this DNA and at the same time lead to a better binding of this site to p50 homodimer as it was determined for duplexes II and VI (Table 1). This event can considerably decrease or block further production of certain proteins being synthesized under normal conditions. It could lead to different scenarios of cell development [68] .
3.6. Methyltransferase SsoII Interaction with the Tg-Containing DNA Duplexes
Further, we investigate the influence of Tg presence on functionality of M.SsoII which also can work as a self- regulated transcription factor. Its transcription regulation is easy to follow as compared with the more complex transcription pathway of NF-κB proteins.
M.SsoII is a bifunctional protein consisting of two domains, C-terminal and N-terminal ones. The C-terminal domain of M.SsoII methylates the second cytosine in the double-stranded sequence 5'-CCNGG-3'/3'-CCNGG-5' where N is any nucleotide [69] . On the other hand, M.SsoII may function as a transcription factor when its another (N-terminal) domain binds to the regulatory site 5'-AGGACAATTTGTCCT-3'/3'-AGGACAAATTGTCCT-5', a 15-bp quasipalindromic sequence [33] . The latter activity is the key feature for functioning of the restriction-modification system SsoII. Briefly, M.SsoII binding to the regulatory site represses transcription of its own gene and stimulates transcription of the gene encoding its cognate restriction endonuclease (R.SsoII). R.SsoII recognizes the same sequence as M.SsoII does and cleaves DNA in case this site remains non-methylated. Typically, DNA of a host cell is fully methylated and only alien DNA (e.g. in case of a phage infection) remains non-modified and thus prone to cleavage by R.SsoII. Therefore, the restriction−modification system SsoII can protect its host cell from phage infection.
In the present research, the influence of thymidine oxidation in the recognition sites on both M.SsoII functions (as a methyltransferase and as a transcription regulator) was studied. Two values were assessed in the case of the methylation site, M.SsoII affinity to DNA duplexes VIII and IX along with efficiency of their methylation by M.SsoII. In the case of the regulatory site, only M.SsoII affinity to DNA duplexes X - XIII was measured (Table 2).
Influence of Tg presence on M.SsoII methytransferase function was assessed using duplexes VIII and IX which carried T or Tg in central degenerated position of M.SsoII methylation site. The classical Scatchard plot [46] was used for determination of the Kd for the following DNA-protein complexes: M.SsoII-duplex VIII and M.SsoII-duplex IX. The M.SsoII was found to bind 3-fold less effectively to the Tg-containing duplex IX.
The methylation efficiency was measured indirectly. At first, DNA duplex was modified by M.SsoII and then R.SsoII was added to the reaction mixture. In the control experiments, R.SsoII was added to DNA duplexes not treated with M.SsoII. We have shown earlier that R.SsoII demonstrated an increased catalytic activity towards a Tg-containing duplex compared to a non-modified control duplex [15] . This feature provides high sensitivity for the analysis performed using R.SsoII. The hydrolysis products were detected by electrophoresis in 20% PAG with 7 M urea. The native duplex VIII was fully protected from the cleavage by R.SsoII after 15-min incubation with M.SsoII. In contrast, the Tg-containing duplex IX was cleaved with high efficiency demonstrating less than 20% methylation (Figure 6). These results show that thymidine oxidation in the sequence 5'-CCTGG-3'/3'- GGACC-5' prevents its methylation by M.SsoII. It can be partially explained by the impaired M.SsoII binding to the oxidized site.
Thus, Tg presence differently influences enzymatic properties of SsoII pair enzymes, M.SsoII and R.SsoII. Since Tg is located in the degenerate position of the methylation site, its base is not expected to form any contacts with the protein. However, local deformations and conformational mobility of the double helix near the Tg residue promotes DNA hydrolysis by R.SsoII, but might prevent M.SsoII from forming proper contacts with the phosphate groups of the DNA backbone.
