Advances in Bioscience and Biotechnology
Vol.4 No.7A2(2013), Article ID:33904,9 pages DOI:10.4236/abb.2013.47A2001
Antibacterial activity of plant methanolic extracts on a field isolate of Pseudomonas syringae pv tomato from the Casablanca region (Morocco)
![]()
1Laboratory of Physiology & Molecular Genetics, Faculty of Sciences Ain Chock, Casablanca, Morocco
2Ibn Rochd University Hospital, Casablanca, Morocco
3Institute for Plant Biochemistry and Photosynthesis (IBVF), CSIC-Universidad de Sevilla, Seville, Spain
Email: *a_soukri@hotmail.com
Copyright © 2013 Bouchra Elkhalfi et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 10 February 2013; revised 12 March 2013; accepted 25 March 2013
Keywords: Bacterial Speck; Bacterial Field Isolate; LOPAT; Pathogenicity Tests; Antibacterial Activity; Methanolic Plant Extracts; Nigella sativa
ABSTRACT
A bacterial field isolate recovered from infected tomato plants in a green-house at Sidi Rehal, a region near Casablanca city (Morocco), was identified as the gammaproteobacterium Pseudomonas syringae pv. tomato DC3000 strain, the causal agent of bacterial speck. The bacterial isolate was characterized by morphological, biochemical and molecular biological tests, its growth curves carried out in various culture media, and its phytopathogenicity verified by infection tests. A screening was performed to evaluate the antibacterial activity of methanolic extracts of 12 selected Moroccan plants against the P. syringae pv. tomato DC3000 isolate, and Agar-well diffusion and Broth microdilution methods were used to determine minimum inhibitory and minimum bactericidal concentrations. Among the methanolic extracts tested, only those of Nigella sativa, Geranuim robertianum, Aizoon canariense and Rubia peregrine showed clear inhibitory and bactericidal activities, although the highest values were achieved with N. sativa, a plant used in Morocco as a spice, condiment and medicinal treatment.
1. INTRODUCTION
The new agricultural strategy, Green Morocco Plan, aims to consolidate the success achieved and to meet new challenges facing Morocco’s competitiveness and opening of markets [1,2]. But in Morocco as in other countries, plant diseases caused by phytopathogenic bacteria are a major problem seen worldwide that impact many valuable agricultural crops, causing damage in yield potential each year [3].
Tomato plant (Solanum lycopersicum L.) is a vegetable well known for its high nutritional value—rich in vitamins P, A and C—and by its favorable influence on kidney function and digestive tract [4,5]. Tomato is one of the most important greenhouse and field-grown vegetables in Morocco. According to Food and Agriculture Organization (FAO) estimation, the annual production in Morocco amounts to 800,000 Tons of fresh tomatoes, most of which is absorbed by the local market cost. Indeed, tomato fields are a potential target of pathogenic bacteria that cause diseases, from little spots on the foliage accented on its leaves to the total defoliation of the plant, with detrimental consequences on photosynthesis and production potential [6,7]. One of these diseases is bacterial speck, the most persistent bacterial disease problem found in tomato-growing plants. Not only decreases yield of plants through foliar necrosis, but it also blemishes the fruits and renders them unsuitable for the fresh market [6]. This pathology is caused by the Gramnegative bacterium Pseudomonas syringae pv. tomato DC3000 strain, which is a member of the gamma subgroup of Proteobacteria [8,9]. Disseminated primarily by water, P. syringae pv. tomato DC3000 has the potential to move through plug greenhouses very quickly; thus, infested seedlings could become an important inoculum source for field epiphytotics [10,11]. Consequently, this bacterium can survive up to 20 years in the crevices and cavities of the coat of tomato seeds causing bacterial speck, a disease characterized by small brown-black specks, 1 - 3 mm in diameter, with a yellow chlorotic halo [12,13].
The pathogenicity of DC3000 strain has been the subject of several scientific studies, and it has emerged as an important organism model in molecular plant pathology because of its genetic tractability [14,15], and its genome has been fully sequenced [16,17]. Other studies showed that DC3000 strain encodes a wide range of proteins that are involved in virulence, suggesting that pathogenicity includes diverse processes such as motility, adhesion, injection of effector proteins into host cells, degradation of host cell walls, production of phytotoxic compounds, iron acquisition, and interference with host defenses, which may promote epiphytic growth as well as actual disease expression [16]. However, most of these studies have focused on molecular aspects of this bacterium, while cellular and physiological aspects have not been as developed.
The purpose of this study was to isolate the causal agent of bacterial speack disease from infected tomato plants in a green-house at Sidi Rehal, a region near Casablanca (Morocco), This bacterium was identified by morphological, biochemical and molecular tests as a field isolate of Pseudomonas syringae pv. tomato DC3000, which was characterized to verify its pathogenicity. Then, growth curves were determined in various culture media. Finally, the antibacterial effect against this pathogenic strain of some plant methanolic extracts was evaluated.
