Advances in Bioscience and Biotechnology
Vol.5 No.2(2014), Article ID:42408,8 pages DOI:10.4236/abb.2014.52012
Meta-analyses of nine polymorphisms of six genes with the risk of schizophrenia
1Zhejiang Provincial Key Laboratory of Pathophysiology, School of Medicine, Ningbo University, Ningbo, China
2Bank of Blood Products, Ningbo No.2 Hospital, Ningbo, China
3Ningbo Addiction Research and Treatment Center, School of Medicine, Ningbo University, Ningbo, China
4Department of Psychiatry, Ningbo Kangning Hospital, Ningbo, China
5The Affiliated Hospital, Ningbo University, Ningbo, China
Email: *dryemeng@yahoo.com.cn, *gaoshugui@sina.com, *duanshiwei@nbu.edu.cn
Copyright © 2014 Xuting Xu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Xuting Xu et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received 29 October 2013; revised 15 December 2013; accepted 1 January 2014
KEYWORDS
Schizophrenia; Meta-Analysis; SNP
ABSTRACT
The aim of this study was to determine whether 9 genetic polymorphisms confered susceptibility to schizophrenia (SCZ). The authors conducted meta-analyses on associations between SCZ and 9 variants of 6 genes including PIK3C3 (432C > T), ABCB1 (C3435T and G2677T), CTLA4 (+49A/G), OLIG2 (rs762178), GAD1 (rs1978340, rs3749034 and rs769395), and GRIN1 (G1001C). A total of 34 case-control studies were involved in our meta-analyses. Our results showed no significant association between all the loci and SCZ. This meta-analysis confirmed a lack of association of SCZ for 9 genetic polymorphisms including GRIN1 G1001C, ABCB1 C3435T and G2677T, CTLA4 + 49A/G, OLIG2 rs762178, GAD1 gene rs1978340, rs3749034 and rs769395, and PIK3C3 432C > T.
1. INTRODUCTION
Schizophrenia (SCZ) affects about 1% of the population in the world. SCZ is a complex mental disorder resulting from the interaction between genetic and environmental factors. Twin studies estimate that the sum of the genetic effects in liability to SCZ is 81% [1,2] in contrast of less than 20% from environmental factors of SCZ [3]. Although a handful of genes have been identified to be associated with SCZ in recent studies [4], replication of these results in other ethnic populations or meta-analyses of available studies is necessary for this high heterogeneous mental disorder [5-7].
Glutamate hypothesis is classical in the pathogenesis of SCZ [8] and provides an explanation of the brain abnormalities associated with SCZ [9]. N-methyl D-aspartate 1 (GRIN1) encodes a critical subunit of N-methyl-Daspartate receptors (NMDAR) that plays a pivotal role in glutamate neurotransmitter system. As a member of the superfamily of ATP-binding cassette (ABC) transporters, ABCB1 was shown to be associated with the metabolic disturbances by antipsychotic drugs [10-12]. The immune system has changed in the SCZ patients such as the complement pathway [13,14]. The interaction between neurodevelopmental immune insults and genetic background will increase the risk for SCZ [15]. As a member of the immunoglobulin superfamily, cytotoxic T-lymphocyte-associated protein 4 (CTLA4) encodes a protein which transmits an inhibitory signal to T cells, and has been shown to be associated with the risk to SCZ [16-18]. Glutamic acid decarboxylase (GAD) enzymes can catalyze glutamate turning to gamma-Aminobutyric acid (GABA). GAD1 encodes one type of glutamic acid decarboxylases (2069816) that can catalyze glutamate turning to GABA. Dysfunction of the GABAergic system is associated with the development of SCZ [19]. Oligodendrocyte lineage transcription factor 2 (OLIG2) encodes the oligodendrocyte transcription factor that was shown with association with SCZ [20-22]. Phosphoinositide- 3-kinase, class III (PIK3C3) was involved in the pathways of phosphoinositide synthesis (PI) [23], and was essential for CNS neuronal homeostasis [24]. Evidences support PIK3C3 as a candidate gene of SCZ [25].
Associations of single-nucleotide polymorphisms (SNPs) in the above six genes with SCZ have been reported in different ethnic populations. Since the allelic frequencies of genes often differ substantially among different ethnic groups, a combined analysis of these studies may help compare the genetic associations in different populations. In the present study, we perform meta-analyses to evaluate the contribution of the polymorphisms of the six genes to SCZ susceptibility in different populations.
