Journal of Modern Physics
Vol.3 No.6(2012), Article ID:20081,7 pages DOI:10.4236/jmp.2012.36059
Nonextensivity and Tsallis Entropy in DNA Fragmentation Patterns by Ionizing Radiation
1Centro de Estudios Avanzados de Cuba (CEAC), Havana, Cuba
2Universidad de las Ciencias Informáticas (UCI), Havana, Cuba
3Instituto Superior de Tecnologías y Ciencias Aplicadas (InSTEC), Havana, Cuba
4Physics Institute, University of São Paulo, São Paulo, Brasil
5CEPESq/UniÍtalo—Italy-Brazilian University Center, São Paulo, Brazil
Email: carlos.mv@cea.cu
Received March 17, 2012; revised April 10, 2012; accepted April 20, 2012
Keywords: Nonextensive statistics; Tsallis entropy; DNA fragment distribution; Theoretical distribution; Fragment minimum length; DNA data analysis
ABSTRACT
Nonextensive statistical mechanics as in Tsallis formalism was used in this study, along with the dynamical Hamiltonian rod-like DNA model and the maximum entropy criteria for Tsallis’ entropy, so as to obtain length distribution of plasmid fragments, after irradiation with very high doses, assuming that the system reaches metaequilibrium. By intensively working out the Grand Canonical Ensemble (used to take into account the variation of the number of base pairs) a simplified expression for Fragment Size Distribution Function (FSDF) was obtained. This expression is dependent on two parameters only, the Tsallis q value and the minimal length of the fragments. Results obtained from fittings to available experimental data were adequate and the characteristic behavior of the shortest fragments was clearly documented and reproduced by the model, a circumstance never verified from theoretical distributions. The results point to the existence of an entropy which characterizes fragmentation processes and depending only on the q entropic index.
1. Introduction
A wide range of studies related to the biological action of ionizing radiation at the cellular level identified the DNA molecule at the top in the hierarchy of possible biological targets. Molecular damages in DNA define the subsequent fate of the cell and could lead to reproductive cell death, apoptosis, mutations and cancer transformations. After treatment of cells by ionizing radiation there is a broad spectrum of radiation-induced damages which can finally lead to the different endpoints. DNA radiation induced effects are mostly: breaking of one strand, called Single Strand Break (SSB), breaking of the two strands, Double Strand Breaks (DSB), nitrogen base damage, and clustered DNA damages (in case of heavy charged particles).
DSB, including the clustered DNA damages, is the most harmful effect since it has a non-negligible probability in inducing cell death, mutation or carcinogenesis. Studying how DSBs occur and the mechanisms by which the cell repairs them, has caught the attention of many investigators, mainly because of many possible applications as e.g. in the improvement of cancer treatment protocols. There are many studies attempting an understanding of DSBs by using several techniques to measure the number and size of fragments after irradiation. The two most successful techniques are pulsed field gel electrophoresis and atomic force microscopy (AFM). The latter being more recent, enabling better resolution, and as such, has been intensively and extensively used.
From a theoretical point of view there are several approaches in obtaining distribution of fragments. One of the most used models is the Random Breaking Model (RBM) [1]. It describes the distribution of fragments at high doses, but fails for shorter fragments, and does not account for the correlation between fragments. Since correlations are present in the formation of DSBs [2], said correlations between the fragments would also be expected. It has already been shown in [3] that long-range correlations between fragments imbedded in a random walk kinematics strongly suggest that DNA constitutes a system driven by nonextensive statistics, with FSDs described by power laws with fractionary exponents (but not by exponential functions as in extensive statistics), where the exponents are functions of both the long-range potential and the system degree of freedom. There is a pioneering work applying nonextensivity when the distribution function is obtained by maximizing Tsallis’ entropy, providing new interesting results at high doses [4]. Monte Carlo based methods yield good data reproducibility for a wide range of doses, but usually needing 5 or more parameters. However, these methods fail in correctly reproducing smaller fragments at high doses [5]. It is noted, for instance, that besides the shortcomings in coping with the region of smaller fragments, the distribution obtained in [4] has a parameter which does not have a precise physical meaning.
Following here this last approach [4], fragment distribution length based on the maximization of Tsallis’ entropy was obtained, by taking into account not only length of the fragments but also their energy values as well.
Firstly, the theoretical background is exposed in the Method section, beginning with the Tsallis’s entropy as a start point for all the subsequent developments. Nonextensive Statistical Mechanics is briefly revisited in order to lay the foundation for its using, and at the end of the section it is proposed a physical model for the DNA fragmentation at very high doses, which allowed us to obtain the FSDFs. In the Results section, the FSDFs are tested with available experimental data from literature, which comprises several types of ionizing particles and at several doses, leading to an empirical simplification of the formula obtained. A parametrization of one of the model’s parameter as function of dose is proposed, by considering plasmids in a HEPES buffer. In Conclusion, the significance and the novelty are highlighted.
2. Method
Tsallis’ entropy (Sq) is a generalization of BoltzmannGibbs entropy (SBG), which has succeeded in the description of many systems previously seeming to elude conventional statistical mechanics. Its form is [6]:
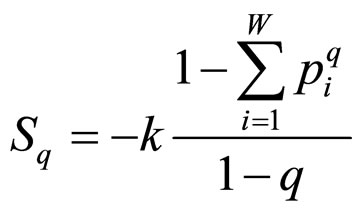 , (1)
, (1)
where q is a real number greater than zero. For q → 1 holds Sq → SBG. The probability that the system is in the ith microstate is pi and k is the Boltzmann’s constant. The value of q can be derived from the dynamics of the system provided exactly known. Alternatively, q could be extracted from experiments by fitting procedures. This entropic form in Equation (1) implies that if A and B are two independent systems, in the sense that , one is led to
, one is led to
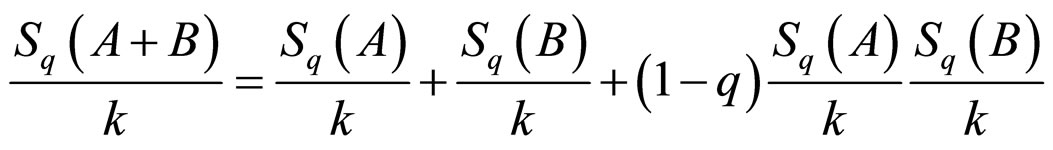 . (2)
. (2)
This formulation implies that the Tsallis’ entropy is generally non-extensive, while that of Boltzmann-Gibbs’ is extensive. Not only the entropy formulation changed, but also the ways by which mean values are calculated. For the sake of simplicity and for our purposes we used the so-called “q-mean value” for the energy,
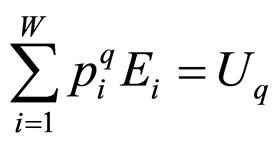 , (3)
, (3)
although this choice in Equation (3) is not unique (in reference [7] a detailed discussion of all possible choices can be found). For the grand canonical ensemble the obtained distribution function is [8]:
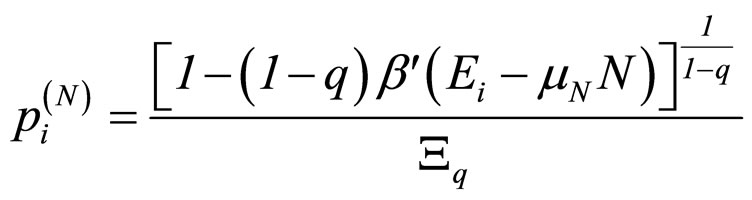 , with (4)
, with (4)
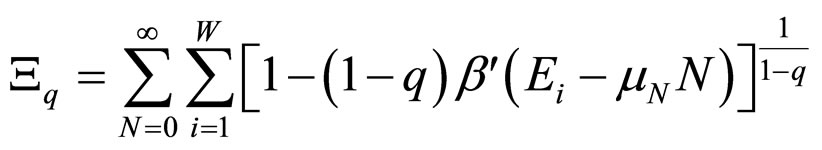 , (5)
, (5)
where β' was used instead of β to remark that β' is not the reciprocal of kT [7].