2. MATERIAL AND METHODS
2.1. Isolation of Pseudomonas syringae pv. Tomato DC3000 Strain from Infected Plant Tissue
Plant tissue samples were washed under running tap water to remove surface soil particles, dust and other contaminants, and then were soaked in saline solution (0.85% w/v NaCl) for 30 min to allow bacterial extraction from leaf-infected tissue into the solution. 100 µl of the resulting bacterial suspension per plate were spread on king’s B medium [18] and incubated 48 h at 26˚C. Isolated colonies were subcultured three times for purification and then transferred to nutrient broth agar with glucose before performing biochemical and physiological tests.
2.2. Identification Tests
The following tests were used to determine a number of morphological and biochemical characters of P. syringae pv tomato DC3000, following recommended bacteriological protocols [19-21]:
1) Gram staining: using bacteriological standard procedures [19].
2) Kligler test: With a sterile needle one a colony of the isolate was took and picked into the center of the agar tube (Kligler medium) till its bottom, then streaked the slant surface. This double inoculation allows bacterial growth both on the aerobic surface and in the largely anaerobic butt of the tube. The Kligler medium was incubated 24 h at 28˚C.
3) Mannitol motility: With a sterile needle a well-isolated colony was took and picked into the medium till 1 cm of the bottom of the tube, keeping the needle in the same trajectory line in the entry as well as in the output of the agar medium. Incubation was performed at 28˚C for 24 h.
4) API 20NE tests (API Laboratory Products of BioMérieux SA, France): All equipments and reagents used were from the manufacturer except plastic Pasteur pipettes (Alpha Laboratories) and oxidase reagent (Difco). The test comports 20 microtubes containing dehydrated substrates which were inoculated with saline bacterial suspension. Inoculation and incubation of the strip were precisely performed following manufacturer’s instructions. Nitrate reduction, indole (TRP) reaction, and glucose fermentation reactions were read after 18 - 24 h incubation. All other results were read at 24 - 48 h. Results for each test were recorded as positive, negative, or weak. A seven digit profile was ascribed and identification was obtained by reference to the Analytical Profile Index.
5) Test of Levan production: It shows the ability of the bacterial isolate to produce polysaccharides on rich sucrose medium [22]. Using a steril loop, levan medium was streaked with the bacteria and incubated at 28˚C for 2 to 5 days.
6) Oxidase test: Is was performed by using the disc method [23]. HIMEDIA disks were placed in a clean slide and selected colonies were transferred onto the disks surface with a Pasteur pipette. The reaction is immediate, the disks become dark purple when the strain is oxidase-positive.
7) Potato rot test: Fresh potato tubers were washed and peeled, alcohol flamed, and sliced into approximately 7 mm - 1 cm width (1slice for each isolate and 1slice for control). The slices were dipped in alcohol and flame, placed in Petri-dishes, then sterile distilled water was added to a depth of half the slice. Nicks were formed in the center of the potato with a sterile tool and inoculate with 100 µl of bacterial suspension. Positive results were indicated by lack of rotting, while rotting suggested negative results [24].
8) Arginine dihydrolase: Inocula were transferred to a sterile tube of arginine dihydrolase broth. The inoculated tube was incubated at 28˚C for 24 h and then preliminary results were determined.
The arginine-dihydrolase enzyme activation needs culture medium acidification, for which the bacterium must first use the glucose present in the medium causing a pH drop (indicated by a medium color change from purple to yellow). Then, cultures were incubated for an additional 24-h period to allow arginine consumption by the bacterium. Final results were obtained after 48 h. Change back to purple from yellow indicates a positive test for arginine dihydrolase.
9) Hypersensitive test in tobacco leaves: Hypersensitivity tests were conducted in tobacco (Nicotiana tabacum L.) according to the method of Klement [25]. Each bacterial isolate was suspended in sterile distilled water and inoculated on the underside of tobacco leaves by injection. Sterile distilled water was used as a negative control. Plants were kept under greenhouse conditions (26˚C during daytime and 18˚C during night time) and observations for a hypersensitive reaction were conducted after 24 h.
2.3. Organisms and Growth Conditions
Unless otherwise stated the Pseudomonas syringae pv tomato cells were grown in LB medium with glucose (3 g/l) and rifampicin (50 µg/ml) at 28˚C with agitation (200 rpm) in the darkness.
2.4. Growth Curves of the Pseudomonas syringae pv. Tomato DC3000 Isolate
The patterns of growth of the bacterial isolate have been analyzed in three culture media, namely LB, LB with glucose (3 g/l) and Brain-Heart broth (BCC). Bacterial culture growth was followed by turbidity analysis with a spectrophotometer (Jenway 6100 spectrophotometer, Dunmow, Essex, UK). 10-ml of overnight cultures were transferred to Erlenmeyer flasks at t = 0. Growth was determined from the change in turbidity at 600 nm for 48 h with continuous shaking (200 rpm) at 28˚C. All growth curve experiments were run in triplicate.