2. METHODS AND MATERIALS
2.1. Publication Search and Data Extraction
Candidate studies for current meta-analyses were retrieved after a search from 2000 to 2013 in the electronic databases including PubMed, Embase, Web of Science, Wanfang database and China National Knowledge Infrastructure (CNKI). The keywords and Medical Subject headings used in the search include “schizophrenia”, together with “polymorphism”, or “allele”, or “genotype” or “SNP”. As shown in Figure 1, SNPs with less than 3 independent case-control studies were excluded from further analysis. SNPs with previous meta-analysis were also discarded from the current study. The retrieved information include the first author’s name, year of publication, ethnic group, number of genotypes and alleles, and total number of cases and controls. Our comprehensive search identified a total of 9 SNPs of 6 genes that
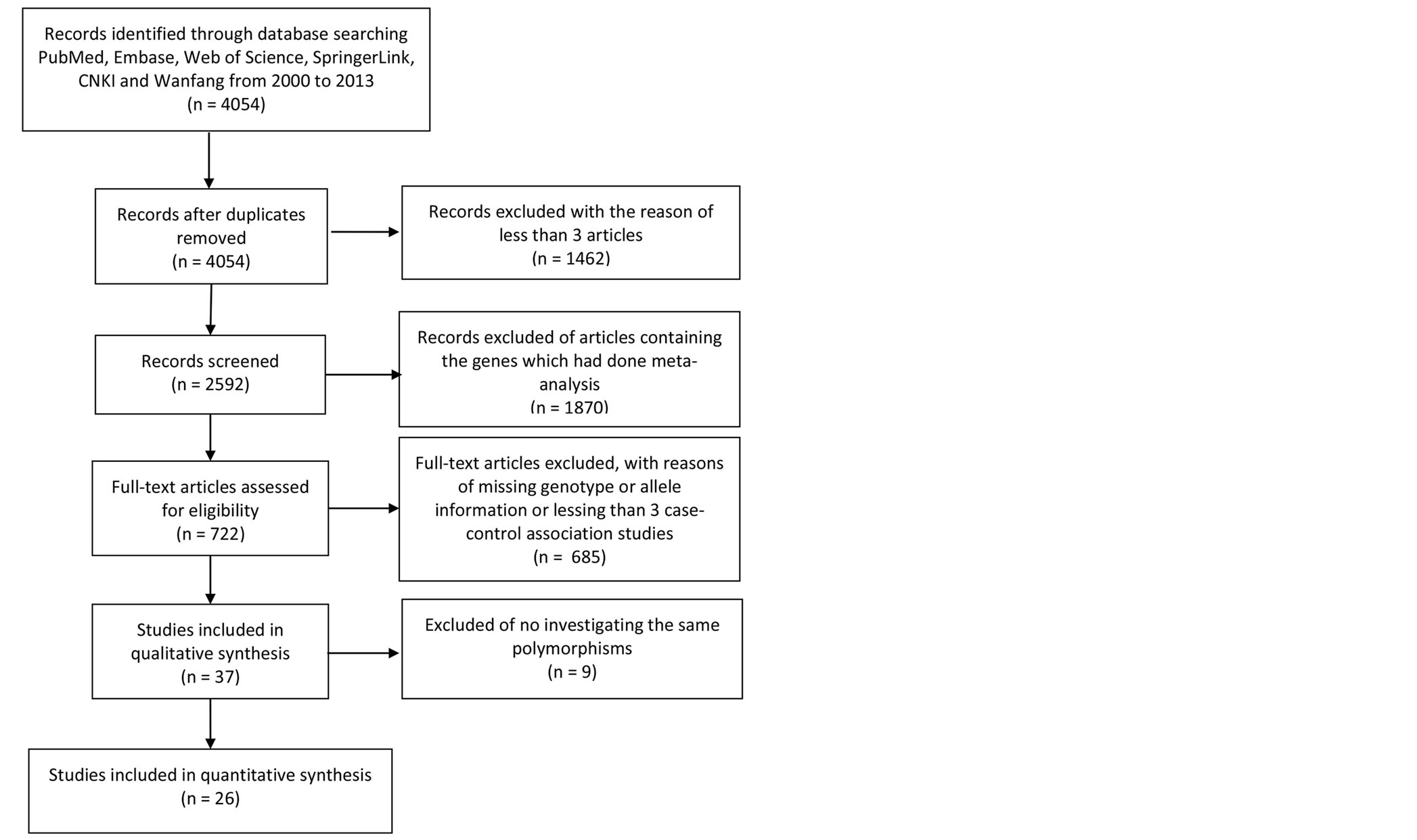
Figure 1. Flow design of meta-analyses statement for trial selection process.
were involved with at least 3 independent genetic studies. These genes comprised, PIK3C3 (432C > T), ABCB1 (C3435T and G2677T), CTLA4 (+49A/G), OLIG2 (rs762178), GAD1 (rs1978340, rs3749034 and rs769395), and GRIN1 (G1001C).
2.2. Statistical Analysis
Meta-analyses were performed using the Review Manager (version 5.0, The Cochrane Collaboration). OR values and 95% CIs of the meta-analyses were demonstrated in the forest plots. Heterogeneity in the meta-analyses was calculated using the Cochran’s Q statistic and I2 test [26]. For the meta-analyses with significant heterogeneity (I2 > 50%), random-effect model was applied instead of the fixed effects model. Publication bias of the studies in the meta-analyses was shown in the funnel plots.
3. RESULTS
As shown in Figure 1, a total of 4054 genetic association studies involving with 200 SCZ candidate genes were initially retrieved from the online databases including PubMed, Embase, Web of Science and Springer. Among these, 1331 studies on 61 genes with previous meta-analyses are discarded for further analysis, and 803 studies without enough genetic information were also excluded. At last, 34 case-control studies from 26 articles among 10,117 SCZ cases and 10,362 controls were included for the current meta-analyses of 9 polymorphisms on 6 genes (Figure 1). The details were showed in Tables 1 and 2.
Our data also demonstrated a moderate heterogeneity of PIK3C3 432C > T (I2 = 42%, Figure 2) and a significant heterogeneity of GRIN1 G1001C (I2 = 79%, Figure 3) and OLIG2 rs762178 polymorphism (I2 = 94%, Figure 3). No evidence of statistical heterogeneity was observed for 8 SNPs, including ABCB1 polymorphisms (C3435T: I2 = 1%; G2677T: I2 = 0%, Figure 2), CTLA4 +49A/G (I2 = 46%. Figure 2), GAD1 polymorphisms (rs1978340: I2 = 0%; rs3749034: I2 = 0%; rs769395: I2 = 0%, Figure 3). The funnel plots of the above SNPs were shown in Figure 4.
