Advances in Bioscience and Biotechnology
Vol.4 No.4A(2013), Article ID:30574,6 pages DOI:10.4236/abb.2013.44A001
Detection of cardiac myosin binding protein-C (cMyBP-C) by a phospho-specific PKD antibody in contracting rat cardiomyocytes
![]()
1Department of Molecular Genetics, Cardiovascular Research Institute Maastricht (CARIM), Maastricht University, Maastricht, The Netherlands
2Department of Human Biology, Nutrition and Toxicology Research Institute Maastricht (NUTRIM), Maastricht University, Maastricht, The Netherlands
3Protein Phosphorylation Unit, Université Catholique de Louvain, Brussels, Belgium
4Department of Cell and Molecular Physiology, Stritch School of Medicine, Loyola University, Chicago, USA
Email: j.luiken@maastrichtuniversity.nl
Copyright © 2013 Ellen Dirkx et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received January 12th, 2013; revised March 6th, 2013; accepted April 11th, 2013
Keywords: Protein Kinase D; Cardiac Myosin Binding Protein-C; Phospho-Specific Antibody; Protein Phosphorylation
ABSTRACT
Protein phosphorylation plays an important role in physiological processes, such as muscle contraction. Phospho-specific antibodies have become powerful tools to study these processes. Cardiac myosin binding protein-C (cMyBP-C) is one of the proteins that make up the contractile apparatus of cardiomyocytes. Phosphorylation of cMyBP-C is essential for normal cardiac function, since dephosphorylation of this protein leads to its degradation and has been associated with cardiomyopathy. One of the upstream kinases, which phosphorylate cMyBP-C, is protein kinase D (PKD). While studying the role of PKD in cMyBP-C phosphorylation, we tried to analyze phosphorylation of PKD with a phospho-specific PKD-Ser744/748 antibody. Contrary to the expected 115 kDa, a signal was found for a 150-kDa protein. By MALDI-TOF mass spectrometry, we identified this protein to be cMyBP-C. These data were confirmed by immunostaining using the p-PKD-Ser744/748 antibody, which displayed a striated pattern similar to the one observed for a regular cMyBP-C antibody. To our knowledge there are no antibodies commercially available for phosphorylated cMyBP-C. Thus, the p-PKDSer744/748 antibody can accelerate research into the role of cMyBP-C phosphorylation in cardiomyocytes.
1. INTRODUCTION
Reversible phosphorylation changes kinetic properties of proteins. As a result, protein kinases can modify the function of a protein in almost every conceivable way. For instance, kinases influence enzymatic activities, movement between subcellular compartments, and proteinprotein interactions [1]. Therefore, protein phosphorylation is now recognized to play a critical role in many physiological processes [1]. Thus, aberrant phosphorylation will disrupt these processes and is expected to be the cause of a number of human diseases. As a result, it has become increasingly important to determine changes in the phosphorylation state of proteins. Phospho-specific antibodies are important and powerful tools to study these processes, because they can detect phosphorylation of a specific site or motif of a protein.
Heart failure in mouse models and in humans has been associated with dephosphorylation and subsequent degradation of cardiac myosin binding protein-C (cMyBP-C) [2-4]. A decrease in cMyBP-C interferes with crossbridge formation that has been demonstrated to depend on cMyBP-C phosphorylation through enzymes such as Ca2+-calmodulin-activated kinase II, protein kinase A and protein kinase C [5]. Also, mutations in this protein, possibly leading to an accelerated degradation of cMyBP-C, have been linked to cardiomyopathy [5,6]. cMyBP-C is thought to facilitate the interaction between myosin and actin by tethering both molecules, but the exact mechanism is hardly understood. The elucidation of the precise function of cMyBP-C phosphorylation is also clinically important because cMyBP-C phosphorylation has been suggested as a biomarker for diagnosing myocardial infarction and as a potential target for therapeutic intervention [5,6]. However, such medical applications await an improved insight in the role of cMyBP-C phosphorylation in cardiac contraction.