Nonextensive statistical mechanics works in the metaequilibrium, i.e. when the system is in a metastable state (including equilibrium as a special case), which is often achieved in stationary regimes or in non-equilibrium organized states. Before the irradiation of the samples all fragments have approximately the same lengths. As doses increase fragments with all possible lengths are produced. At very high doses the most likely fragments are the smaller, while the remaining show a very low or nearly null probability. In this paper it is assumed that the system reaches metaequilibrium during irradiation, when it organizes itself by gathering energy with radiation.
Our system is considered as an ensemble of plasmids, each one having N base pairs closely related to its length through L = Na, where a is the distance between base pairs; under normal conditions a = 0.34 nm. In the fragmentation process at high doses, energy is distributed among the system’s constituents faster than when low doses are involved. This fact allows considering solutions as if plasmids were imbedded in a thermal reservoir at an effective temperature. Furthermore, recalling that one is dealing with a highly excited system, energy necessary to remove a base pair from the plasmid is considered, a kind of base pair chemical potential, constant and independent of the base pairs specificities. This circumstance shows why the grand canonical ensemble is most appropriate. Thus, the distribution should be the same as in Equation (4). To find p(N) (the probability to find a fragment with N base pairs) it is necessary to compute energy values of the plasmids, which can be obtained from the DNA coupled road model [9] without considering folding motion (mostly valid for supercoiled plasmids). The basic ingredient of this road like model consists in approaching the DNA as a set of coupled disks (which are the base pairs) with longitudinal (un) and rotational (φn) degrees of freedom. In this way, one is able to take into account the size of the fragments as well as their energies.
The Hamiltonians are specified by the following expressions, where the sum runs through all values of the base pairs; M denotes the mass of each base pair and I refers to their moment of inertia. Thus,
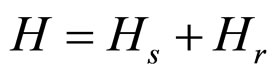 , (6)
, (6)
 , (7)
, (7)
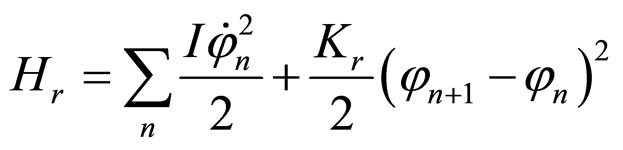 , (8)
, (8)
where Ks is the longitudinal stiffness constant and Kr is the stiffness associated with rotations. Solutions obtained by solving the dynamical equations are plane waves. Plasmids in metaequilibrium are constantly excited by radiation from the environment, generating wave trains traveling in both directions. They are expected to overlap, forming standing waves. For the longitudinal case
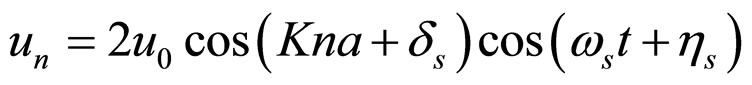 . (9)
. (9)
For rotational coordinates a similar expression is obtained; K is the wave number (K = 2π/ma) and m ranges in principle from 2 to 2N. Substituting the stationary solutions in the Hamiltonian and transforming summations into integrals, plus assuming that the plasmid is a closed structure and also that for n > 12 (the resolution of the AFM, approximately) holds sin(π/n) ≈ π/n, we obtain
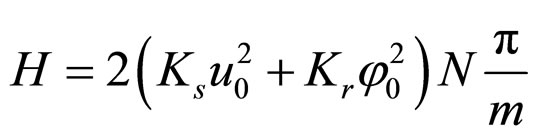 . (10)
. (10)
However, the system cannot absorb an arbitrarily large amount of energy. There should be a specific mmin, as well as given values for the amplitudes and stiffness constants, so that the plasmid is broken into a number of fragments approximately equal to the number of nodes 2N/mmin, and this would occur when δs – δr = π. The longitudinal and rotational waves must be in phase or in counter phase, since nodes must match in order to fulfill the equilibrium condition: u → 0 and φ → 0, simultaneously. Therefore, the energy in Equation (10) have to be equal to the number of nodes mentioned above multiplied by the dissociation energy of two consecutive base pairs μN. Substituting in Equation (5), and integrating in L instead of summing in N it is obtained
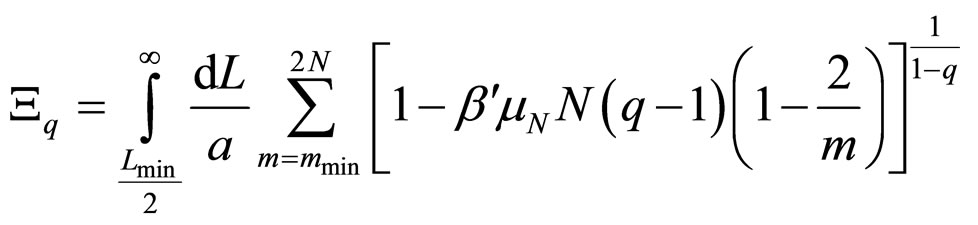 , (11)
, (11)
where Lmin/2 is the length of the shortest fragment. As 2/m decreases swiftly to zero, it can be neglected when compared to unity, thus allowing obtaining the probability distribution
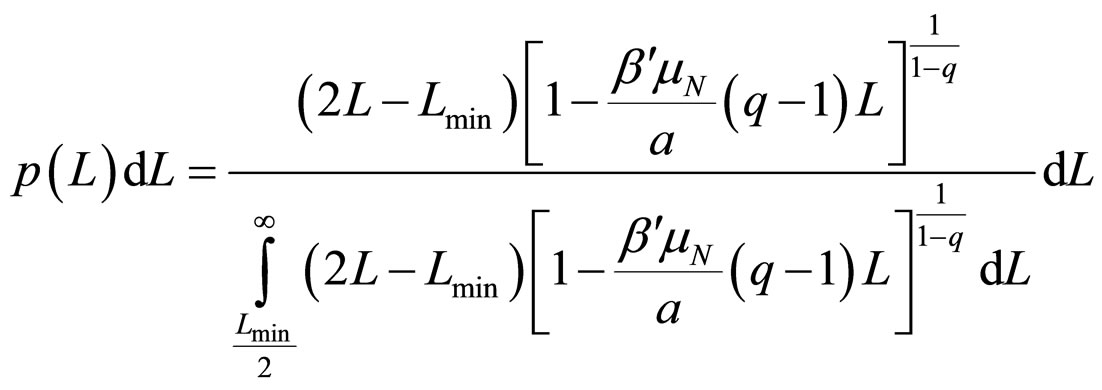 , (12)
, (12)
where q must be in the range (1 - 3/2).