2.5. Molecular Identification
1) DNA isolation by the phenol-chloroform method: A phenol-chloroform method was used for the extraction of genomic DNA from the P. syringae pv tomato isolate. Five-ml of bacterial suspension were transferred to sterile microcentrifuge tubes and spun 10 min at 5000 g, followed by the addition of 467 µl TE buffer (SigmaAldrich), 30 µl of 10% (w/v) SDS (Sigma-Aldrich) and 3 µl of proteinase K (20 mg/ml, Sigma-Aldrich). Samples were briefly mixed and then incubated for 45 min at 37˚C and 15 min at 42˚C. An equal volume of phenolchloroform-isoamyl alcohol (25:24:1) mixture (SigmaAldrich) was added, carefully mixed and spun for 2 min. Then, the upper aqueous phase was transferred to a new tube and DNA was precipitated by adding 1/10 volume of 0.1 M sodium acetate and 0.6 volumes of isopropanol. After gently mixing until DNA precipitation, the slimy white material was spooled onto a glass rod and then washed with 70% (v/v) ethanol (Sigma-Aldrich), air dried and finally resupended in 100 - 200 µl TE buffer. After DNA has dissolved its concentration was estimated by measuring absorbance at 260 nm after diluting 10 µl of the DNA solution into 1 ml of TE (1:100 dilution).
2) Primers design: A pair of specific primers for the 5’- and 3’-ends of the P. syringae pv. tomato DC3000 gap1 gene (PSPTO_1287, gi|28868497), encoding the glyceraldehyde-3-phosphate dehydrogenase 1 isoform (GAPDH1), were designed using Primer Premier 5.0 program (Biosoft International).
Forward 5’-GGAATTCCATATGACTCTCCGTATCGCAATC-3’
Reverse 5’-CGGGATCCTTACTCGGCGTTATGCAGCGC-3’
3) PCR amplification and agarose gel electrophoresis: PCR amplification of the gap1 gene from the P. syringae pv tomato isolate was carried out with the pair of specific oligonucleotides above described, using its genomic DNA (ca. 1 µg per reaction) as a template. The reaction mixture contained polymerase buffer, 80 nM of each primer, 0.8 mM of each deoxynucleotide, 1.5 mM MgCl2, and 1 unit of GoTaq DNA Polymerase (Promega). Amplification was performed in a DNA thermal cycler (PerkinElmer Cetus, Norwalk, CT) programmed for 2 min at 95˚C (initial step), 35 cycles of 40 s at 94˚C, 40 s at 60˚C and 1 min at 72˚C, and a final step of 7 min at 72˚C. The amplification products were analyzed by electrophoresis in a 0.8% (w/v) agarose gels according to Sambrook [26] and detected by staining with ethidium bromide.
2.6. Pathogenicity Tests
For the pathogenicity tests, tomato plants leaves were inoculated by injection with a bacterial suspension of the P. syringae pv tomato isolate (108 colony forming units (CFU)/ml). After inoculation, detached leaves were incubated on Petri plates containing 20 ml of King’s B solid medium with rifampicin (50 µg/ml) at 28˚C. Disease development on tomato leaves was evaluated 7 days after inoculation. Re-isolations and identification were made from the diseased plants. Sterile distilled water was used as a negative control.
2.7. Plant Material and Preparation of Methanolic Extracts
The plant material used in this study consisted of aerial and/or underground parts of the followimg plant species collected from the Casablanca region in Morocco: Santolina chamaecypara, Rubia peregrina, Pisenlit (Taraxacum sp.), Mesembryentenum cristallinum, Geranuim robertianum, Eucalyptus cladocalyx, Dittrichia graveolens, Clematis cirrhosa, Celtis australis, Aizoon canariense, Anagallis arvensis, and Nigella sativa (Table 1). In all cases, the air-dried plant material was ground into fine powder and extraction was performed at room temperature by maceration with 80% (v/v) methanol/distilled water. After filtration, the extracts were evaporated to dryness in vacuo and weighed. The resulting residue was then dissolved in 80% methanol and used for the bacterial susceptibility tests.
2.8. Antibacterial Activity Tests
The antibacterial activity of the crude plant extracts was determined by the agar well diffusion method according to the National Committee for Clinical Laboratory. Standards (NCCLS) [27]. Petri plates containing 20 ml of LB solid medium were seeded with samples of 24 h broth cultures of the bacterial isolate. Wells (5 mm diameter) were cut into the agar and 50 µl of the plant extracts were tested at a concentration of 100 mg/ml. The inoculum size was standardized at approximately 108 CFU/ml and incubation was performed at 28˚C for 24 h. The assessment of antibacterial activity was based on measurement of the diameter of the clear inhibition zone formed around the wells.