Protein kinase D1 (PKD1) has been shown to participate in many signaling cascades. The 115 kDa PKD1 belongs to a serine/threonine kinase family and contains an N-terminal regulatory domain and a C-terminal kinase domain [7]. Its activity, phosphorylation of downstream targets in the signaling cascade, depends on its phosphorylation state as a number of residues can be (de) phosphorylated, giving rise to specific reaction profiles. In the heart it has been demonstrated to affect contractility, metabolism and remodeling [5,8-10]. Recently, PKD (in addition to the above mentioned kinases) has been demonstrated to phosphorylate cMyBP-C [10]. However, the relevance of this cMyBP-C modification is still a matter of research, which has been hampered by the lack of adequate tools to detect phosphorylated cMyBP-C.
During a series of experiments aimed at determining the role of PKD in cMyBP-C phosphorylation we observed an aberrant band in western blots probed with a phospho-PKD-Ser744-748 antibody (p-PKD-Ser744/748 antibody). Further investigations showed that this phospho-PKD “specific” antibody recognized cMyBP-C phosphorylation, making this antibody an important tool in further research into the function of cMyBP-C.
2. EXPERIMENTAL PROCEDURES
2.1. Materials
Antibodies directed against phospho-PKD/PKC-μ (Ser916) (Antibody #2051), phospho-PKD/PKC-μ (Ser744/748) (Antibody #2054), PKD/ PKC-μ (Antibody #2052) and caveolin-3 were obtained from Cell Signaling (Beverly, USA). The caveolin-3 antibody (Catalog # 10420) was obtained from BD Transduction LaboratoriesTM (Breda, The Netherlands) and the MYBPC3 (cMyBP-C) antibody (K-16) (Catalog # sc-50115) from Santa Cruz Biotechnology, Inc. (Santa Cruz, USA). The phosphospecific cMyBP-C-Ser315 antibody was generated by Dr. Sadayappan [11]. The Titin antibody (mouse monoclonal, T12) was a gift from Prof. Furst, (Potsdam, Germany) and the Desmin antibody (rabbit polyvalent) was a gift from Dr. Schaart, (Maastricht, The Netherlands). Secondary antibodies used for western blotting were: anti-rabbit IgG, HRP-linked antibody (Antibody #7074) obtained form Cell Signaling (Beverly, USA) and polyclonal rabbit anti-mouse immunoglobulins/HRP (Catalog # P 0161) obtained from Dakocytomation (Denmark). Secondary antibodies used for the immunostaining (goat antimouseFITC (#ab7064) and goat anti-rabbit-FITC (#ab7086) were purchased from Abcam (Cambridge, UK), and rabbit anti-goat-FITC (#F025002) from Dakocytomation (Denmark). Oligomycin and phenylephrine were purchased from Sigma-Aldrich (St. Louis, USA).
2.2. Isolation and Treatment of Adult Rat Cardiomyocytes
Cardiomyocytes were isolated from male Lewis rats (200 - 250 g) (obtained from Charles River) using a Langendorff perfusion system and a Krebs Henseleit bicarbonate medium equilibrated with a 95% O2/5% CO2 gas phase at 37˚C as previously described [12]. Cardiomyocytes were incubated for 15 min at 37˚C with continuous shaking either with 5 mmol/L oligomycin or 50 mmol/L phenylephrine, or were electrically stimulated for 6 min using a commercially available IONOPTIX stimulator (4 Hz, 40 V, pulse duration 5 ms).
2.3. Immunoblotting
Proteins were separated by electrophoresis and transferred to nitrocellulose membranes for Western Blotting. The membranes were probed with the relevant primary antibody. The protein bands were visualized using enhanced chemiluminescence and immunoblot intensities and analyzed by densitometry, as described previously [8].