3. Results
The most accurate experimental results for length distributions of fragments are reported in the form of histograms with 50 nm width bars. Fittings performed to the experimental data showed that β'μN/a → −∞, although there is a wide range of values for which the fitting results change slightly. Calculating this limit in Equation (12) leads to the final simplified equation for the Fragment Size Distribution Function (FSDF), which only depends on the Tsallis q value and on the minimal length of the fragments (Lmin/2):
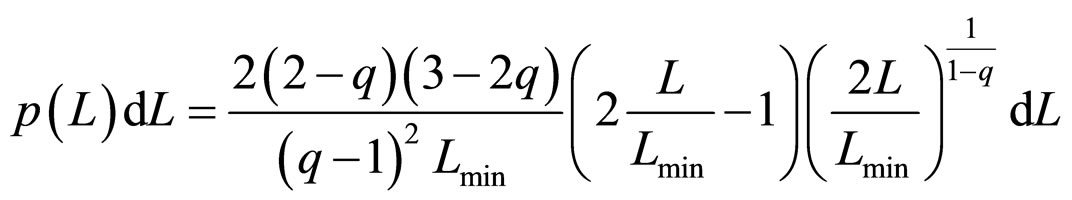 , (13)
, (13)
Substituting Lmin = p1, and q = p2, the fitting was performed for a total of 20 experimental results reported in the literature. It should be noted that the fragment size pattern obtained for a given dose also depends on the solution where the plasmids were diluted. The experiments analyzed here were carried out by using two different solutions: water and HEPES buffer. They differ significantly as to the diffusion length of the free radicals created in the ionization of the medium, being much higher in water. Therefore, one needs a much lower dose in water to achieve effects similar to those with the HEPES buffer.
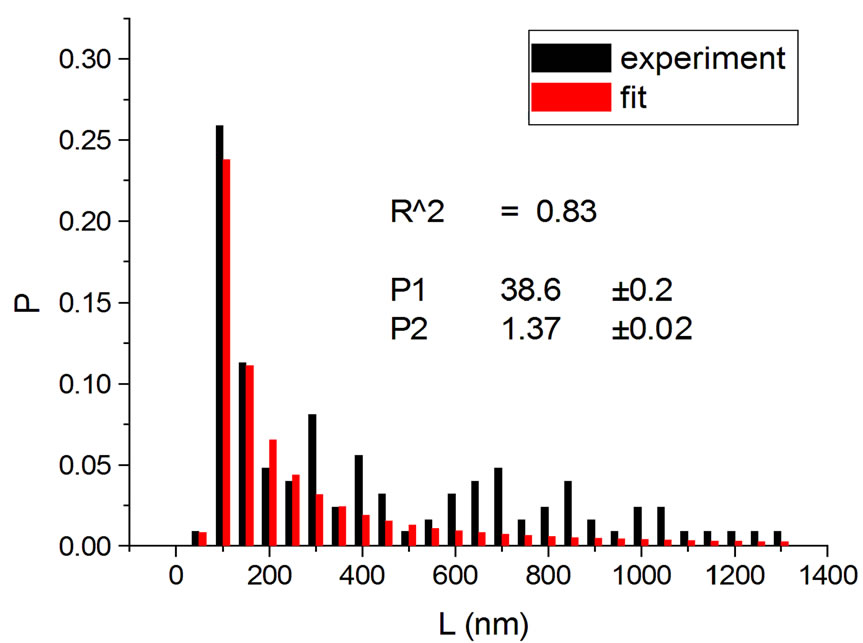
Figure 1, panel a, shows the experimental distribution of fragments and the theoretical fitting for irradiation of plasmids (80% in supercoiled conformation) with 12C ions at 8 Gy in water [10]. There is, in panel b, another experiment with Ni ions at 3000 Gy, with supercoiled plasmids in HEPES buffer [5]. R2 is the squared correlation coefficient. It can be seen that the model is capable of reproducing the initial peak of the experimental distribution.
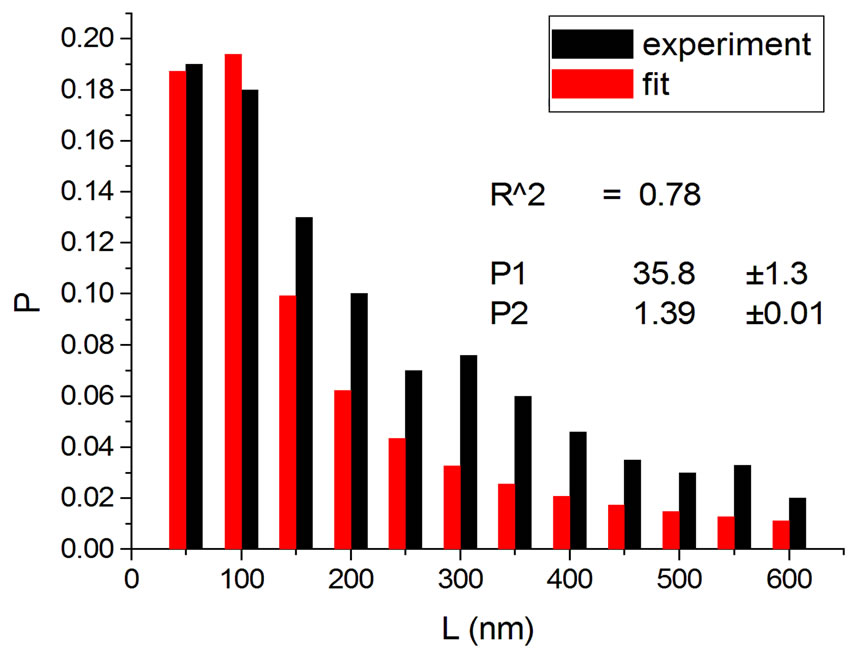
Figures 2 and 3 show results for irradiations with electrons and neutrons, respectively, at several doses.
 (a)
(a)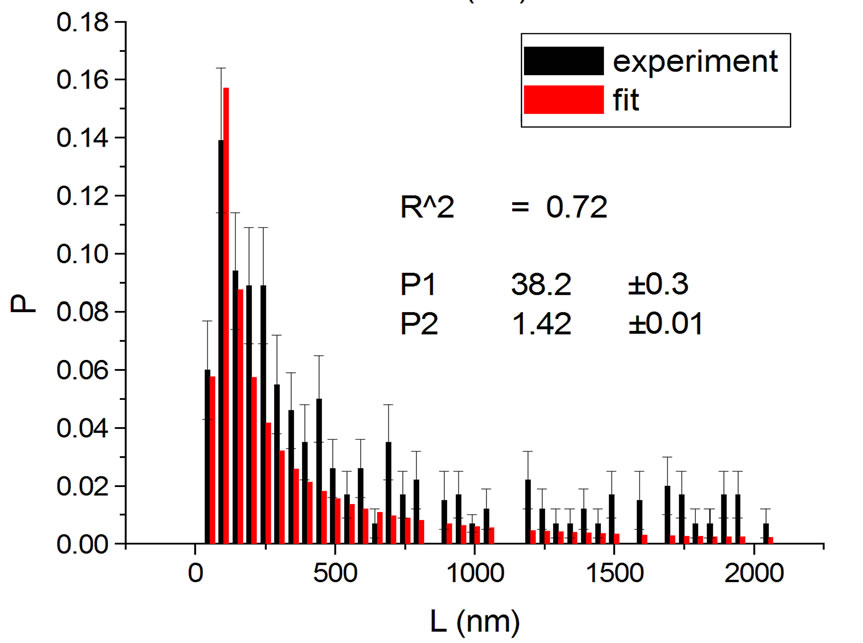 (b)
(b)
Figure 1. Irradiation with 12C ions at 8 Gy in water [10] (a); with Ni ions at 3000 Gy in HEPES buffer [5] (b).