2.9. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
Plant extracts that exhibited considerable activity were 1/2 diluted, and the MIC was determined by adopting the tube dilution method. A 1-ml aliquot of the bacterial suspension (1.5 × 106 CFU/ml) was inoculated into each tube. Control tubes were inoculated with the same volume of 80% methanol in sterile distilled water. All tubes
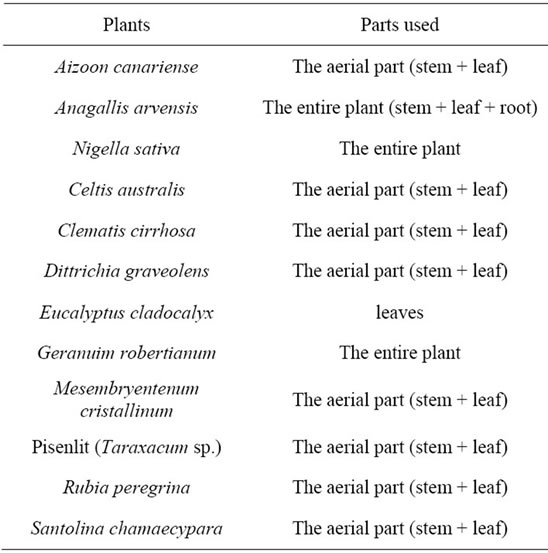
Table 1. Parts of plants used for the study.
were incubated at 28˚C for 24 h. The lowest concentration that did not allow any detectable bacterial growth when compared with the control was considered as the minimum inhibitory concentration (MIC). Cultures of those tubes that showed no visible growth were plated on solid medium, and incubated at 28˚C for 24 h. The minimum bactericidal concentration (MBC) was considered as the lowest concentration of the plant extracts that could not allow the appearance of a single bacterial colony.
3. RESULTS & DISCUSSION
3.1. Plant Disease Symptoms and Isolation of the Phytopathogenic Bacterium
Bacterial speck is one of the most common bacterial diseases of growing tomato plants. All plant samples used in the present study presented the symptoms of Bacterial speck on fruits and leaves, appearing as small black lesions surrounded by a yellow halo. After thorough washing, this material was used to isolate the pathogenic bacterial strain. The resulting bacterial suspension was spread on King’s B medium plates. After 24 h incubation at 28˚C, circular bacterial colonies that showed strong green fluorescence under UV light were observed on the medium. Both microbiological features and plant disease symptoms are identical to those described in other studies on DC3000 strain pathogenicity [6,13,28].
3.2. Identification of the Bacterial Strain
According to the description of Bergey’s Manual of Systematic Bacteriology [29], the bacterial isolate appeared to be a Pseudomonas syringae strain. Thereby, a set of specific identification tests were carried out, as described by Lelliott and Stead [22]. Table 2 summarizes the results obtained. Detection of fluorescence on King’s B medium [18] was very useful as a first identification step of P. syringae, all obtained colonies showed a fluorescent pigmentation; besides, bacterial cells were Gram negative, motile, Kligler and mannitol negative, and oxidatively metabolized glucose. Moreover, the profiles of the isolates in API 20NE test after 48 h suggested that the bacterium was most likely a fluorescent Pseudomonas strain. For more accuracy, the LOPAT test was carried out (Table 2) and revealed that the bacterial isolates produced Levan-positive colonies on nutrient agar medium (supplemented with 5%, w/v, sucrose) and exhibit positive hypersensitive reaction in the presence of tobacco leaves. However, no response was observed in potato soft rot oxidase and arginine dihydrolase activity tests. All these identifying features show that the isolates correspond to P. syringae pv. tomato DC3000. Similar strategy and microbiological tests were used by Milijašević et al. for the identification of DC3000 field isolates from
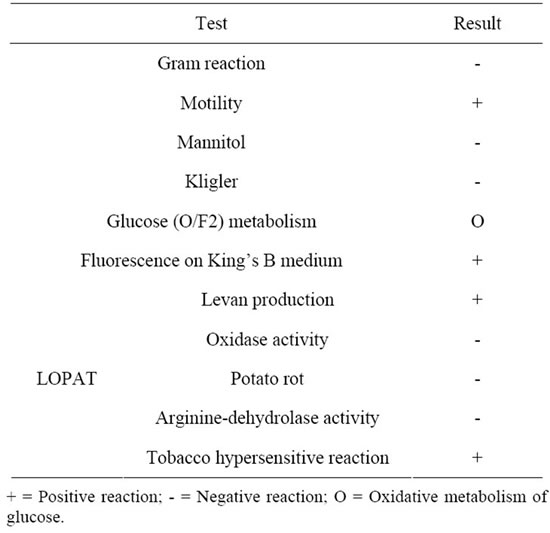
Table 2. Morphological, biochemical and physiological features of the bacterial isolate.
Eastern Europe [30].