To investigate the Tg impact on the transcription regulation by M.SsoII, 31-bp DNA duplexes X - XIII were used (Table 2). As shown by chemical footprinting with hydrazine, two thymidine residues in the palindromic regulatory site form direct contacts with M.SsoII [70] . They are located symmetrically, each one in the proper half-site. These residues were substituted with Tg (Table 2). As the regulatory site is palindromic, two M.SsoII molecules normally bind to one site [45] . However, the protein dimer can assemble the proper complex only when the DNA is long enough (60 bp and more) [39] . The 31-bp duplexes used in the present work allow to detect only the intermediate complex where one M.SsoII molecule binds to one DNA duplex ― either to the “left” half or to the “right” half of the regulatory site [34] [70] [71] . These two complexes have equal molecular mass and therefore cannot be distinguished by EMSA. Thus, the Kd for the duplex X is the result for two different DNA-protein complexes. The apparent Kd values (Table 2) demonstrate that M.SsoII affinity to the duplexes XI and XII is 2-fold lower than this value for the duplex X. This result suggests that the Tg presence ruins the half of the regulatory site and only the other part of the site remains recognizable for the protein. Indeed, there is no detectable binding of M.SsoII to duplex XIII which contains Tg in both half-sites even at 240-fold protein excess over DNA. The apparent Kd values for the duplexes XI and XII are almost equal demonstrating that M.SsoII binding does not discriminate the “left” and the “right” halves of the regulatory site.
Taken together, these data show that M.SsoII affinity to the regulatory site is more sensitive to oxidative stress than M.SsoII affinity to the methylation site. Since M.SsoII represses transcription of its own gene and stimulates transcription of the R.SsoII gene, oxidation of thymidine residue in the regulatory site is likely to disrupt such regulation and therefore result in producing more M.SsoII molecules and less R.SsoII ones. Increasing concentration of M.SsoII may compensate its low activity towards the oxidized methylation sites. However,
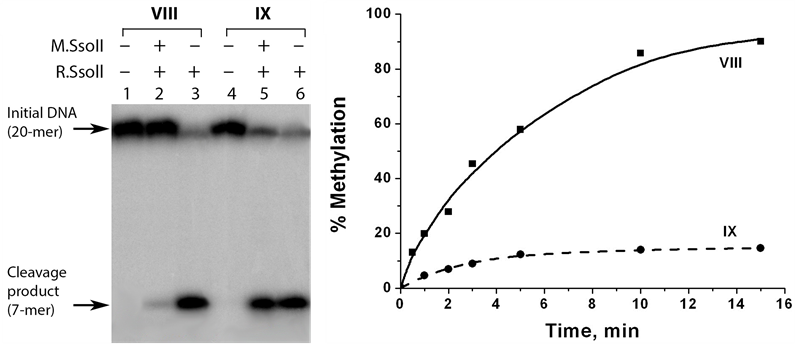
Figure 6. Analysis of methylation of DNA duplexes VIII and IX by М.SsoII. The efficiency of the DNA methylation is assessed on the basis of the DNA protection from hydrolysis by R.SsoII. (a) Analysis of the reaction mixtures was performed by electrophoresis in 20% PAG with 7 М urea followed by radioautography. Lanes 1 and 4, the initial duplexes VIII and IX (32Р-labeled at the “top” strands). Lanes 2 and 5, duplexes VIII and IX treated with M.SsoII and then with R.SsoII. Lanes 3 and 6, non-methylated duplexes VIII and IX hydrolysed by R.SsoII. The reaction mixtures contained 0.35 µM DNA, 92 nM M.SsoII, and 4.8 µM R.SsoII. DNA methylation and DNA hydrolysis were carried out at 37˚C for 15 min and 1 h, respectively; (b) Typical kinetic curves for methylation of duplexes VIII and IX. DNA methylation was performed at 37˚C and stopped at different time points within 15 min. DNA hydrolysis was carried at 37˚C for 1 h.
methylation of non-oxidized sites would be highly promoted under such conditions while DNA cleavage by R.SsoII would be less likely. Thus, we can speculate that the restriction-modification system SsoII would not protect its host cell from phage infection under oxidative stress conditions.
4. Conclusion
Single Tg residue incorporated in the different positions of 17-bp DNA duplexes was shown to destabilize the double helix in a sequence independent manner. In contrast, the extent of the alterations in the CD spectra induced by Tg depends on the number of the A/T pairs flanking the damaged site. According to MD simulations data, the Tg lesion exhibits an increased hydrophobic surface relative to thymidine, thus supporting the weakened base stacking and partly extrahelical position of the Tg base. The Tg containing DNA duplexes were used to study the damage impact on the recognition and functioning several site-specific DNA-binding proteins. p50/ p50 and p65/p65 homodimers of NF-κB were shown to have different affinity to the oxidized κB site depending on the Tg position. The changes in binding to Tg-containing DNA as compared with a non-modified one are more significant in the case of p65/p65. The difference of NF-κB dimers behavior towards oxidative damages in DNA probably has some input into biological processes. The data obtained show that M.SsoII binding with the regulatory site is more sensitive to the presence of Tg residue than its binding with the methylation site. We have demonstrated that M.SsoII does not act as a transcription factor under conditions of oxidative stress, in contrast to NF-κB.