No evidence of an association with SCZ obvious for all the 9 SNPs including PIK3C3 (432C > T: OR = 0.93, 95% CI = 0.75 - 1.15, P = 0.50; Figure 2), ABCB1 (G2677T: OR = 1.17, 95% CI = 0.85 - 1.63, P = 0.34; C3435T: OR = 1.20, 95% CI = 0.92 - 1.57, P = 0.17; Figure 2), CTLA4 (+49A/G: OR = 0.89, 95% CI = 0.76 - 1.04, P = 0.15; Figure 2), GAD1 (rs1978340: OR = 1.03, 95% CI = 0.84 - 1.26, P = 0.79; rs3749034: OR = 0.94, 95% CI = 0.78 - 1.15, P = 0.57; rs769395: OR = 0.96, 95% CI = 0.78 - 1.17, P = 0.68; Figure 3), OLIG2 rs762178 (OR = 0.99, 95% CI = 0.57 - 1.72, P = 0.97, Figure 3) and GRIN1 G1001C (OR = 1.00, 95% CI = 0.63 - 1.58, P = 1.0). No obvious publication bias was
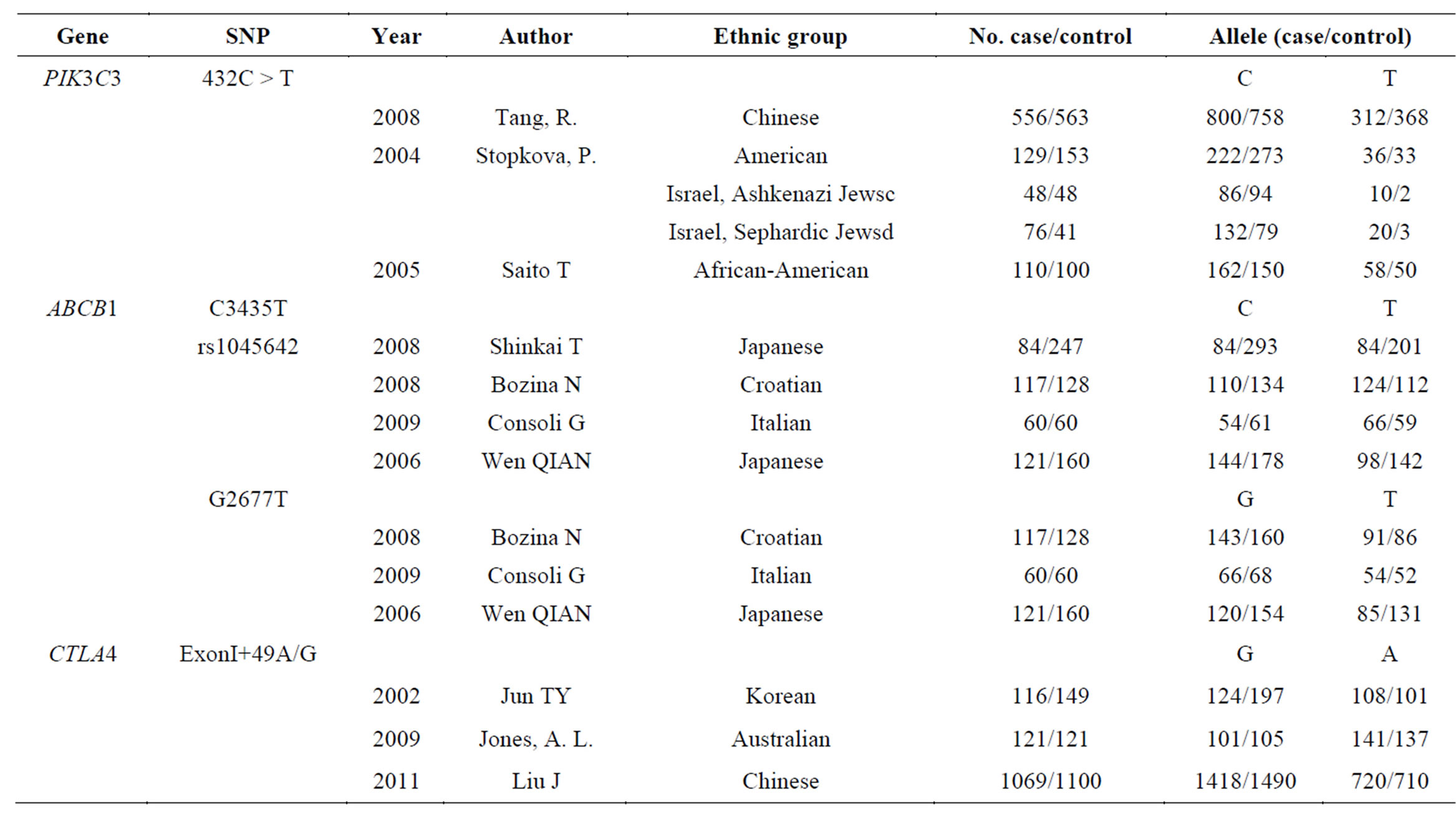
Table 1. The detailed data of the enrolled SNPs (PIK3C3 432C > T, ABCB1 C3435T, ABCB1 G2677T, and CTLA4 +49A/G).