3.3. Pathogenicity Tests
The pathogenicity of each field isolate should normally be checked before assigning it to a pathovar [31]. Disease development on tomato leaves was evaluated 7 days after inoculation (108 cfu/ml )with the P. syringae pv tomato DC3000 isolates. All infected plants showed the same first symptoms (moist spots), and after 5 days the spots became dark-brown with chlorotic halos, corresponding to the typical symptoms of bacterial speck. No symptoms appeared on control leaves injected with distilled water. All re-isolated bacteria were identified as P. syringae pv. tomato DC3000 strains and eventually stored at −20˚C. Similar results of pathogenicity tests on tomato leaves, pepper leaves, and immature tomato and pepper fruits were reported by Arsenijević and Jovanović [32].
Likewise, data by Milijašević et al. [30] indicate similar effects of other pathogenic DC3000 isolates on plant infected tissues after 4 days.
3.4. Molecular Identification
In order to confirm the identity of our isolate, the complete coding region of the gap1 gene (gi|28868497) was amplified by PCR from genomic DNA from the isolates (Figure 1), using specifics primers designed after the complete genome sequence of the Pseudomonas syringae pv. tomato DC3000 strain EMBL/Gen-Bank databases: http://www.ncbi.nlm.nih.gov/genome/?term=txid223283
As shown in Figure 1, a single fragment of approximately 1.0 kb, the expected size for the gap1 gene of P.
syringae pv. tomato DC3000, was amplified from the bacterial isolate, while the blank control was negative. The amplified DNA fragment was subcloned and sequenced, exhibiting a nucleotidic sequence virtually identical to that of DC3000 gap1 gene available in public databases (data not shown). Overall, these molecularbiological results corroborate those obtained by conventional microbiological methods.
3.5. Growth Curves of Cultures of the Pseudomonas syringae pv Tomato Isolate
The P. syringae pv. tomato isolate was cultured in three liquid media (LB, LB plus glucose and BCC) in order to determine optimal growth conditions (Figure 2). The isolate grows better in LB plus glucose medium than in the others, exhibiting a clear reduction of the lag phase (5 h vs. 8 h) and a faster exponential phase. These results are in agreement with previous reports claiming that glucose requirement in culture media is an widespread feature among pathogenic bacteria [33,34].
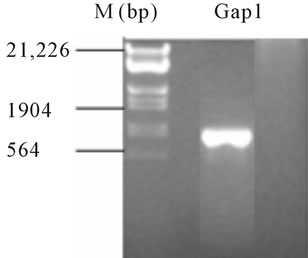
Figure 1. PCR amplification of the full ORF of a gap gene encoding the glycolytic GAPDH-1 isoform using a pair of specific primers for P. syringae pv. Tomato DC3000 strain, and genomic DNA of the bacterial isolate as a template. A single PCR product of ca. 1 kb was amplified.
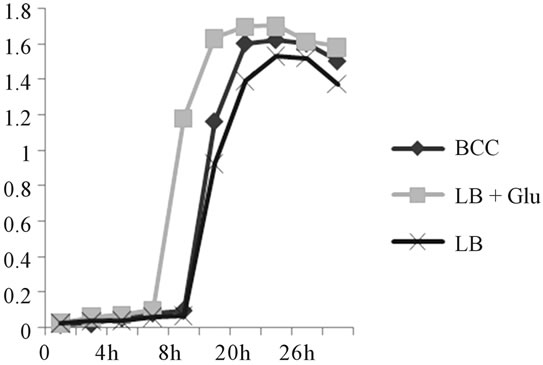
Figure 2. Growth curves of the Pseudomonas syringae pv. tomato isolate in various bacteriological culture media.
3.6. Antibacterial Activity Screening of Plant Extracts
Bacterial diseases of plants are often very difficult to eradicate, especially in biological agriculture, but to control them few effective strategies can be adopted. Medicinal plants are among the richest bio-resources of the drugs currently used to treat bacterial and other infections [35,36]. In the present work, 12 plant species found in central Morocco (Table 1), and chosen for their antiseptic and medicinal properties [37], were used to obtain crude methanolic extracts. These plant extracts were screened for bactericidal/bacteriostatic activities against the DC3000 isolate and some of them were found in vitro to have the phytochemical potential to control bacterial speck. Table 3 shows that methanolic extracts of the studied plants have widely diverse inhibitory effects on the bacterial growth of the DC3000 isolate, when assayed by the agar well-diffusion method (0.6 - 4.0 cm diameters of inhibition zones). Although these results revealed the antibacterial potential of most plant extracts tested, only four extracts showed a substantial antibacterial activity: the maximum antibacterial activity was clearly shown by the Nigella sativa (Ranunculaceae) extract followed by, in decreasing order, Geranuim robertianum, Aizoon canariense and Rubia peregrine. Other methanolic plant extracts showed very low inhibitory effects while the 80% methanol solution used as a control in these experiments no effect at all (see Table 3).