 (a)
(a)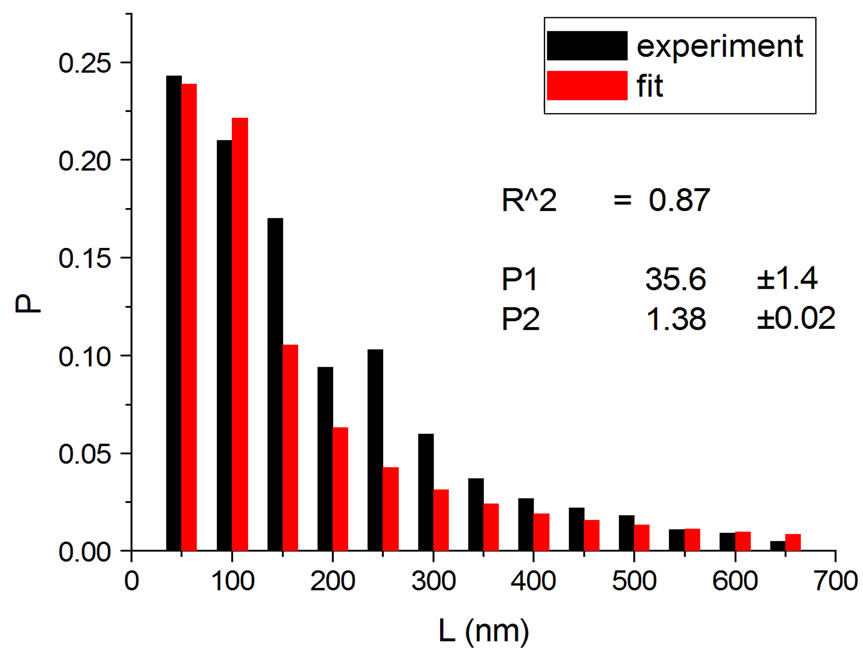 (b)
(b)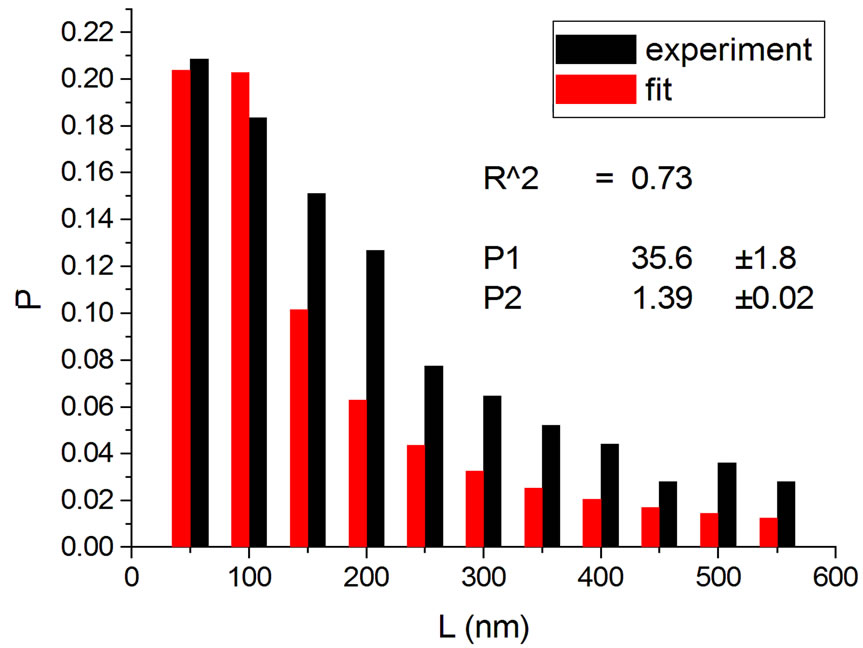 (c)
(c) (d)
(d)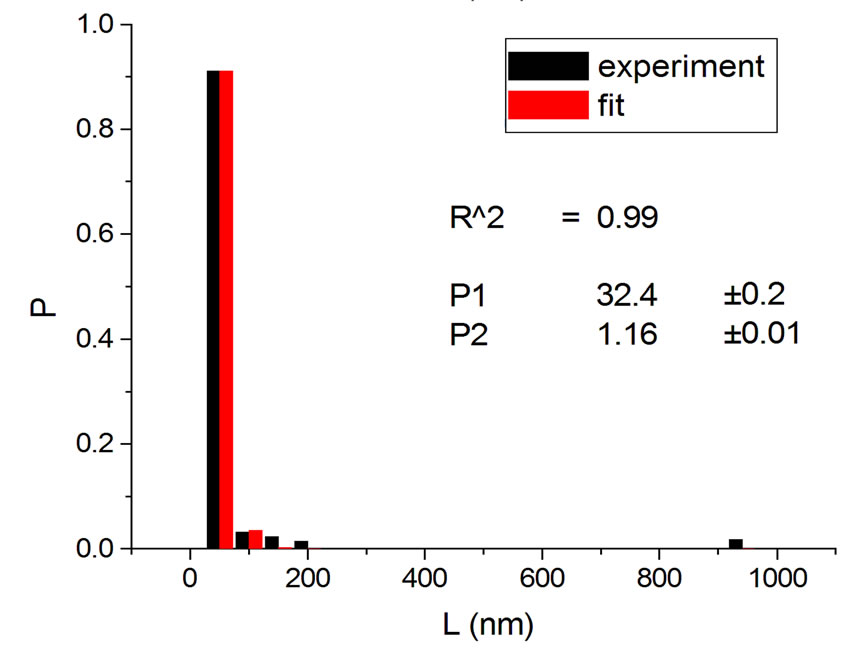 (e)
(e)
Figure 2. Irradiations with electrons at 5000 Gy [4] (a); 6000 Gy [11] (b); 7000 Gy [4] (c); 8000 Gy [12] (d); and 10,000 Gy [12] (e).