Acknowledgements
We thank Prof. Anna Karyagina, Prof. Alain Israёl, and Prof. Neil Perkins for kindly providing us with plasmids encoded SsoII restriction-modification enzymes, p50 and p65 subunits of NF-κB transcription factor. This research was supported by Russian Scientific Foundation, grant N 14-14-00489 (the interaction of NF-κB protein with modified DNA fragments) and Russian Foundation for Basic Research, grant N 13-04-00165 (the synthesis of DNA fragments, physico-chemical properties and interaction with SsoII protein).
Cite this paper
Elena A.Kubareva,FanYang,Alexandra Yu.Ryazanova,Nina G.Dolinnaya,Andrei V.Golovin,Nikolai V.Molochkov,Elena A.Romanova,Elizaveta A.Karpova,Timofei S.Zatsepin,Tatiana S.Oretskaya, (2015) Thymidine Glycol: The Effect on DNA Structure and DNA Binding by Site-Specific Proteins. Natural Science,07,491-509. doi: 10.4236/ns.2015.711051
References
- 1. Frenkel, K., Goldstein, M.S. and Teebor, G.W. (1981) Identification of the cis-Thymine Glycol Moiety in Chemically Oxidized and Gamma-Irradiated Deoxyribonucleic Acid by High-Pressure Liquid Chromatography Analysis. Biochemistry, 20, 7566-7571. http://dx.doi.org/10.1021/bi00529a035
- 2. Teebor, G., Cummings, A., Frenkel, K., Shaw, A., Voituriez, L. and Cadet, J. (1987) Quantitative Measurement of the Diastereoisomers of cis Thymidine Glycol in Gamma-Irradiated DNA. Free Radical Research Communications, 2, 303-309. http://dx.doi.org/10.3109/10715768709065296
- 3. Wallace, S.S. (2002) Biological Consequences of Free Radi-cal-Damaged DNA Bases. Free Radical Biology & Medicine, 33, 1-14. http://dx.doi.org/10.1016/S0891-5849(02)00827-4
- 4. Adelman, R., Saul, R.L. and Ames, B.N. (1988) Oxidative Damage to DNA: Relation to Species Metabolic Rate and Life Span. Proceedings of the National Academy of Sciences of the United States of America, 85, 2706-2708. http://dx.doi.org/10.1073/pnas.85.8.2706
- 5. Zuo, S., Boorstein, R.J. and Teebor, G.W. (1995) Oxidative Damage to 5-Methylcytosine in DNA. Nucleic Acids Research, 23, 3239-3243. http://dx.doi.org/10.1093/nar/23.16.3239
- 6. Cathcart, R., Schwiers, E., Saul, R.L. and Ames, B.N. (1984) Thymine Glycol and Thymidine Glycol in Human and Rat Urine: A Possible Assay for Oxidative DNA Damage. Pro-ceedings of the National Academy of Sciences of the United States of America, 81, 5633-5637. http://dx.doi.org/10.1073/pnas.81.18.5633
- 7. Their, R., Brüning, T., Kocher, K., Blaszkewicz, M., Makropoulos, V., Sundberg, A. and Bolt, H.M. (1999) Determination of Urinary Thymidine Glycol Using Affinity Chromatography, HPLC and Post-Column Reaction Detection: A Biomarker of Oxidative DNA Damage upon Kidney Transplantation. Archives of Toxicology, 73, 479-484. http://dx.doi.org/10.1007/s002040050638
- 8. Makropoulos, W., Kocher, K., Heintz, B., Schwarz, E.R., Mertens, P.R. and Stefanidis, I. (2000) Urinary Thymidine Glycol as a Biomarker for Oxidative Stress after Kidney Transplantation. Renal Failure, 22, 499-510. http://dx.doi.org/10.1081/JDI-100100891
- 9. Lowe, F.J., Luettich, K. and Gregg, E.O. (2013) Lung Cancer Biomarkers for the Assessment of Modified Risk Tobacco Products: An Oxidative Stress Perspective. Biomarkers, 18, 183-195. http://dx.doi.org/10.3109/1354750X.2013.777116