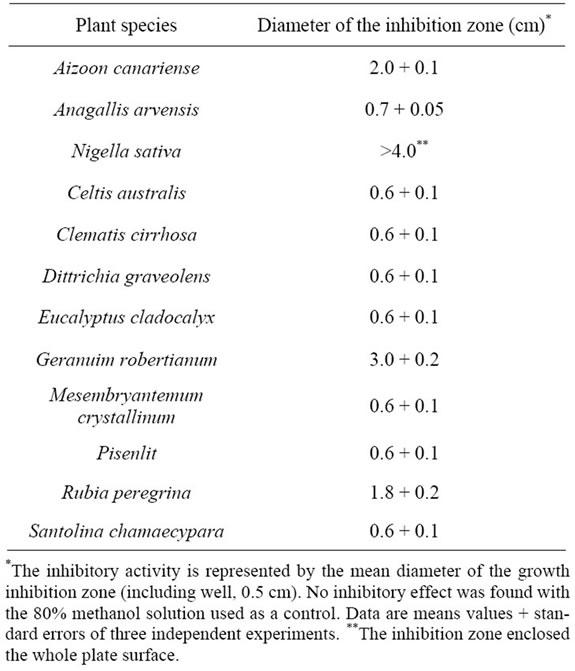
Table 3. Growth inhibitory activity of plant methanolic extracts on the Pseudomonas syringae pv tomato isolate.
These findings are in agreement with a previous report [38], which showed that Gram-negative strains are in general more resistant than Gram-positive ones in welldiffusion tests. This higher resistance could be due to the external lipopolysaccharide wall that surrounds the peptidoglycan cell wall of the formers [39] or to the occurrence of membrane accumulation mechanisms.
3.7. Determination of MIC and MBC Values for Antibacterial Plant Extracts
The minimum inhibitory (MIC) and bactericidal (MBC) concentrations, two parameters that respectively quantify the bacteriostatic and bactericidal potential of bioactive compounds, were determined using dry extracts and the serial dilution method for the 4 plant extracts with substantial antibacterial activitiy against the DC3000 isolate. As is shown in Table 4, the obtained values were in the ranges of 1 - 4 mg/ml for MIC and 1 - 9 mg/ml for MBC, and on the whole are in agreement with the inhibitory activities shown above. The Nigella sativa extract exhibited by far the strongest bacteriostatic and bactericidal effects on the P. syringae pv. tomato DC3000 isolate. Noteworthy, a similar bactericidal effect was demonstrated on both pathogenic and environmental bacteria causing food spoilage [40]. Several other studies have described the antibacterial activity of N. sativa black seeds and its crude extracts [41-43]. In others studies, it has also been used as a natural therapeutic agent in the treatment of fever, common cold, and rheumatic diseases [41]. Also, it can be used as an anti-inflammatory, anti-oxidant and hypoglycemic agent [44-48]. Finally, N. sativa is a traditional condiment of the Old World since classical times; and its black seeds were extensively used to flavour food.
To confirm the biocontrol potential of the N. sativa extract, its antibacterial activity against the DC3000 isolate was evaluated by analyzing its effect on the bacterial growth in liquid medium. When changes in A600 were monitored in the presence of plant extract (1 mg dry extract/ml) added at time 0 or after 5 h of culture (beginning of exponential phase), significant detrimental effects
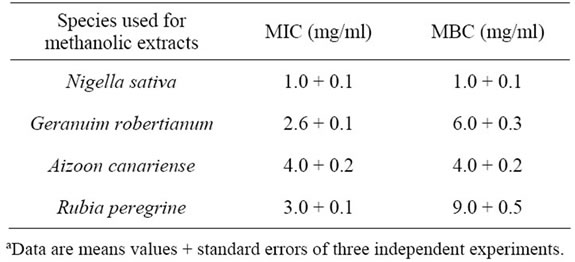
Table 4. Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of selected plant extracts on the P. syringae pv. tomato field isolateª.
were observed on the bacterial growth when compared with the control culture, namely an extended lag phase (1 h longer) and a significantly reduced growth rate during the exponential phase (data not shown). These findings show the value of the Nigella extract for the biological control of a bacterial pest, being in agreement with previous studies performed with other pathogenic bacterial strains of environmental and clinical relevance [49-51].
The strong antibacterial activity of the Nigella sativa extract demonstrated in this study is most probably due to its main bioactive component thymoquinone, as was reported in several studies [52,53]. This active principle acts as an antimicrobial agent against environmental and medical relevant Gram positive and negative bacteria [54].
4. CONCLUSION
In this study we presented an effective method for detection and identification of the phytopathogenic bacterium P. syringae pv. tomato DC3000 and a screening on the antibacterial effect that 12 methanolic plant extracts had on it. To our knowledge, this is the first time that the effectiveness of the N. sativa extract as antimicrobial agent against a phytopathogenic bacterium, the DC3000 strain in this case, was established. It could be considered, therefore, as a potential biocontrol agent for the bacterial speck of tomato, a widely spread plant disease that causes significant economic losses worldwide.