 (a)
(a)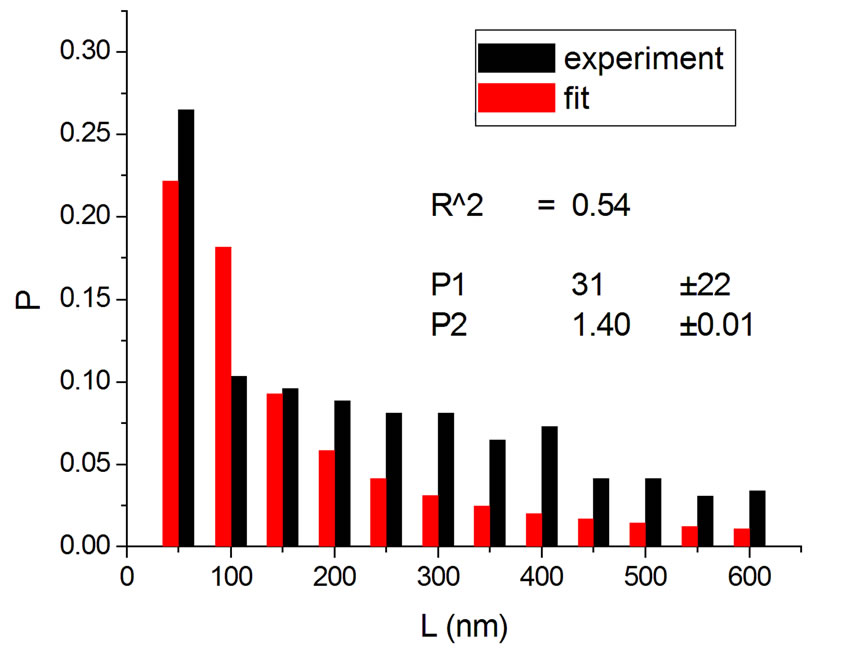 (b)
(b)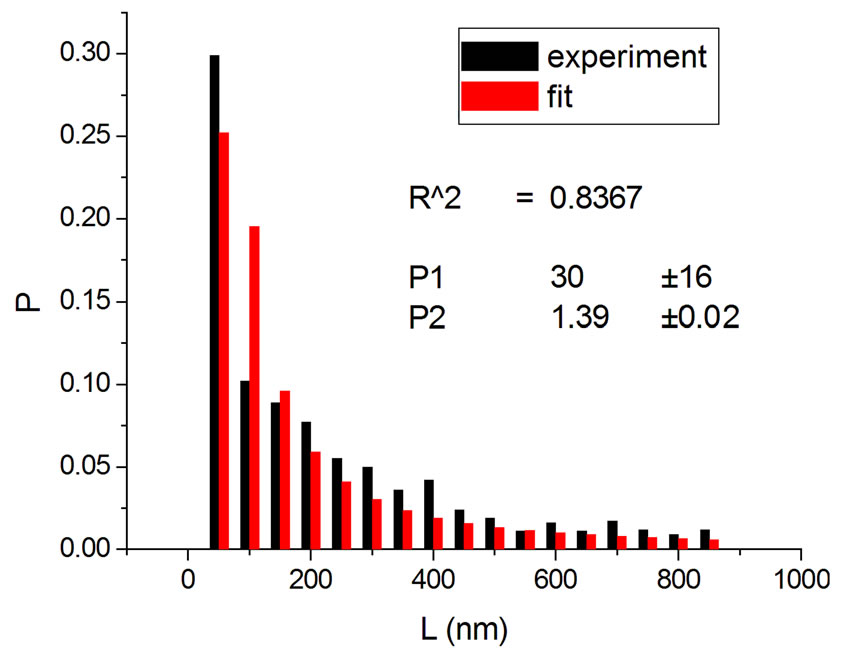 (c)
(c)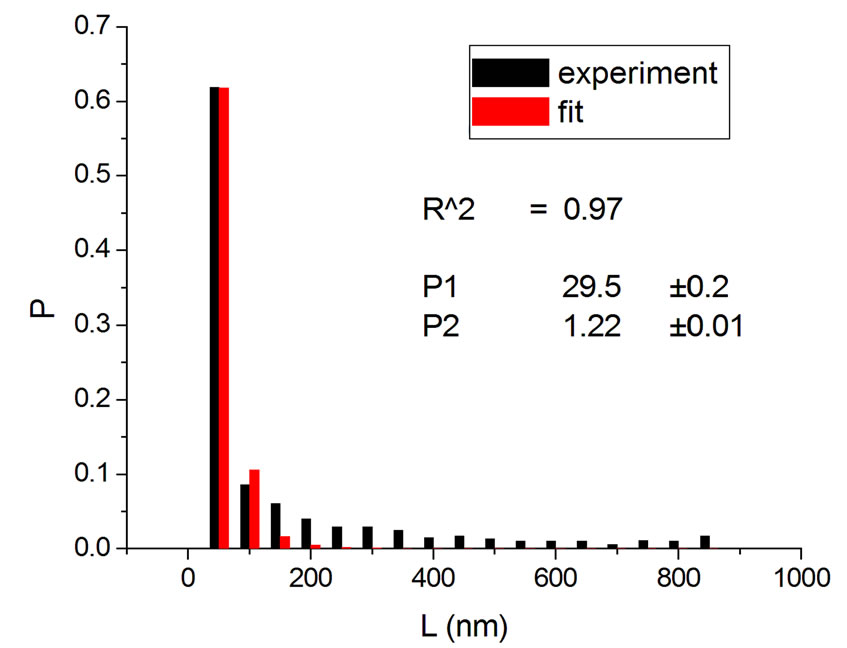 (d)
(d)
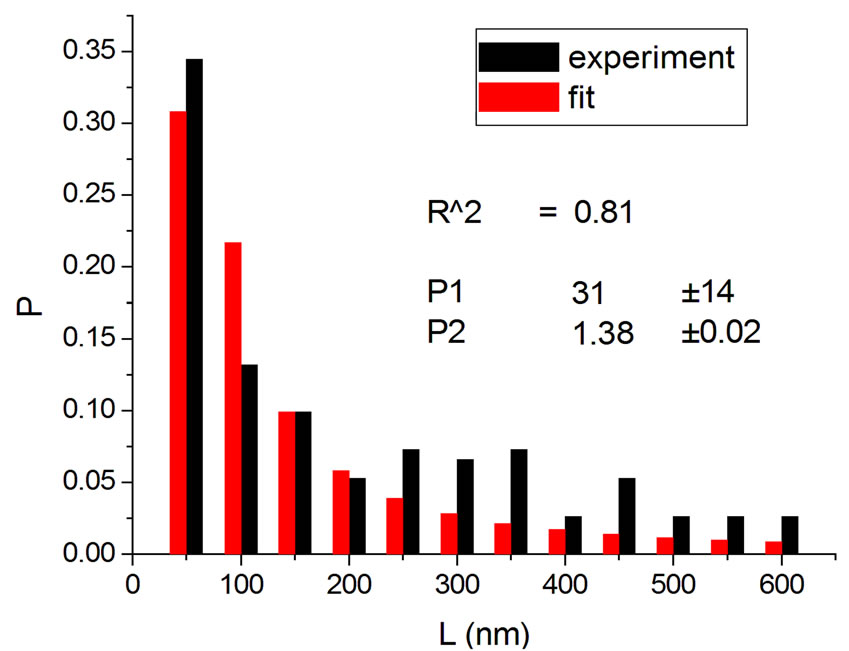
Figure 3. Irradiations with neutrons at 5000 Gy [4] (a); 7000 Gy [4] (b); 7500 Gy [13] (c); and 10,000 Gy [13] (d).
Figure 4 shows the results at 10,000 Gy for gammas and Ar ions. In order to appraise to which extent fitting quality varies with dose, the dose range was divided into five equal intervals and within each of these the average value of R2 was calculated. Figure 5 shows how the fitting is linearly improved with the increasing doses, indicating that the model functions better when doses are higher.
The behavior of Lmin with dose in a total of 13 experiments with better resolution (bar width equal to 50 nm) was studied, and those performed with buffer were chosen. For dose values pertaining to more than one experiment, an average of Lmin was computed. This allowed obtaining a parametrization in the form of a linear function with R2 = 0.65 (Figure 6):
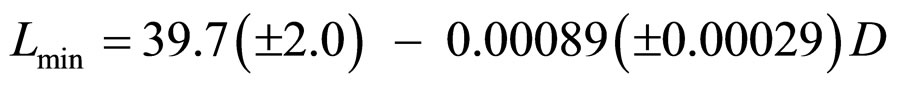 , (14)
, (14)
where D is the dose.
If one substitutes the parametrization in Equation (14) into Equation (13), aiming at achieving new fittings, the obtained values of q are very close to the original, except in one case. This can be seen in Figures 7 and 8 where the experiments were listed on the horizontal axis; significantly, only the 11th experiment differs from the original. The error bars correspond to the uncertainty provided by the fitting.
Another result obtained by fitting reveals that in 70% of the cases q is in the interval q = 1.40 ± 0.03, independently of the type of ionizing particles. The remaining 30% is distributed almost uniformly from 1.16 to 1.37, as shown by the histogram in Figure 9. Therefore, the parameter q is predominant in a narrow interval. Related to the q values, an interesting feature emerge; that of the entropy Sq, associated with the distribution of the fragments by length and energy, when considering the distribution function in Equation (4) and the limit β'μN → −∞ (a must be positive and must not vanish). One can observe in Figures 10 and 11 that for q values around those predominantly obtained by fitting, the entropy is almost independent of the remaining two parameters (Lmin and a). The results point to the existence of entropy characterizing the fragmentation process and depending strongly on the q entropic index and weakly on the others parameters.