2.4. 2D-Gel-Electrophoresis
Total protein (50 µg) was loaded for the first dimension. Iso-electric focusing (IEF) was preformed on an IPG PHOR electrophoresis unit (Amersham Biosciences) at 20˚C. IEF was performed using the following program: 500 V for 1 h, 1000 V for 1 h, 1000 - 8000 V for 2 h and a final step of 8000 V for 56,250 Vh. The second dimension run was carried out on 12.5% polyacrylamide gels. Electrophoresis was conducted at a constant voltage of 200 V for 5 h in a 24 cm Dodeca Cell (Bio-Rad, Veenendaal, The Netherlands) [13]. Proteins were stained with Sypro Ruby. Gel images were obtained with a FX Molecular Imager (Bio-Rad, Veenendaal, The Netherlands).
2.5. In-Gel Digestion
Protein spots were excised from gels using an automated spot cutter (Bio-Rad, Veenendaal, the Netherlands) and processed on a MassPREP digestion robot (Waters, Manchester UK). A solution of 50 mmol/L ammonium bicarbonate in 50% (v/v) acetonitrile was used for destaining. Cysteines were reduced with 10 mmol/L DTT in 100 mmol/L NH4CO3 for 30 min followed by alkylation with 55 mmol/L iodoacetamide in 100 mmol/L NH4CO3 for 20 min. Spots were washed with 100 mmol/L NH4CO3 to remove excess reagents and were subsequently dehydrated with 100% acetonitrile. Trypsin (6 ng/µl) in 50 mmol/L NH4CO3 was added to the gel plug and incubation was preformed at 37˚C for 5 h. Peptides were extracted in 30 μl of 1% (v/v) formic acid/2% (v/v) acetonitrile in water for 30 min at room temperature. A second extraction was performed using 24 μl of 50% (v/v) acetonitrile in water for 20 min at room temperature [13].
2.6. Mass Spectrometry
For MALDI-TOF mass spectrometry 1.5 µl of peptide mixture and 0.5 µl matrix solution (2.5 mg/ml α-cyano- 4-hydroxycinnamic acid in 50% acetonitrile/0.1% TFA) was spotted automatically onto a 96 well-format target plate. Spots were allowed to air dry for homogeneous crystallization. Spectra were obtained using a MALDILR mass spectrometer (Waters). The instrument was operated in positive reflector mode. Acquisition mass range was 800 - 4000 Da. The instrument was calibrated on 10 - 12 reference masses from a tryptic digest of alcohol dehydrogenase. In addition, a near point lockmass correction for each sample spot was performed using adrenocorticotropic hormone fragment 18 - 39 (MH + 2465.199) to achieve maximum mass accuracy. Typically 120 shots were combined and background subtracted. A peptide mass list was generated for the subsequent database search [13].
2.7. Database Search
The peptide mass list was searched with the Mascot search engine (http://www.matrixscience.com) against the Swiss-Prot database (http://expasy.ch/sprot) for protein identification. One missed cleavage was tolerated; carbamidomethylation was set as a fixed modification and oxidation of methionine as an optional modification. The peptide mass tolerance was set to 100 ppm. No restrictions were made on the protein molecular mass and the isoelectric point. A Mascot probability Score was calculated with following formula: –10*Log(P), where P is the probability that the observed match is a random event. A protein was regarded as identified with a significant Mascot probability score, when the. protein scored greater than 67 (P < 0.05) and when there were at least five matched peptides assigned to the identified protein.
2.8. Immunostaining
Isolated rat cardiomyocytes were immunostained with several antibodies. To determine the integrity status of the cells, antibodies against desmin and titin were applied. Cells were fixed by a short immersion in ice-cold methanol. After air-drying the cells were incubated for 1 h at RT with 1/20 horse serum. After a short rinse primary antibodies were applied (Titin 1/10, Desmine 1/25, cMyBP-C 1/10 and p-PKD-Ser744/748 1/20) and the cells were incubated overnight at 4˚C. After washing the cells in PBS for 2 times 10 min, they were again blocked with 1/20 horse serum and after a short rinse in PBS incubated with the FITC-labeled secondary antibodies (goat-anti-mouse, goat-anti-rabbit, and rabbit anti-goat) (1/1000 dilution) for 1 h at RT. The secondary antibodies were labeled with FITC (GaR, RaG). Then the cells were washed in PBS for 30 min with several changes of washing buffer. Finally, the cells were mounted in 50% glycerol containing DAPI.