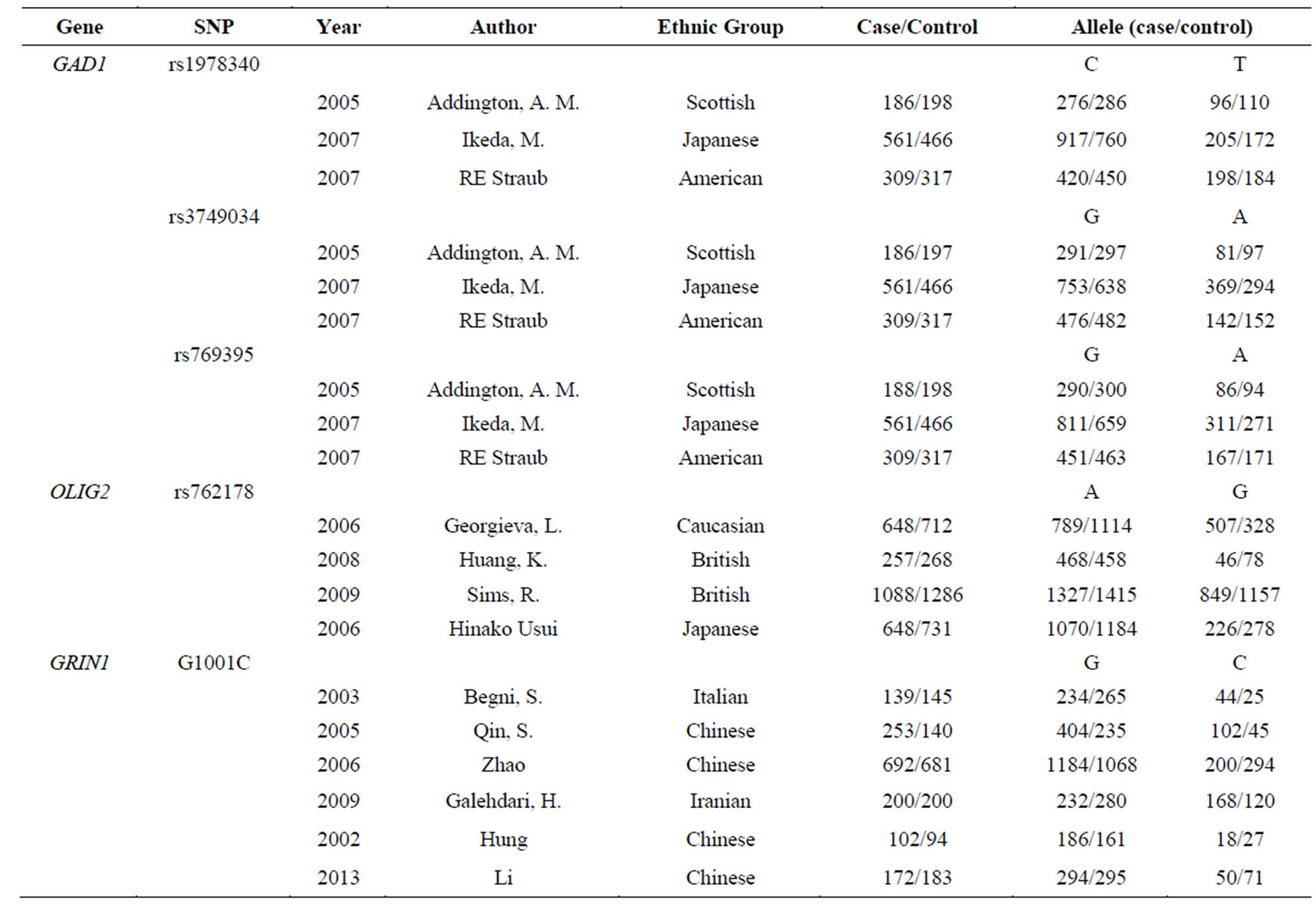
Table 2. The detailed data of the enrolled SNPs (GAD1 rs1978340, GAD1 rs3749034, GAD1 rs769395, OLIG2 rs762178 and GRIN1 G1001C).

Figure 2. Forest Plot for the relationship between SNPs (PIK3C3 432C > T, ABCB1 C3435T, ABCB1 G2677T, and CTLA4 + 49A/G) and SCZ in the meta-analysis.
observed in the funnel plots for the 9 meta-analyses (Figure 4). Moreover, subgroup meta-analyses by ethnicity of GRIN1 G1001C polymorphism in the Chinese population was failed to find the association between GRIN1 G1001C and the risk of SCZ (OR = 0.74, 95% CI = 0.52 - 1.07, P = 0.11).
4. DISUSSION
With the advancement in the genotyping technologies, the number of association studies is soaring in order to harvest the genetic variants underlying SCZ. Meta-analysis as a tool has been widely applied for a comprehensive analysis to overcome the defection of small sample size. Instead of focusing hot genes including COMT, DRD2 and BDNF, we aimed to check the ignored SNPs without being summarized in the previous meta-analysis. Through a comprehensive filtration starting from 1267 genes in 4054 literatures, 9 variants of 6 genes from 34 studies were included in the current meta-analyses.