4. Conclusion
A novel approach has been established in order to compute fragment length distribution at very high doses. A new Fragment Size Distribution Function was obtained using a nonextensive grand canonical ensemble, with good fitting results of experimental data at different doses. This is the first time a FSDF for DNA under radiation is obtained departing from a physical model and with a potential-like decay for long fragments without the a priori conjecture of having this kind of behavior.
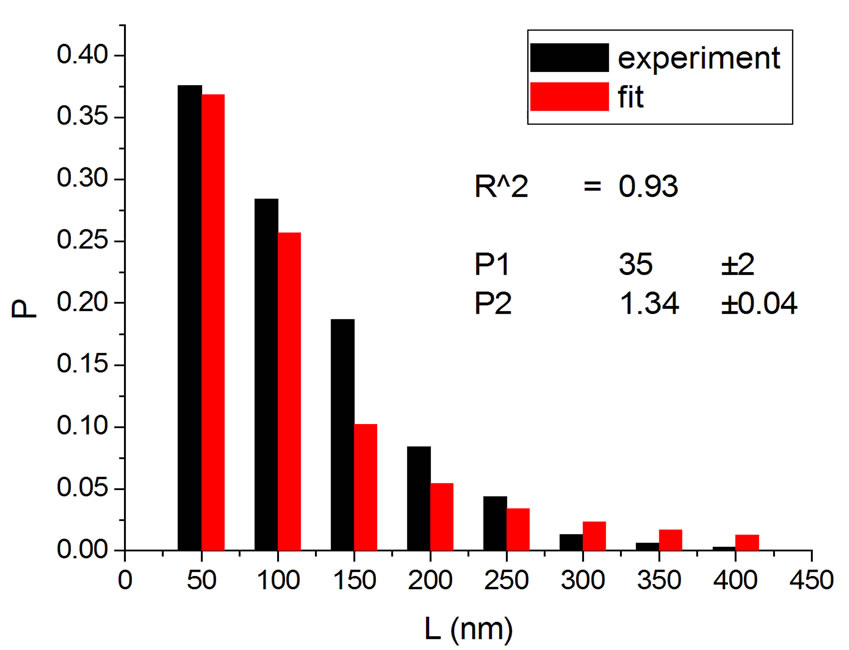 (a)
(a)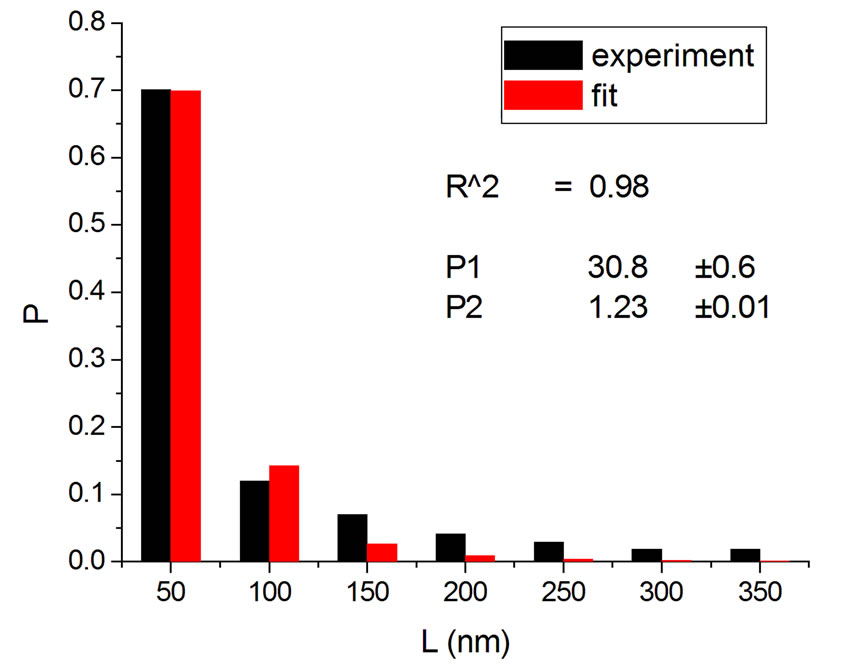 (b)
(b)
Figure 4. Irradiation at 10,000 Gy with gamma (a) [14] and Ar ions (b) [14].
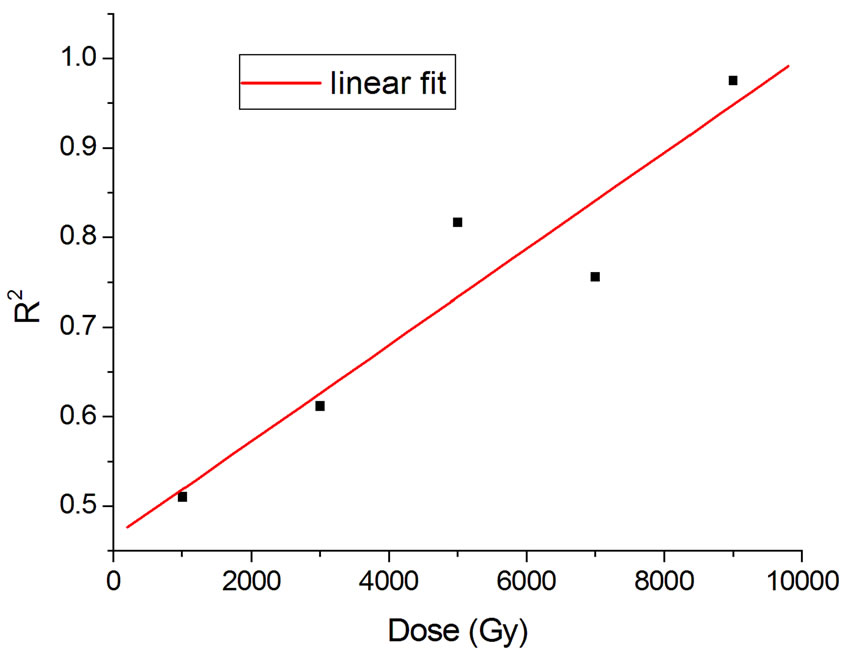
Figure 5. Linear fit with  = 0.88.
= 0.88.
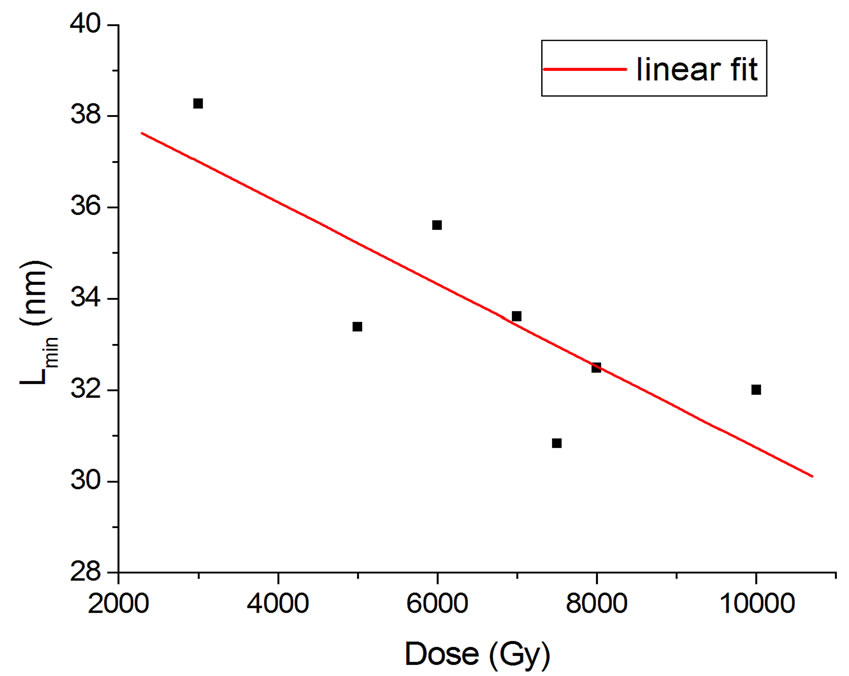
Figure 6. Lmin parametrization in Equation (16).