3. RESULTS AND DISCUSSION
Stimulation of rat cardiomyocytes by contraction (electrical field stimulation, EFS), oligomycin (oli) or phenylephrine (PE) resulted in PKD-Ser916 phosphorylation (PKD visible as a 115 kDa band) (Figure 1). Probing of the same blots with a phospho-specific PKD-Ser744/748 antibody showed that only phenylephrine was able to phosphorylate the PKD-Ser744/748 site (at 115 kDa) (Figure 1). This is in agreement with previous observations which showed that oligomycin-treatment of cardiomyocytes did not result in translocation of various PKCs [8] and showed that this type of PKD activation is independent of Ser744/748 phosphorylation. However, using the phospho-specific PKD-Ser744/748 antibody, in all three conditions (EFS, oli and PE treatment), a stronger signal of approximately 150 kDa was detected (Figure 1). This surprising observation suggested that the phospho-PKD Ser744/748 antibody does not detect phosphorylation of PKD (115 kDa), but rather phos-
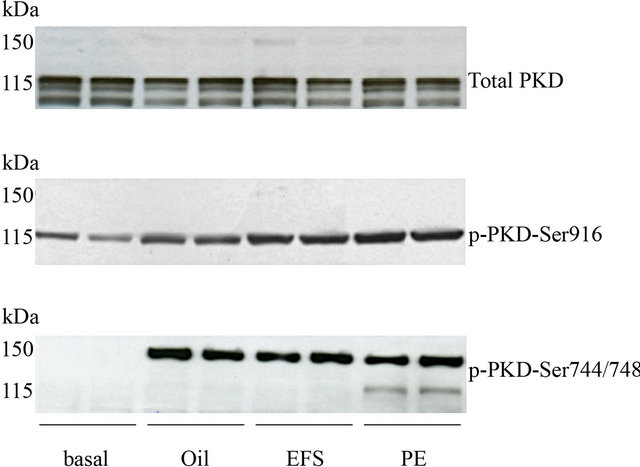
Figure 1. Phosphorylation of PKD upon different stimuli. Isolated cardiomyocytes were treated with 5 µmol/L oligomycin (oli) or 50 µmol/L phenylephrine (PE) for 15 min, or subjected to electric field stimulation (EFS) and were compared to nonstimulated cells. Samples were immunoblotted against total PKD protein expression (115 kDa), p-PKD-Ser916 (115 kDa), pPKD-Ser744/748 (150 kDa).
phorylation of another protein with a molecular mass of 150 kDa. To identify the 150-kDa protein band, detected by the phospho-PKD-Ser744/748 antibody, lysates from control and oligomycin-treated cardiomyocytes were subjected to 2D-gel-electrophoresis, followed by probing with the phospho-PKD-Ser744/748 antibody. First, lysates from control and oligomycin-treated cardiomyocytes were dissolved in urea buffer and subjected to western blotting and probed with p-PKD-Ser744/748 antibody to confirm PKD-Ser744/748 phosphorylation and its location on the gel (PE treatment was used as a control) (Figure 2(a)). Next, total protein (50 µg) was loaded from oligomycin-treated cardiomyocytes for the 2D-gel electrophoresis. The protein spots appearing after Sypro Ruby staining (Figure 2(b)) were compared with the protein pattern from the immunoblotting (Figure 2(c)), and the matching spots were excised from the gel and combined for analysis. Further analysis was done with a MALDI-TOF mass spectrometer. The isolated protein was identified as cMyBP-C with a mascot score of 110 for the western blot gel and a mascot score of 170 for the 2D gel (data not shown). Furthermore, we observed that, after cMyBP-C immunoprecipitation of oligomycinstimulated cardiomyocytes, the p-PKD-Ser744/748 antibody detected selectively cMyBP-C phosphorylation (150 kDa) and not PKD phosphorylation (115 kDa) (Figure 2(d)). To verify these data, immuno-staining on isolated cardiomyocytes for cMyBP-C with the p-PKD-Ser744/ 748 antibody were performed. Staining for titin and desmin (used as control) clearly revealed the intact striated
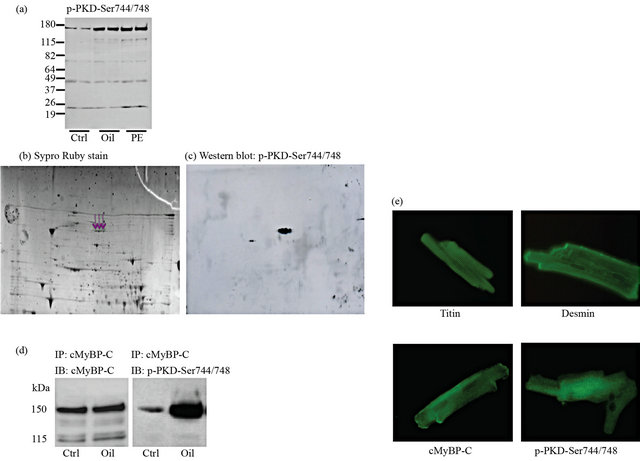