Based on case-control association studies for the susceptibility of SCZ, our meta-analyses found no evidence of significant associations between the 9 SNPs and SCZ. Encoding by GRIN1 (NR1) gene, NR1 subunit was the member of the NMDA receptor, which functioned as a glutamate-gated cation channel [27,28]. Mice model of SCZ expressing only 5% of normal levels of the essential NMDAR1 (NR1) displayed behavioral abnormalities [29]. The transcription of cloning of GRIN1 gene in the chicken showed higher activity of the 5'-flanking region retinal neurons and neuronally-differentiated PC12 cells [30]. G1001C located in the promoter region of GRIN1 seems to alter a consensus sequence for the p50 subunit of the transcription factor NF-kB [31]. Via N-methyl-Daspartate (NMDA)-receptor activation, glutamate regulates synaptic activation of NF-kappa B which would act as pivotal regulators of activity-dependent inhibitory and excitatory neuronal function regulating synaptic plasticity and memory [32,33]. Furthermore, another study showed that the NF-κB site positively regulated the GRIN1 promoter during neuronal differentiation via interacting mainly with Sp, a transcription factor [34].
The association analysis of the SNPs in GRIN1 gene with SCZ has been evaluated in Italian, Chinese and Iranian populations with inconsistent outcomes. Zhao’s study indicated GRIN1 1001C as a protective factor in Chinese population, while other studies including 1 study in Italian population showed GRIN1 1001C as a risky factor. A meta-analysis with exclusion of Zhao’s study (OR = 1.15, 95% CI = 0.74 - 1.78, P = 0.53) or Begin’s study (OR = 0.90, 95% CI = 0.56 - 1.45, P = 0.66) showed no significant association between GRIN1 1001C and SCZ. In addition, a significant genetic inte-
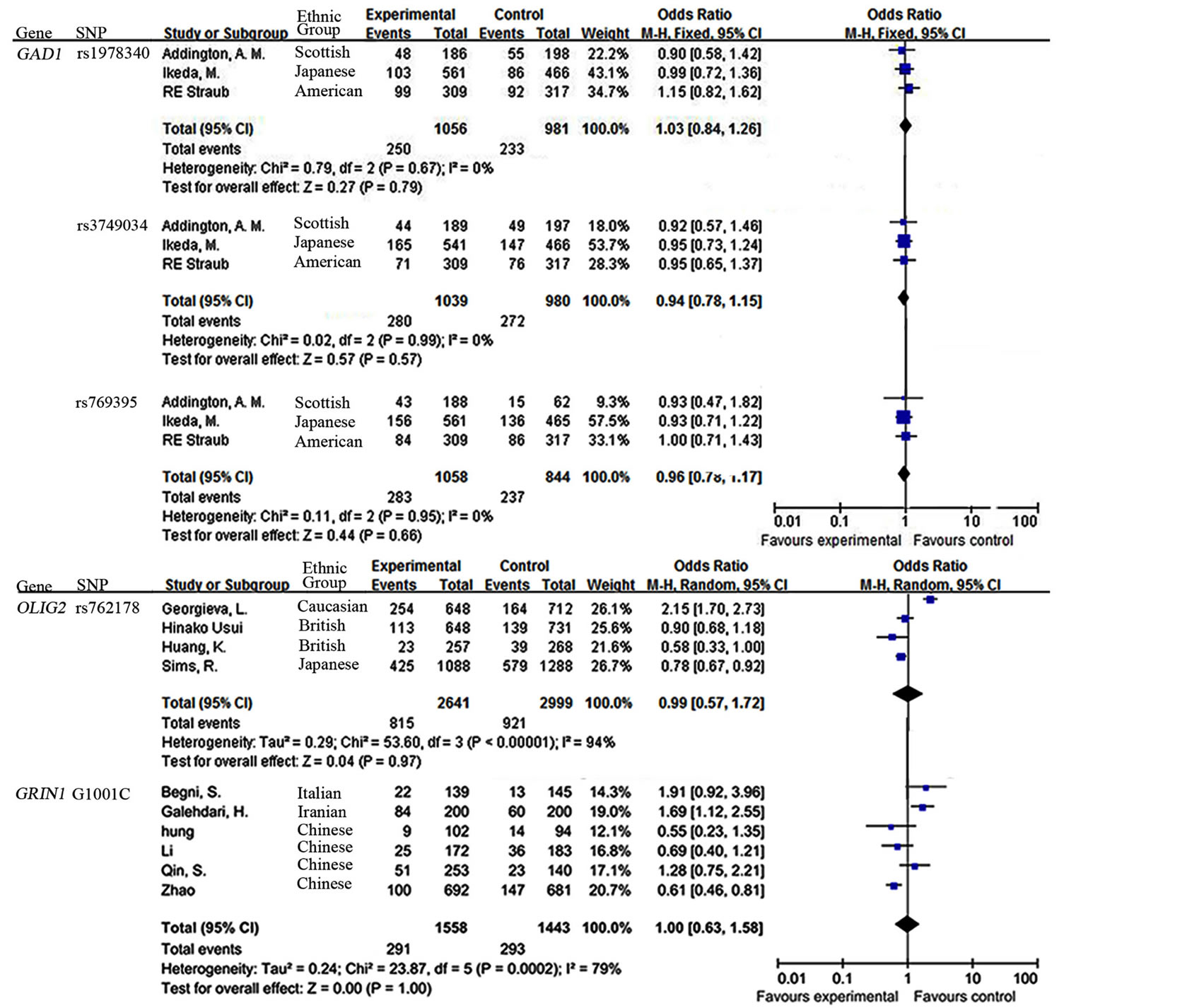
Figure 3. Forest Plot for the relationship between SNPs (GAD1 rs1978340, GAD1 rs3749034 and GAD1 rs769395, OLIG2 rs762178, and GRIN1 G1001C) and SCZ in the meta-analysis.
raction between the G1001C in the GRIN1 gene and the T4197C and T5988C polymorphisms in the GRIN2B gene implied that the combined effects might be involved in the etiology of schizophrenia [34].