5. ACKNOWLEDGEMENTS
Support of the Collaborative grant A1/043076/11 of the Spanish AECID (MAEC), and Moroccan CNRST (URAC42) is acknowledged. Also, we gratefully acknowledge the contribution financial support of Mr Abderrahim Zakari director of Bioprocessing SARL.
![]()
![]()
REFERENCES
- Akesbi, N. (2006) Évolution et perspectives de l’agriculture marocaine. Rapport 50 Ans De Développement Humain Et Perspectives, 2025, 85-198.
- The Ministry of Agriculture and Maritime Fisheries of Morocco (2010) Moroccan agriculture in number.
- Stavrinides, J. (2009) Origin and evolution of phytopathogenic bacteria. Caister Academic Press, Norfolk.
- Hernández Suárez, M., Rodríguez Rodríguez, E.M. and Díaz Romero, C. (2008) Chemical composition of tomato (Lycopersicon esculentum) from Tenerife, the Canary Islands. Food Chemistry, 106, 1046-1056. doi:10.1016/j.foodchem.2007.07.025
- Olaniyi, J.O., Akanbi, W.B., Adejumo, T.A. and Akande, O.G. (2010) Growth fruit yield and nutritional quality of tomato varieties. African Journal of Food Science, 4, 398-402.
- Bashan, Y. and De-Bashan, L.E. (2002) Protection of tomato seedlings against infection by Pseudomonas syringae pv. tomato by using the plant growth-promoting bacterium Azospirillum brasilense. Applied and Environmental Microbiology, 68, 2637-2643. doi:10.1128/AEM.68.6.2637-2643.2002
- Pérez-García, A., Cánovas, F.M., Gallardo, F., Hirel, B. and Vicente, A. (1995) Differential expression of glutamine synthetase isoforms in tomato detached leaflets infected with Pseudomonas syringae pv. tomato. MPMIMolecular Plant Microbe Interactions, 8, 96-103. doi:10.1094/MPMI-8-0096
- Das, S., Ramm, M., Kochanowski, H. and Basu, S. (1994) Structural studies of the side chain of outer membrane lipopolysaccharide from Pseudomonas syringae pv. coriandricola W-43. Journal of Bacteriology, 176, 6550- 6557.
- Kvitko, B.H., et al. (2009) Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathogens, 5, e1000388. doi:10.1371/journal.ppat.1000388
- Rico, A. and Preston, G.M. (2008) Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Molecular Plant-Microbe Interactions, 21, 269-282. doi:10.1094/MPMI-21-2-0269
- Schneider, R.W. and Grogan, R.G. (1977) Tomato leaf trichomes, a habitat for resident populations of Pseudomonas syringae pv tomato DC3000. Phytopathology, 67, 898-902. doi:10.1094/Phyto-67-898
- Bashan, Y., Okon, Y. and Henis, Y. (1985) Detection of cutinases and pectic enzymes during infection of tomato by Pseudomonas syringae pv. tomato. Phytopathology, 75, 940. doi:10.1094/Phyto-75-940
- Cuppels, D.A. and Elmhirst, J. (1999) Disease development and changes in the natural Pseudomonas syringae pv. tomato populations on field tomato plants. Plant Disease, 83, 759-764. doi:10.1094/PDIS.1999.83.8.759
- Fouts, D.E., et al. (2002) Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proceedings of the National Academy of Sciences, 99, 2275-2280. doi:10.1073/pnas.032514099
- Wright, C.A. and Beattie, G.A. (2004) Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 101, 3269-3274. doi:10.1073/pnas.0400461101
- Buell, C.R., et al. (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proceedings of the National Academy of Sciences, 100, 10181-10186. doi:10.1073/pnas.1731982100
- Feil, H., et al. (2005) Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proceedings of the National Academy of Sciences of the United States of America, 102, 11064-11069. doi:10.1073/pnas.0504930102
- King, E.O., Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. Journal of Laboratory and Clinical Medicine, 44, 301-307.
- Bergey, D.H. and Holt, J.G. (1994) Bergey’s manual of determinative bacteriology. Lippincott Williams & Wilkins, Philadelphia.
- Boone, D.R., Brenner, D.J., Vos, P.D., Garrity, G.M., Goodfellow, M., Staley, J.T. and Krieg, N.R. (2007) Bergey’s manual of systematic bacteriology: The proteobacteria: Part A introductory essays; part B the gammaproteobacteria; part C the alpha-, beta-, delta-, and epsilonproteobacteria. Springer, Berlin.
- Klement, Z. and Rudolph, K. (1990) Methods in phytobacteriology. Akadémiai Kiadó.
- Lelliott, R.A. and Stead, D.E. (1987) Methods for the diagnosis of bacterial diseases of plants. Blackwell Scientific Publications, Oxford.
- Kovacs, N. (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction.