Figure 2. Detection of cMyBP-C with the p-PKD-Ser744/748 antibody. Isolated rat cardiomyocytes were treated with 5 µmol/L oligomycin for 15 min at 37˚C. Incubations were terminated by centrifugation and subsequent suspension of the cells in urea buffer or sample buffer. (a) Western blotting was performed to confirm the phosphorylation pattern with the p-PKD-Ser744/748 antibody (phenylephrine stimulation was used as a control); (b) 50 µg of protein was separated by 2D-gel electrophoresis. Afterwards the gel was stained with Sypro Ruby stain to visualize all protein spots; (c) The protein spots appearing after immunoblotting, using the phospho-PKD-Ser744/48 antibody, were excised on a glass plate from the Sypro Ruby stained gel (the excised spots were indicated with arrows) and processed on a MassPREP digestion robot. Further analysis was done with a MALDI-TOF mass spectrometer; (d) Isolated cardiomyocytes were treated with 5 µmol/L oligomycin (oli) whereafter cMyBP-C was immunoprecipitated. Samples were immunoblotted against cMyBP-C and p-PKD-Ser744/748; (e) immunostaining was performed with antibodies against titin, desmin, cMyBP-C and p-PKD-Ser744/748.
structure of the cardiomycytes, proving the structural integrity of the cells. The cMyBP-C antibody (from Santa Cruz), as expected, reveals also a striated pattern (Figure 2(e)). The striation is not as sharp as for desmin and titin, which may be due to the fact that not all cMyBP-C proteins are attached to the contractile apparatus. The p-PKD-Ser744/748 antibody also provides a striated picture, much like the cMyBP-C antibody (Figure 2E). Other PKD antibodies gave a diffuse pattern (data not shown). Thus, these results demonstrate that the p-PKDSer744/748 antibody is able to detect cMyBP-C in isolated rat cardiomyocytes.
Knowing that the phospho-PKD-Ser744/748 antibody recognizes a short sequence of amino acids within the activation loop of PKD containing both Ser744 and Ser748 residues, we reasoned that a loop with a similar amino acid sequence could be expected to be present within cMyBP-C. Indeed, cMyBP-C possesses a phosphomotif around Ser315/320 (in rat) and Ser302/307 (in mouse) with strong homology to the Ser744/748 site of PKD (Figure 3(a)). Also in mouse cardiomyocytes, the p-PKD-Ser744/748 antibody detects phosphorylation of cMyBP-C-Ser302/307 (Figure 3(b)), indicating that the PKD antibody binds to this particular phospho-motif on cMyBP-C. To verify this, we used a “home-made” phospho-specific cMyBP-C antibody directed against cMyBPC-Ser302 in mouse [11]. Also with this antibody, we detected phosphorylation of cMyBP-C-Ser315 in contracting rat cardiomyocytes (Figure 3(c)). These findings are in agreement with previous data, which showed that the region around Ser315 in rat, that is similar to the region around Ser302 in mouse, is a phosphorylation site of cMyBP-C (Figure 3(a)) [5,14,15]. These data indicate that the p-PKD-Ser744/748 antibody is able to recognize this sequence, with considerable homology between PKD and cMyBP-C if both serine residues are phosphorylated.