The results of our meta-analysis do not provide support for the association of the rest 8 polymorphisms with SCZ. Under a moderate risk of AD (OR = 1.2), power analysis showed G1001C of GRIN1 (40.6%), C3435T of ABCB1 (49.9%), G2677T of ABCB1 (34.7%), 49A/G of CTLA4 (89.6%), rs1978340 of GAD1 (71.8%), rs3749034 of GAD1 (75.7%), rs769395 of GAD1 (74.9%), 432C > T of PIK3C3 (68.3%) and rs762178 of OLIG2 (99.6%). Among those, 3 SNPs lacked power for the meta-analyses. It might be the reason why we fail to find the evidence between the 9 polymorphisms of SCZ.
There were several limitations in our meta-analyses. Firstly, sample size was rather small, which might lead to a mistake in finding markers with small effects on SCZ. Future studies with larger sample size and a rigorous study design may help prevent a lack of statistical power.
Secondly, SCZ is a complex disorder that has been classified into several complex subtypes in clinical. The case samples in the involved studies didn’t provide enough subtype information which may introduce hidden stratification in our meta-analyses. Lastly, lack of enough information and available studies, the subgroup analysis of the stratifying variable including gender and ethnicity was unable to be performed in our studies.
In conclusion, our meta-analyses demonstrated a lack of association between SCZ and 9 variants of 6 genes including PIK3C3 (432C > T), ABCB1 (C3435T and G2677T), CTLA4 (+49A/G), OLIG2 (rs762178), GAD1 gene (rs1978340, rs3749034 and rs769395) and GRIN1 (G1001C).
ACKNOWLEDGEMENTS
The research was supported by the grants from National Natural Science Foundation of China (31100919, 81371469), Natural Science Foundation of Zhejiang Province (LR13H020003), Ningbo social de
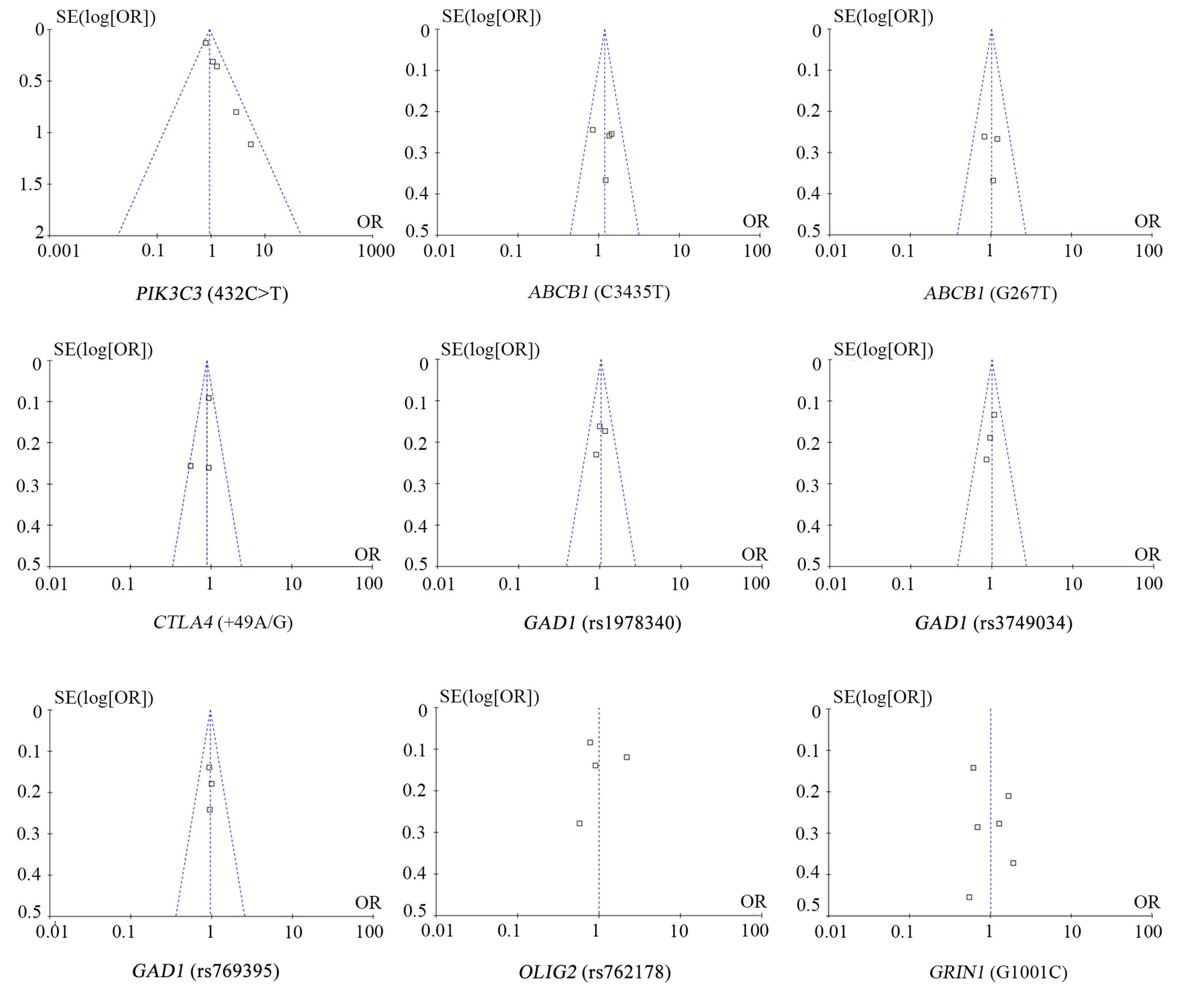
Figure 4. Funnel plot for the relationship between SNPs (432C > T, C3435T, G2677T, +49A/G, rs1978340, rs3749034, rs769395, rs762178, and G1001C) and SCZ in the meta-analysis.