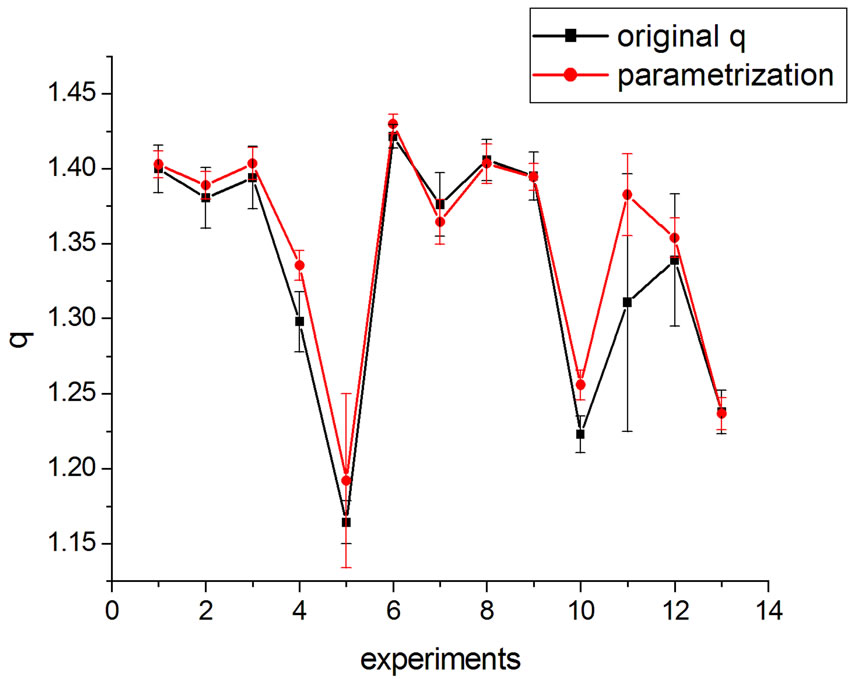
Figure 7. q variation with parametrization in Equation (16).
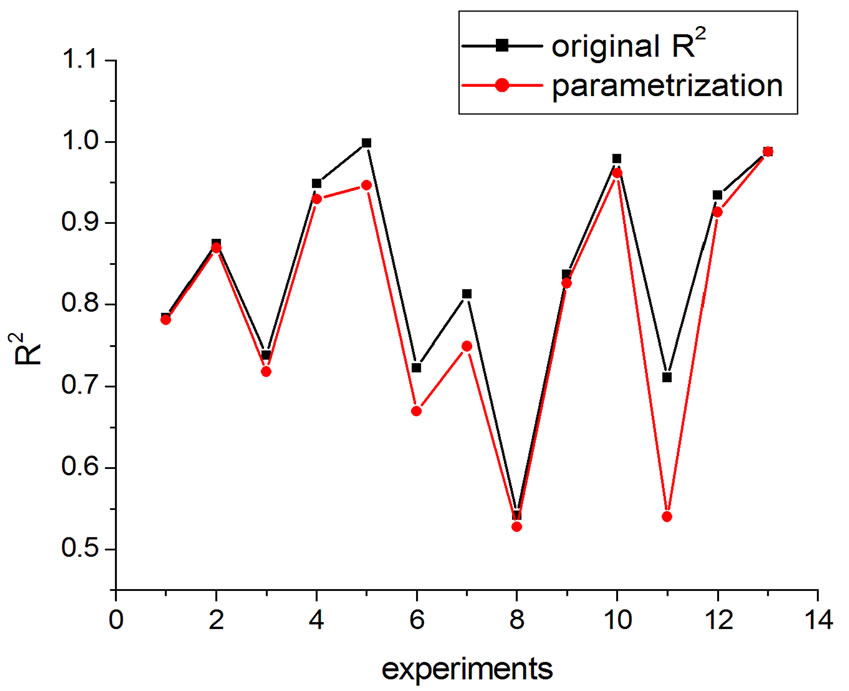
Figure 8. R2 variation with parametrization in Equation (16).
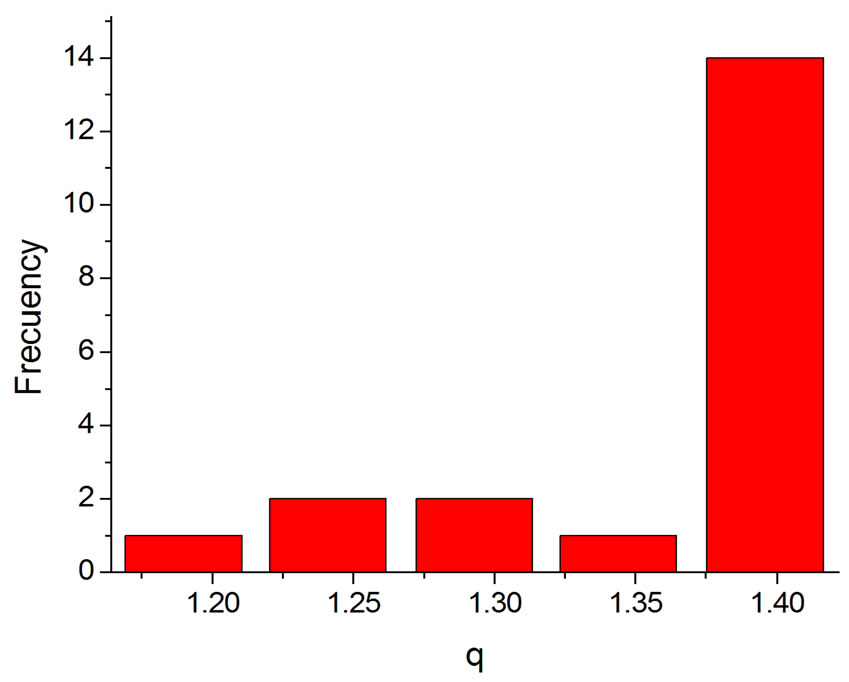
Figure 9. Histogram of the q values obtained by fitting.