4. CONCLUSION
In this study we describe the observation that a phospho-specific antibody originally directed against PKDSer744/748, can be used to detect phosphorylation of cMyBP-C in contracting cardiomyocytes. So far, no antibodies against phospho-cMyBP-C are commercially available. Although this p-PKD-Ser744/748 antibody was not designed to detect cMyBP-C it can contribute to defining the role of cMyBP-C phosphorylation and, more in general, the function of cMyBP-C. Since phosphorylation of cMyBP-C is markedly decreased in human heart failure and experimental models [5,16,17], this may open new ways for early diagnosis and preventive treatment of cardiac pathologies.

Figure 3. The p-PKD-Ser744/748 antibody might recognize a phospho-region around cMyBP-C-Ser315/320. (a) Comparison of the PKD-Ser744/748 sequence with Ser315/ 320 of cMyBP-C in rat and cMyBP-C-Ser302/307 in mouse; (b) Detection of cMyBP-C phosphorylation with the phospho-PKD-Ser744/748 antibody in mouse cardiomyocytes. Mouse Cardiomyocytes were treated with 5 µmol/L oligomycin for 15 min at 37˚C (ctrl = control, oli = oligomycin); (c) Isolated rat cardiomyocytes were electric field stimulated (EFS) and subjected to western blot to detect phosphorylation of cMyBP-C-Ser315 with a “homemade” phospho-specific antibody.
5. ACKNOWLEDGEMENTS
This study was supported by the transnational University Limburg and the European Community (Integrated Project LSHM-CT-2004-005272, Exgenesis). DV is “Collaborateur Logisitique” of the FRS-FNRS (Belgium).
REFERENCES
- Cohen, P. (2002) The origins of protein phosphorylation. Nature Cell Biology, 4, E127-E130. doi:10.1038/ncb0502-e127
- Decker, R.S., Decker, M.L., Kulikovskaya, I., Nakamura, S., Lee, D.C., Harris, K., Klocke, F.J. and Winegrad, S. (2005) Myosin-binding protein c phosphorylation, myofibril structure and contractile function during low-flow ischemia. Circulation, 111, 906-912. doi:10.1161/01.CIR.0000155609.95618.75
- Jacques, A.M., Copeland, O., Messer, A.E., Gallon, C.E., King, K., McKenna, W.J., Tsang, V.T. and Marston, S.B. (2008) Myosin-binding protein c phosphorylation in normal, hypertrophic and failing human heart muscle. Journal of Molecular and Cellular Cardiology, 45, 209- 216. doi:10.1016/j.yjmcc.2008.05.020
- Sadayappan, S., Osinska, H., Klevitsky, R., Lorenz, J.N., Sargent, M., Molkentin, J.D., Seidman, C.E., Seidman, J. G. and Robbins, J. (2006) Cardiac myosin-binding protein c phosphorylation is cardioprotective. Proceedings of the National Academy of Sciences of the United States of America, 103, 16918-16923. doi:10.1073/pnas.0607069103
- Barefield, D. and Sadayappan, S. (2010) Phosphorylation and function of cardiac myosin-binding protein c in health and disease. Journal of Molecular and Cellular Cardiology, 48, 866-875. doi:10.1016/j.yjmcc.2009.11.014
- Schlossarek, S., Mearini, G. and Carrier, L. (2011) Cardiac myosin-binding protein c in hypertrophic cardiomyopathy: Mechanisms and therapeutic opportunities. Journal of Molecular and Cellular Cardiology, 50, 613- 620. doi:10.1016/j.yjmcc.2011.01.014