velopment research projects (2012C50032), and K. C. Wong Magna Fund in Ningbo University.
REFERENCES
- Fowler, T., Zammit, S., Owen, M.J. and Rasmussen, F. (2012) A population-based study of shared genetic variation between premorbid IQ and psychosis among male twin pairs and sibling pairs from Sweden. Archives of General Psychiatry, 69, 460-466. http://dx.doi.org/10.1001/archgenpsychiatry.2011.1370
- van Haren, N.E., Rijsdijk, F., Schnack, H.G., et al. (2012) The genetic and environmental determinants of the association between brain abnormalities and schizophrenia: the schizophrenia twins and relatives consortium. Biological Psychiatry, 71, 915-921. http://dx.doi.org/10.1016/j.biopsych.2012.01.010
- Sullivan, P.F., Kendler, K.S. and Neale, M.C. (2003) Schizophrenia as a complex trait: Evidence from a metaanalysis of twin studies. Archives of General Psychiatry, 60, 1187-1192. http://dx.doi.org/10.1001/archpsyc.60.12.1187
- Ding, C.L., Zhou, X., Wang, B.J., Ding, M. and Pang, H. (2012) Association and its forensic significance between COMT gene and schizophrenia. Journal of Forensic Medicine, 28, 299-304.
- Duan, S., Gao, R., Xing, Q., et al. (2005) A family-based association study of schizophrenia with polymorphisms at three candidate genes. Neuroscience Letters, 379, 32-36. http://dx.doi.org/10.1016/j.neulet.2004.12.040
- Duan, S., Yin, H., Chen, W., et al. (2005) No association between the serotonin 1B receptor gene and schizophrenia in a case-control and family-based association study. Neuroscience Letters, 376, 93-97. http://dx.doi.org/10.1016/j.neulet.2004.11.028
- Duan, S., Xu, Y., Chen, W., et al. (2004) No association between the promoter variants of tumor necrosis factor alpha (TNF-alpha) and schizophrenia in Chinese Han population. Neuroscience Letters, 366, 139-143. http://dx.doi.org/10.1016/j.neulet.2004.05.039
- Sakurai, K., Toru, M., Yamakawa-Kobayashi, K. and Arinami, T. (2000) Mutation analysis of the N-methylD-aspartate receptor NR1 subunit gene (GRIN1) in schizophrenia. Neuroscience Letters, 296, 168-170. http://dx.doi.org/10.1016/S0304-3940(00)01599-8
- Labrie, V., Wang, W., Barger, S.W., Baker, G.B. and Roder, J.C. (2010) Genetic loss of D-amino acid oxidase activity reverses schizophrenia-like phenotypes in mice. Genes, Brain and Behavior, 9, 11-25. http://dx.doi.org/10.1111/j.1601-183X.2009.00529.x
- Jovanovic, N., Bozina, N., Lovric, M., Medved, V., Jakovljevic, M. and Peles, A.M. (2010) The role of CYP- 2D6 and ABCB1 pharmacogenetics in drug-naive patients with first-episode schizophrenia treated with risperidone. European Journal of Clinical Pharmacology, 66, 1109-1117. http://dx.doi.org/10.1007/s00228-010-0850-1
- Nikisch, G., Baumann, P., Oneda, B., et al. (2011) Cytochrome P450 and ABCB1 genetics: Association with quetiapine and norquetiapine plasma and cerebrospinal fluid concentrations and with clinical response in patients suffering from schizophrenia. A pilot study. Journal of Psychopharmacology, 25, 896-907. http://dx.doi.org/10.1177/0269881110389208
- Kuzman, M.R., Medved, V., Bozina, N., Grubisin, J., Jovanovic, N. and Sertic, J. (2011) Association study of MDR1 and 5-HT2C genetic polymorphisms and antipsychotic-induced metabolic disturbances in female patients with schizophrenia. Pharmacogenomics Journal, 11, 35- 44. http://dx.doi.org/10.1038/tpj.2010.7
- Singh, B., Bera, N.K., Nayak, C.R. and Chaudhuri, T.K. (2011) Immunomodulation in schizophrenia: A study among the Indian schizophrenia patients of Siliguri, West Bengal. Asian Journal of Psychiatry, 4, 277-283. http://dx.doi.org/10.1016/j.ajp.2011.08.002
- Li, Y., Zhou, K., Zhang, Z., et al. (2012) Label-free quantitative proteomic analysis reveals dysfunction of complement pathway in peripheral blood of schizophrenia patients: Evidence for the immune hypothesis of schizophrenia. Molecular BioSystems, 8, 2664-2671. http://dx.doi.org/10.1039/c2mb25158b
- Michel, M., Schmidt, M.J. and Mirnics, K. (2012) Immune system gene dysregulation in autism and schizophrenia. Developmental Neurobiology, 72, 1277-1287. http://dx.doi.org/10.1002/dneu.22044