- 10. Brown, K.L., Adams, T., Jasti, V.P., Basu, A.K. and Stone, M.P. (2008) Interconversion of the cis-5R,6S- and trans- 5R,6R-Thymine Glycol Lesions in Duplex DNA. Journal of the American Chemical Society, 130, 11701-11710. http://dx.doi.org/10.1021/ja8016544
- 11. Basu, A.K., Loechler, E.L., Leadon, S.A. and Essigmann, J.M. (1989) Genetic Effects of Thymine Glycol: Site-Specific Mutagenesis and Molecular Modeling Studies. Journal of the American Chemical Society, 86, 7677-7681. http://dx.doi.org/10.1073/pnas.86.20.7677
- 12. Belousova, E.A., Maga, G., Fan, Y., Kubareva, E.A., Romanova, E.A., Lebedeva, N.A., Oretskaya, T.S. and Lavrik, O.I. (2010) DNA Polymerases Beta and Lambda Bypass Thymine Glycol in Gapped DNA Structures. Biochemistry, 49, 4695-4704. http://dx.doi.org/10.1021/bi901792c
- 13. Aller, P., Duclos, S., Wallace, S.S. and Doublie, S. (2011) A Crystallographic Study of the Role of Sequence Context in Thymine Glycol Bypass by a Replicative DNA Polymerase Serendipitously Sheds Light on the Exonuclease Complex. Journal of Molecular Biology, 412, 22-34. http://dx.doi.org/10.1016/j.jmb.2011.07.007
- 14. Dolinnaya, N.G., Kubareva, E.A., Romanova, E.A., Trikin, R.M. and Oretskaya, T.S. (2013) Thymidine Glycol: The Effect on DNA Molecular Structure and Enzymatic Processing. Biochimie, 95, 134-147. http://dx.doi.org/10.1016/j.biochi.2012.09.008
- 15. Yang, F., Romanova, E., Kubareva, E., Dolinnaya, N., Gajdos, V., Burenina, O., Fedotova, E., Ellis, J.S., Oretskaya, T., Hianik, T. and Thompson, M. (2009) Detection of DNA Damage: Effect of Thymidine Glycol Residues on the Thermodynamic, Substrate and Interfacial Acoustic Properties of Oligonucleotide Duplexes. The Analyst, 134, 41-51. http://dx.doi.org/10.1039/B806604N
- 16. Wan, F. and Lenardo, M.J. (2010) The Nuclear Signaling of NF-κB: Current Knowledge, New Insights, and Future Perspectives. Cell Research, 20, 24-33. http://dx.doi.org/10.1038/cr.2009.137
- 17. McCool, K.W. and Miyamoto, S. (2012) DNA Damage-Dependent NF-κB Activation: NEMO Turns Nuclear Signaling Inside Out. Immunological Reviews, 246, 311-326. http://dx.doi.org/10.1111/j.1600-065X.2012.01101.x
- 18. Gerondakis, S., Banerjee, A., Grigoriadis, G., Vasanthakumar, A., Gugasyan, R., Sidwell, T. and Grumont, R.J. (2012) NF-κB Subunit Specificity in Hemopoiesis. Immunological Reviews, 246, 272-285. http://dx.doi.org/10.1111/j.1600-065X.2011.01090.x
- 19. Mincheva-Tasheva, S. and Soler, R.M. (2013) NF-κB Signaling Pathways: Role in Nervous System Physiology and Pathology. Neuroscientist, 19, 175-194. http://dx.doi.org/10.1177/1073858412444007
- 20. Oh, H. and Ghosh, S. (2013) NF-κB: Roles and Regulation in Different CD4+ T-Cell Subsets. Immunological Reviews, 252, 41-51. http://dx.doi.org/10.1111/imr.12033
- 21. Niederberger, E. and Geisslinger, G. (2013) Proteomics and NF-κB: An Update. Expert Review of Proteomics, 10, 189-204. http://dx.doi.org/10.1586/epr.13.5
- 22. O’Dea, E. and Hoffmann, A. (2010) The Regulatory Logic of the NF-κB Signaling System. Cold Spring Harbor Perspectives in Biology, 2, a000216. http://dx.doi.org/10.1101/cshperspect.a000216
- 23. Ghosh, G., Wang, V.Y., Huang, D.B. and Fusco, A. (2012) NF-κB Regulation: Lessons from Structures. Immuno- logical Reviews, 246, 36-58. http://dx.doi.org/10.1111/j.1600-065X.2012.01097.x