- Rykx, A., De Kimpe, L., Mikhalap, S., Vantus, T., Seufferlein, T., Vandenheede, J.R. and Van Lint, J. (2003) Protein kinase D: A family affair. FEBS Letters, 546, 81- 86. doi:10.1016/S0014-5793(03)00487-3
- Luiken, J.J., Vertommen, D., Coort, S.L., Habets, D.D., El Hasnaoui, M., Pelsers, M.M., Viollet, B., Bonen, A., Hue, L., Rider, M.H. and Glatz, J.F. (2008) Identification of protein kinase D as a novel contraction-activated kinase linked to GLUT4-mediated glucose uptake, independent of AMPK. Cell Signal, 20, 543-556. doi:10.1016/j.cellsig.2007.11.007
- Fielitz, J., Kim, M.S., Shelton, J.M., Qi, X., Hill, J.A., Richardson, J.A., Bassel-Duby, R. and Olson, E.N. (2008) Requirements of protein kinase d1 for pathological remodeling. Proceedings of the National Academy of Sciences of the United States of America, 105, 3059-3063. doi:10.1073/pnas.0712265105
- Bardswell, S.C., Cuello, F., Rowland, A.J., Sadayappan, S., Robbins, J., Gautel, M., Walker, J.W., Kentish, J.C. and Avkiran, M. (2010) Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. The Journal of Biological Chemistry, 285, 5674- 5682. doi:10.1074/jbc.M109.066456
- Sadayappan, S., Gulick, J., Klevitsky, R., Lorenz, J.N., Sargent, M., Molkentin, J.D. and Robbins, J. (2009) Cardiac myosin-binding protein c phosphorylation is cardiaoprotective. Circulation, 119, 1253-1262. doi:10.1161/CIRCULATIONAHA.108.798983
- Luiken, J.J., van Nieuwenhoven, F.A., America, G., van der Vusse, G.J. and Glatz, J.F. (1997) Uptake and metabolism of palmitate by isolated cardiac myocytes from adult rats: Involvement of sarcolemmal proteins. The Journal of Lipid Research, 38, 745-758.
- Bouwman, F., Renes, J. and Mariman, E. (2004) A combination of protein profiling and isotopomer analysis using matrix-assisted laser desorption/ionization-time of flight mass spectrometry reveals an active metabolism of extracellular matrix of 3T3-L1 adipocytes. Proteomics, 4, 3855-3863. doi:10.1002/pmic.200400861
- Wu, S.C. and Solaro, R.J. (2007) A novel regulator of both phosphorylation and dephosphorylation of cardiac sarcomeric proteins. The Journal of Biological Chemistry, 282, 30691-30698. doi:10.1074/jbc.M703670200
- Mohamed, A.S., Dignam, J.D. and Schlender, K.K. (1998) Cardiac myosin-binding protein C (MyBP-C): Identification of protein kinase A and protein kinase C phosphorylation sites. Archives of Biochemistry and Biophysics, 358, 313-319. doi:10.1006/abbi.1998.0857
- El-Armouche, A., Pohlmann, L., Schlossarek, S., Starbatty, J., Yeh, Y.H., Nattel, S., Dobrev, D., Eschenhagen, T. and Carrier, L. (2007) Decreased phosphorylation levels of cardiac myosin -binding protein-c in human and experimental heart failure. Journal of Molecular and Cellular Cardiology, 43, 223-229. doi:10.1016/j.yjmcc.2007.05.003
- Sadayappan, S., Gulick, J., Osinska, H., Martin, L.A., Hahn, H.S., Dorn 2nd, G.W., Klevitsky, R., Seidman, C.E., Seidman, J.G. and Robbins, J. (2005) Cardiac myosin-binding protein c phosphorylation and cardiac function. Circulation Research, 97, 1156-1163. doi:10.1161/01.RES.0000190605.79013.4d