Interestingly, the theoretical distribution was able to reproduce the initial peak, sometimes present in irradiation with heavy ions (Figure 1); hence, it does not merely reproduce the potential-like decays, constituting thus an improvement of the former models. The present study also found that information on the fragmentation process, via Sq, depends heavily on q, while dependence on the remaining parameters, as Lmin and a, is considerably weaker, irrespective of what kind of particle is involved in the irradiation, provided q is approximately higher than 1.37 (occurrence of 70% in all the examined cases). Although the model improves the data description as doses increase, it is clear that the necessity to analyze
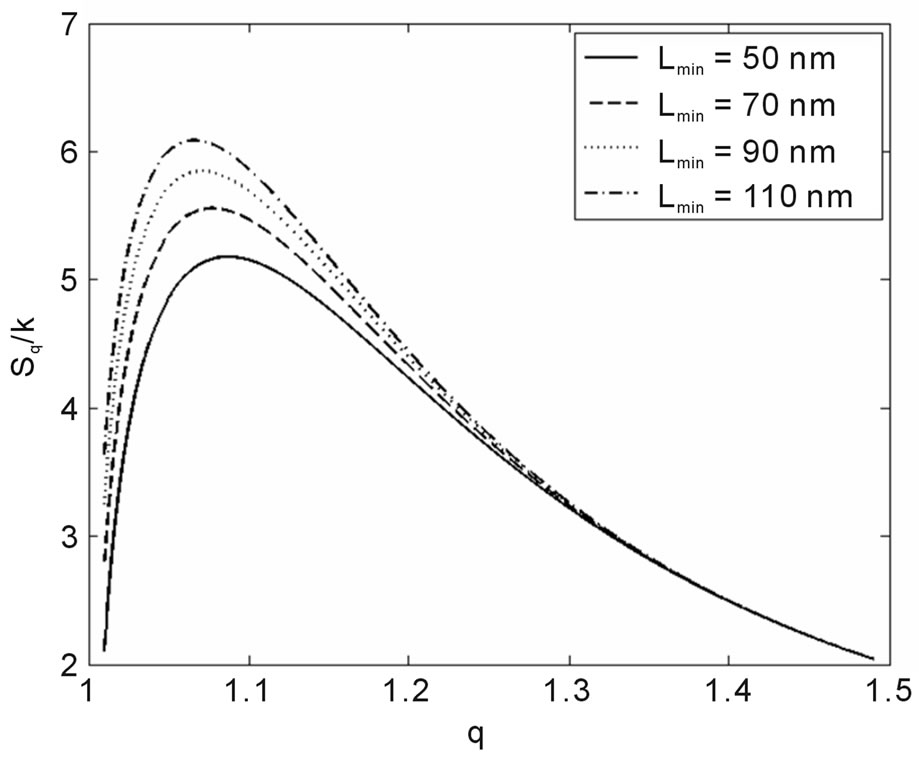
Figure 10. Sq/k vs. q for the distance between base pairs a = 0.34 nm and several Lmin.
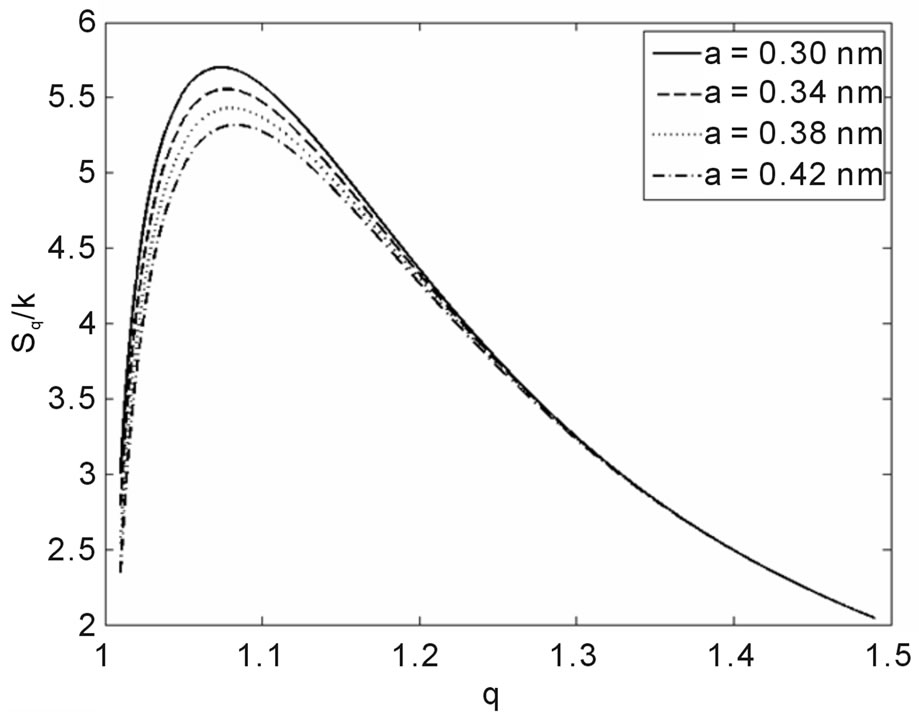
Figure 11. Sq/k vs. q for Lmin = 70 nm and several values of the base pairs separation a.
more experiments remains. It should be noted, notwithstanding, that the parametrization in Equation (14) for HEPES buffer works quite well with the data so far available, where only one fitting parameter (q) was necessary.
REFERENCES
- K. Fuquan, et al., “Analysis of Length Distribution of Short DNA Fragments Induced by 7Li Ions Using the Random-Breakage Model,” Chinese Science Bulletin, Vol. 50, No. 9, 2005, pp. 841-844. doi:10.1360/982004-827
- M. Löbrich, P. K. Cooper and B. Rydberg, “Non-random Distribution of DNA Double-Strand Breaks Induced by Particle Irradiation,” International Journal of Radiation Biology, Vol. 70, No. 5, 1996, pp. 493-503. doi:10.1080/095530096144680
- F. A. Castro, O. Rodriguez and J. D. T. Arruda-Neto. “Present Status of Radiation Interaction with DNAStrand-Break Cross-Section and Fragment-Size Distributions,” Radiation Effects and Defects in Solids, Vol. 162, No. 3-4, 2007, pp. 237-245. doi:10.1080/10420150601134616
- O. Sotolongo-Costa, et al., “A Non Extensive Approach for DNA Breaking by Ionizing Radiation,” 2002.
- T. Elsässer, et al., “Biophysical Modeling of Fragment Length Distributions of DNA Plasmids after X and Heavy-Ion Irradiation Analyzed by Atomic Force Microscopy,” Radiation Research, Vol. 169, No. 6, 2008, pp. 649-59. doi:10.1667/RR1028.1
- C. Tsallis, F. Baldovin, R. Cerbino and P. Pierobon. “Introduction to Nonextensive Statistical Mechanics and Thermodynamics,” 2003.
- C. Tsallis, R. Mendes and A. Plastino, “The Role of Constraints within Generalized Nonextensive Statistics,” Physica A, Vol. 261, No. 3-4, 1998, pp. 534-554. doi:10.1016/S0378-4371(98)00437-3
- S. Curilef, “Generalized Statistical Mechanics for the N Body Quantum Problem—Ideal Gases,” Zeitschrift Für Physik B Condensed Matter, Vol. 100, No. 3, 1995, pp. 433-440.
- L. V. Yakushevich, “Nonlinear Physics of DNA,” 2nd Edition, Wiley-VCH Verlag-GmbH & Co. KGaA, Weinheim, 2004.
- S. Li, et al., “Atomic Force Microscopy Measurement of DNA Fragment Induced by Heavy Ions,” Chinese Physics Letters, Vol. 22, No. 4, 2005, pp. 1010-1013. doi:10.1088/0256-307X/22/4/064
- D. Pang, J. E. Rodgers, B. L. Berman, S. Chasovskikh and A. Dritschilo, “Spatial Distribution of Radiation-Induced Double-Strand Breaks in Plasmid DNA as Resolved by Atomic Force Microscopy,” Radiation Research, Vol. 164, No. 6, 2005, pp. 755-765. doi:10.1667/RR3425.1
- J. D. T. Arruda-Neto, et al., “Personal communication,” Journal of Biological Physics, in press.
- D. Pang, B. L. Berman, S. Chasovskikh, J. E. Rodgers and A. Dritschilo, “Investigation of Neutron-Induced Damage in DNA by Aromic Force Microscopy: Experimental Evidence of Clustered DNA Lesions,” Radiation Research, Vol. 150, No. 6, 1998, pp. 612-618. doi:10.2307/3579883
- D. Pang, et al., “Radiation-generated Short DNA Fragments May Perturb Non-homologous End-joining and Induce Genomic Instability,” Journal of Radiation Research, Vol. 52, No. 3, 2011, pp. 309-319. doi:10.1269/jrr.10147