- Liu, J., Li, J., Li, T., et al. (2011) CTLA-4 confers a risk of recurrent schizophrenia, major depressive disorder and bipolar disorder in the Chinese Han population. Brain, Behavior, and Immunity, 25, 429-433. http://dx.doi.org/10.1016/j.bbi.2010.10.024
- Jones, A.L., Holliday, E.G., Mowry, B.J., et al. (2009) CTLA-4 single-nucleotide polymorphisms in a Caucasian population with schizophrenia. Brain, Behavior, and Immunity, 23, 347-350. http://dx.doi.org/10.1016/j.bbi.2008.09.008
- Jun, T.Y., Pae, C.U., Chae, J.H., Bahk, W.M., Kim, K.S. and Han, H. (2002) Polymorphism of CTLA-4 gene at position 49 of exon 1 may be associated with schizophrenia in the Korean population. Psychiatry Research, 110, 19-25. http://dx.doi.org/10.1016/S0165-1781(02)00031-8
- Blum, B.P. and Mann, J.J. (2002) The GABAergic system in schizophrenia. International Journal of Neuropsychopharmacology, 5, 159-179. http://dx.doi.org/10.1017/S1461145702002894
- Katsel, P., Davis, K.L. and Haroutunian, V. (2005) Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: A gene ontology study. Schizophrenia Research, 79, 157- 173. http://dx.doi.org/10.1016/j.schres.2005.06.007
- Sims, R., Hollingworth, P., Moskvina, V., et al. (2009) Evidence that variation in the oligodendrocyte lineage transcription factor 2 (OLIG2) gene is associated with psychosis in Alzheimer's disease. Neuroscience Letters, 461, 54-59. http://dx.doi.org/10.1016/j.neulet.2009.05.051
- Schurman, S.H., Sharon, N., Goldschmidt, R.A. and Scanlon, E.F. (1990) Improved detection of metastases to lymph nodes and estrogen receptor determination. Archives of Surgery, 125, 179-182. http://dx.doi.org/10.1001/archsurg.1990.01410140057010
- Fruman, D.A., Meyers, R.E. and Cantley, L.C. (1998) Phosphoinositide kinases. Annual Review of Biochemistry, 67, 481-507. http://dx.doi.org/10.1146/annurev.biochem.67.1.481
- Wang, L., Budolfson, K. and Wang, F. (2011) Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience, 172, 427-442. http://dx.doi.org/10.1016/j.neuroscience.2010.10.035
- Tang, R., Zhao, X., Fang, C., et al. (2008) Investigation of variants in the promoter region of PIK3C3 in schizophrenia. Neuroscience Letters, 437, 42-44. http://dx.doi.org/10.1016/j.neulet.2008.03.043
- Coory, M.D. (2010) Comment on: Heterogeneity in metaanalysis should be expected and appropriately quantified. International Journal of Epidemiology, 39, 932. http://dx.doi.org/10.1093/ije/dyp157
- Ishii, T., Moriyoshi, K., Sugihara, H., et al. (1993) Molecular characterization of the family of the N-methyl-Daspartate receptor subunits. Journal of Biological Chemistry, 268, 2836-2843.
- Monyer, H., Sprengel, R., Schoepfer, R., et al. (1992) Heteromeric NMDA receptors: Molecular and functional distinction of subtypes. Science, 256, 1217-1221. http://dx.doi.org/10.1126/science.256.5060.1217
- Mohn, A.R., Gainetdinov, R.R., Caron, M.G. and Koller, B.H. (1999) Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell, 98, 427-436. http://dx.doi.org/10.1016/S0092-8674(00)81972-8
- Zarain-Herzberg, A., Lee-Rivera, I., Rodriguez, G. and Lopez-Colome, A.M. (2005) Cloning and characterization of the chick NMDA receptor subunit-1 gene. Molecular Brain Research, 137, 235-251. http://dx.doi.org/10.1016/j.molbrainres.2005.03.006
- Qin, S., Zhao, X., Pan, Y., et al. (2005) An association study of the N-methyl-D-aspartate receptor NR1 subunit gene (GRIN1) and NR2B subunit gene (GRIN2B) in schizophrenia with universal DNA microarray. European Journal of Human Genetics, 13, 807-814. http://dx.doi.org/10.1038/sj.ejhg.5201418
- Guerrini, L., Blasi, F. and Denis-Donini, S. (1995) Synaptic activation of NF-kappa B by glutamate in cerebellar granule neurons in vitro. Proceedings of the National Academy of Sciences of USA, 92, 9077-9081. http://dx.doi.org/10.1073/pnas.92.20.9077
- O’Mahony, A., Raber, J., Montano, M., et al. (2006) NFkappaB/Rel regulates inhibitory and excitatory neuronal function and synaptic plasticity. Molecular and Cellular Biology, 26, 7283-7298. http://dx.doi.org/10.1128/MCB.00510-06
- Liu, A., Hoffman, P.W., Lu, W. and Bai, G. (2004) NFkappaB site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. Journal of Biological Chemistry, 279, 17449-17458. http://dx.doi.org/10.1074/jbc.M311267200
NOTES
*Corresponding authors.