- 24. Gilmore, T.D. (2006) Introduction to NF-κB: Players, Pathways, Perspectives. Oncogene, 25, 6680-6684. http://dx.doi.org/10.1038/sj.onc.1209954
- 25. Saccani, S., Pantano, S. and Natoli, G. (2003) Modulation of NF-κB Activity by Exchange of Dimers. Molecular Cell, 11, 1563-1574. http://dx.doi.org/10.1016/S1097-2765(03)00227-2
- 26. Kabe, Y., Ando, K., Hirao, S., Yoshida, M. and Handa, H. (2005) Redox Regulation of NF-κB Activation: Distinct Redox Regulation between the Cytoplasm and the Nucleus. Antioxidants & Redox Signaling, 7, 395-403. http://dx.doi.org/10.1089/ars.2005.7.395
- 27. Kunsch, C., Ruben, S.M. and Rosen, C.A. (1992) Selection of Optimal Κ-B/Rel DNA-Binding Motifs: Interaction of Both Subunits of NF-κB with DNA Is Required for Transcriptional Activation. Molecular and Cellular Biology, 12, 4412-4421. http://dx.doi.org/10.1128/MCB.12.10.4412
- 28. Tisne, C., Delepierre, M. and Hartmann, B. (1999) How NF-κB Can Be Attracted by Its Cognate DNA. Journal of Molecular Biology, 293, 139-150. http://dx.doi.org/10.1006/jmbi.1999.3157
- 29. Tisne, C., Hartmann, B. and Delepierre, M. (1999) NF-κ B Binding Mechanism: A Nuclear Magnetic Resonance and Modeling Study of a GGG → CTC Mutation. Biochemistry, 38, 3883-3894. http://dx.doi.org/10.1021/bi982402d
- 30. Wecker, K., Bonnet, M.C., Meurs, E.F. and Delepierre, M. (2002) The Role of the Phosphorus BI-BII Transition in Protein-DNA Recognition: The NF-κB Complex. Nucleic Acids Research, 30, 4452-4459. http://dx.doi.org/10.1093/nar/gkf559
- 31. Huang, D.B., Phelps, C.B., Fusco, A.J. and Ghosh, G. (2005) Crystal Structure of a Free κB DNA: Insights into DNA Recognition by Transcription Factor NF-κB. Journal of Molecular Biology, 346, 147-160. http://dx.doi.org/10.1016/j.jmb.2004.11.042
- 32. Hailer-Morrison, M.K., Kotler, J.M., Martin, B.D. and Sugden, K.D. (2003) Oxidized Guanine Lesions as Modulators of Gene Transcription. Altered p50 Binding Affinity and Repair Shielding by 7,8-Dihydro-8-oxo-2’-deoxyguanosine Lesions in the NF-κB Promoter Element. Biochemistry, 42, 9761-9770. http://dx.doi.org/10.1021/bi034546k
- 33. Karyagina, A., Shilov, I., Tashlitskii, V., Khodoun, M., Vasilev, S., Lau, P.C. and Nikolskaya, I. (1997) Specific Binding of SsoII DNA Methyltransferase to Its Promoter Region Provides the Regulation of SsoII Restriction-Modification Gene Expression. Nucleic Acids Research, 25, 2114-2120. http://dx.doi.org/10.1093/nar/25.11.2114
- 34. Shilov, I., Tashlitsky, V., Khodoun, M., Vasil’ev, S., Alekseev, Y., Kuzubov, A., Kubareva, E. and Karyagina, A. (1998) DNA-Methyltransferase SsoII Interaction with Own Promoter Region Binding Site. Nucleic Acids Research, 26, 2659-2664. http://dx.doi.org/10.1093/nar/26.11.2659
- 35. Konarev, P.V., Kachalova, G.S., Ryazanova, A.Y., Kubareva, E.A., Karyagina, A.S., Bartunik, H.D. and Svergun, D.I. (2014) Flexibility of the Linker between the Domains of DNA Methyltransferase SsoII Revealed by Small-Angle X-Ray Scattering: Implications for Transcription Regulation in SsoII Restriction-Modification System. PLoS ONE, 9, e93453. http://dx.doi.org/10.1371/journal.pone.0093453
- 36. Smith, D.B. and Johnson, K.S. (1988) Single-Step Purification of Polypeptides Expressed in Escherichia coli as Fusions with Glutathione S-Transferase. Gene, 67, 31-40. http://dx.doi.org/10.1016/0378-1119(88)90005-4
- 37. Tanaka, H., Vickart, P., Bertrand, J.R., Rayner, B., Morvan, F., Imbach, J.L., Paulin, D. and Malvy, C. (1994) Sequence-Specific Interaction of Alpha-Beta-Anomeric Double-Stranded DNA with the p50 Subunit of NFκB: Application to the Decoy Approach. Nucleic Acids Research, 22, 3069-3074. http://dx.doi.org/10.1093/nar/22.15.3069
- 38. Thi, H.L., Zatsepin T.S., Schierling B., Volkov, E.M., Wende, W., Pingoud, A., Kubareva, E.A. and Oretskaya T.S. (2011) Restriction Endonuclease SsoII with Photoregulated Activity — A “Molecular Gate” Approach. Bioconjugate Chemistry, 22, 1366-1373. http://dx.doi.org/10.1021/bc200063m
- 39. Ryazanova, A.Y., Winkler, I., Friedhoff, P., Viryasov, M.B., Oretskaya, T.S. and Kubareva, E.A. (2011) Crosslinking of (Cytosine-5)-DNA Methyltransferase SsoII and Its Complexes with Specific DNA Duplexes Provides an Insight into Their Structures. Nucleosides Nucleotides & Nucleic Acids, 30, 632-650. http://dx.doi.org/10.1080/15257770.2011.584339
- 40. Cantor, C.R., Warshaw, M.M. and Shapiro, H. (1970) Oligonucleotide Interactions. 3. Circular Dichroism Studies of the Conformation of Deoxyoligonucleotides. Biopolymers, 9, 1059-1077. http://dx.doi.org/10.1002/bip.1970.360090909
- 41. Perez, A., Marchan, I., Svozil, D., Sponer, J., Cheatham III, T.E., Laughton, C.A. and Orozco, M. (2007) Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of Alpha/Gamma Conformers. Biophysical Journal, 92, 3817-3829. http://dx.doi.org/10.1529/biophysj.106.097782
- 42. Dupradeau, F.Y., Pigache, A., Zaffran, T., Savineau, C., Lelong, R., Grivel, N., Lelong, D., Rosanski, W. and Cieplak, P. (2010) The R.E.D. Tools: Advances in RESP and ESP Charge Derivation and Force Field Library Building. Physical Chemistry Chemical Physics, 12, 7821-7839. http://dx.doi.org/10.1039/c0cp00111b
- 43. Gordon, M.S. and Schmidt, M.W. (2005) Advances in Electronic Structure Theory: GAMESS a Decade Later. In: Dykstra, C.E., Frenking, G., Kim, K.S. and Scuseria, G.E., Eds., Theory and Applications of Computational Chemistry: The First Forty Years, Elsevier, Amsterdam, 1167-1189. http://dx.doi.org/10.1016/b978-044451719-7/50084-6
- 44. Vorob’eva, O.V., Kariagina, A.S., Volkov, E.M., Viriasov, M.B., Oretskaia, T.S. and Kubareva, E.A. (2002) An Analysis of Methyltransferase SsoII-DNA Contacts in the Enzyme-Substrate Complex. Bioorganicheskaya Khimiya, 28, 402-410.
- 45. Ryazanova, A.Y., Kubareva, E.A., Grman, I., Lavrova, N.V., Ryazanova, E.M., Oretskaya, T.S. and Hianik, T. (2011) The Study of the Interaction of (Cytosine-5)-DNA Methyltransferase SsoII with DNA by Acoustic Method. The Analyst, 136, 1227-1233. http://dx.doi.org/10.1039/c0an00545b
- 46. Scatchard, G. (1949) The Attractions of Proteins for Small Molecules an Ions. Annals of the New York Academy of Sciences, 51, 660-672. http://dx.doi.org/10.1111/j.1749-6632.1949.tb27297.x
- 47. Uporova, T.M., Kartashova, I.M., Skripkin, E.A., Lopareva, E. and Nikol’skaia, I.I. (1985) Restriction Endonucleases from Shigella sonnei 47. Voprosy Meditsinskoi Khimii, 31, 131-136.
- 48. Kung, H.C. and Bolton, P.H. (1997) Structure of a Duplex DNA Containing a Thymine Glycol Residue in Solution. The Journal of Biological Chemistry, 272, 9227-9236. http://dx.doi.org/10.1074/jbc.272.14.9227
- 49. Brown, K.L., Roginskaya, M., Zou, Y., Altamirano, A., Basu, A.K. and Stone, M.P. (2010) Binding of the Human Nucleotide Excision Repair Proteins XPA and XPC/HR23B to the 5R-Thymine Glycol Lesion and Structure of the cis-(5R,6S) Thymine Glycol Epimer in the 5'-GTgG-3' Sequence: Destabilization of Two Base Pairs at the Lesion Site. Nucleic Acids Research, 38, 428-440. http://dx.doi.org/10.1093/nar/gkp844
- 50. Siggers, T., Chang, A.B., Teixeira, A., Wong, D., Williams, K.J., Ahmed, B., Ragoussis, J., Udalova, I.A., Smale, S.T. and Bulyk, M.L. (2012) Principles of Dimer-Specific Gene Regulation Revealed by a Comprehensive Characterization of NF-κB Family DNA Binding. Nature Immunology, 13, 95-102. http://dx.doi.org/10.1038/ni.2151
- 51. Metelev, V.G., Kubareva, E.A. and Oretskaya, T.S. (2013) Regulation of Activity of Transcription Factor NF-κB by Synthetic Oligonucleotides. Biochemistry (Moscow), 78, 867-878. http://dx.doi.org/10.1134/s0006297913080026
- 52. Sakurai, H., Chiba, H., Miyoshi, H., Sugita, T. and Toriumi, W. (1999) IκB Kinases Phosphorylate NF-κB p65 Subunit on Serine 536 in the Transactivation Domain. The Journal of Biological Chemistry, 274, 30353-30356. http://dx.doi.org/10.1074/jbc.274.43.30353
- 53. Yu, Z., Zhang, W. and Kone, B.C. (2002) Histone Deacetylases Augment Cytokine Induction of the iNOS Gene. Journal of the American Society of Nephrology, 13, 2009-2017. http://dx.doi.org/10.1097/01.ASN.0000024253.59665.F1
- 54. Schwabe, R.F., Schnabl, B., Kweon, Y.O. and Brenner, D.A. (2001) CD40 Activates NF-κB and c-Jun N-Terminal Kinase and Enhances Chemokine Secretion on Activated Human Hepatic Stellate Cells. Journal of Immunology, 166, 6812-6819. http://dx.doi.org/10.4049/jimmunol.166.11.6812
- 55. Benezra, M., Chevallier, N., Morrison, D.J., MacLachlan, T.K., El-Deiry, W.S. and Licht, J.D. (2003) BRCA1 Augments Transcription by the NF-κB Transcription Factor by Binding to the Rel Domain of the p65/RelA Subunit. The Journal of Biological Chemistry, 278, 26333-26341. http://dx.doi.org/10.1074/jbc.M303076200
- 56. Schwabe, R.F. and Sakurai, H. (2005) IKKbeta Phosphorylates p65 at S468 in Transactivaton Domain 2. FASEB Journal, 19, 1758-1760.
- 57. Buss, H., Handschick, K., Jurrmann, N., Pekkonen, P., Beuerlein, K., Muller, H., Wait, R., Saklatvala, J., Ojala, P.M., Schmitz, M.L., Naumann, M. and Kracht, M. (2012) Cyclin-Dependent Kinase 6 Phosphorylates NF-κB P65 at Serine 536 and Contributes to the Regulation of Inflammatory Gene Expression. PLoS ONE, 7, e51847. http://dx.doi.org/10.1371/journal.pone.0051847
- 58. Ferguson, K.A. (1964) Starch-Gel Electrophoresis-Application to the Classification of Pituitary Proteins and Polypeptides. Metabolism, 13, 985-1002. http://dx.doi.org/10.1016/S0026-0495(64)80018-4
- 59. Muller, C.W., Rey, F.A., Sodeoka, M., Verdine, G.L. and Harrison, S.C. (1995) Structure of the NF-κB p50 Homodimer Bound to DNA. Nature, 373, 311-317. http://dx.doi.org/10.1038/373311a0
- 60. Ghosh, G., van Duyne, G., Ghosh, S. and Sigler, P.B. (1995) Structure of NF-κB p50 Homodimer Bound to a κB Site. Nature, 373, 303-310. http://dx.doi.org/10.1038/373303a0
- 61. Chen, Y.Q., Ghosh, S. and Ghosh, G. (1998) A Novel DNA Recognition Mode by the NF-κB p65 Homodimer. Nature Structural Biology, 5, 67-73. http://dx.doi.org/10.1038/nsb0198-67
- 62. Sengchanthalangsy, L.L., Datta, S., Huang, D.B., Anderson, E., Braswell, E.H. and Ghosh, G. (1999) Characterization of the Dimer Interface of Transcription Factor NFκB p50 Homodimer. Journal of Molecular Biology, 289, 1029-1040. http://dx.doi.org/10.1006/jmbi.1999.2823
- 63. Wurster, S.E., Bida, J.P., Her, Y.F. and Maher III, L.J. (2009) Characterization of Anti-NF-κB RNA Aptamer-Binding Specificity in Vitro and in the Yeast Three-Hybrid System. Nucleic Acids Research, 37, 6214-6224. http://dx.doi.org/10.1093/nar/gkp670
- 64. McTigue, M.A., Williams, D.R. and Tainer, J.A. (1995) Crystal Structures of a Schistosomal Drug and Vaccine Target: Glutathione S-Transferase from Schistosoma japonica and Its Complex with the Leading Antischistosomal Drug Praziquantel. Journal of Molecular Biology, 246, 21-27. http://dx.doi.org/10.1006/jmbi.1994.0061
- 65. Sheehan, D., Meade, G., Foley, V.M. and Dowd, C.A. (2001) Structure, Function and Evolution of Glutathione Transferases: Implications for Classification of Non-Mammalian Members of an Ancient Enzyme Superfamily. The Bio- chemical Journal, 360, 1-16. http://dx.doi.org/10.1042/bj3600001
- 66. Romanenkov, A.S., Ustyugov, A.A., Zatsepin, T.S., Nikulova, A.A., Kolesnikov, I.V., Metelev, V.G., Nikulova, A.A., Kolesnikov, I.V., Metelev, V.G., Oretskaya, T.S. and Kubareva, E.A. (2005) Analysis of DNA-Protein Interactions in Complexes of Transcription Factor NF-κB with DNA. Biochemistry (Moscow), 70, 1212-1222. http://dx.doi.org/10.1007/s10541-005-0249-2
- 67. Smale, S.T. (2012) Dimer-Specific Regulatory Mechanisms within the NF-κB Family of Transcription Factors. Immunological Reviews, 246, 193-204. http://dx.doi.org/10.1111/j.1600-065X.2011.01091.x
- 68. Hayden, M.S. and Ghosh, S. (2008) Shared Principles in NF-κB Signaling. Cell, 132, 344-362. http://dx.doi.org/10.1016/j.cell.2008.01.020
- 69. Nikolskaya, I.I., Lopatina, N.G., Suchkov, S.V., Kartashova, I.M. and Debov, S.S. (1984) Sequence Specificity of Isolated DNA-Cytosine Methylases from Shigella sonnei 47 Cells. Biochemistry International, 9, 771-781.
- 70. Vorob’eva, O.V., Vasil’ev, S.A., Kariagina, A.S., Oretskaia, T.S. and Kubareva, E.A. (2000) Analysis of Contacts between DNA and Protein in a Complex of SsoII Methyltransferase-Promoter Region of the Gene for the SsoII Restriction-Modification System. Molekuliarnaia Biologiia, 34, 1074-1080.
- 71. Romanenkov, A.S., Kisil, O.V., Zatsepin, T.S., Yamskova, O.V., Karyagina, A.S., Metelev, V.G., Oretskaia, T.S. and Kubareva, E.A. (2006) DNA-Methyltransferase SsoII as a Bifunctional Protein: Features of the Interaction with the Promoter Region of SsoII Restriction-Modification Genes. Biochemistry (Moscow), 71, 1341-1349. http://dx.doi.org/10.1134/s0006297906120091
NOTES
*Corresponding author.