- Ignjatov, M., Milovsević, M., Nikolić, Z., Vujaković, M. and Petrović, D. (2007) Characterization of Pseudomonas savastanoi pv. glycinea isolates from Vojvodina. Phytopathologia Polonica, 43-54
- Klement, Z. (1963) Rapid detection of the pathogenicity of phytopathogenic pseudomonads.
- Maniatis, T. (1989) Molecular cloning: A laboratory manual. In: Sambrook, J., Fritsch, E.F. and Maniatis, T., Eds., Cold Spring Harbor Laboratory Press, New York.
- Wikler, M.A. (2009) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard. Clinical and Laboratory Standards Institute.
- Preston, G.M. (2001) Pseudomonas syringae pv. tomato: The right pathogen, of the right plant, at the right time. Molecular Plant Pathology, 1, 263-275. doi:10.1046/j.1364-3703.2000.00036.x
- Vos, P., et al. (2009) Bergey’s manual of systematic bacteriology: Volume 3, the firmicutes. Springer, Berlin.
- Milijasevic, S., Todorovic, B., Rekanovic, E., Potocnik, I. and Gavrilovic, V. (2009) Races and hosts of Pseudomonas syringae pv. tomato in Serbia. Archives of Biological Sciences, 61, 93-103. doi:10.2298/ABS0901093M
- Bultreys, A. and Kaluzna, M. (2010) Bacterial cankers caused by Pseudomonas syringae on stone fruit species with special emphasis on the pathovars syringae and morsprunorum race 1 and race 2. Journal of Plant Pathology, 92, S1-S21
- Arsenijević, M. and Jovanović, O. (1993) Pseudomonas syringae pv. tomato parazit rasada paradajza.
- Friedemann, T.E. (1939) Metabolism of pathogenic bacteria growing under aerobic conditions in carbohydraterich culture media. Proceedings of the Society for Experimental Biology and Medicine, 40, 505-509.
- Jacobs, J.M., Babujee, L., Meng, F., Milling, A. and Allen, C. (2012) The in planta transcriptome of ralstonia solanacearum: Conserved physiological and virulence strategies during bacterial wilt of tomato. mBio, 3, e00114. doi:10.1128/mBio.00114-12
- Kothari, V., Naraniwal, M. and Gupta, A. (2011) Effect of certain phytochemicals on Aeromonas hydrophila. Research Journal of BioTechnology, 2, 20-25
- Nanasombat, S. and Lohasupthawee, P. (2005) Antibacterial activity of crude ethanolic extracts and essential oils of spices against salmonellae and other enterobacteria. KMITL Science and Technology Journal, 5, 527-538.
- Gupta, V.K., Singh, G.D., Singh, S. and Kaul, A. (2010) Medicinal plants: Phytochemistry, pharmacology and therapeutics, Volume 1. Daya Publishing House, Delhi.
- Adwan, K. and Abu-Hasan, N. (1998) Gentamicin resistance in clinical strains of Enterobacteriaceae associated with reduced gentamicin uptake. Folia Microbiologica, 43, 438-440. doi:10.1007/BF02818588
- Lopez, P., Sanchez, C., Batlle, R. and Nerin, C. (2005) Solid-and vapor-phase antimicrobial activities of six essential oils: Susceptibility of selected foodborne bacterial and fungal strains. Journal of Agricultural and Food Chemistry, 53, 6939-6946. doi:10.1021/jf050709v
- Arici, M., Sagdic, O. and Gecgel, U. (2005) Antibacterial effect of Turkish black cumin (Nigella sativa L.) oils. Grasas y Aceites, 56, 259-262. doi:10.3989/gya.2005.v56.i4.90
- Akgul, A. (1989) Antimicrobial activity of black cumin (Nigella sativa L.) essential oil. Gazi Universitesi Eczacilik Fakultesi Dergisi, 6, 63-68.
- Khan, M.A.U., Ashfaq, M.K., Zuberi, H.S., Mahmood, M.S. and Gilani, A.H. (2003) The in vivo antifungal activity of the aqueous extract from Nigella sativa seeds. Phytotherapy Research, 17, 183-186. doi:10.1002/ptr.1146
- Salman, M.T., Khan, R.A. and Shukla, I. (2008) Antimicrobial activity of Nigella sativa Linn. seed oil against multi-drug resistant bacteria from clinical isolates. Natural Product Radiance, 7, 10-14.
- Nair, S.C., Salomi, M.J., Panikkae, B. and Panikkar, K.R. (1991) Modulatory effects of Crocus sativus and Nigella sativa extracts on cisplatin-induced toxicity in mice. Journal of Ethnopharmacology, 31, 75-83. doi:10.1016/0378-8741(91)90146-5
- Bamosa, A.O., Ali, B.A. and Sowayan, S.A. (1997) Effect of oral ingestion Nigella sativa seeds on some blood parameters. Saudi Pharmaceutical Journal, 5, 126-129.
- Al-Ghamdi, M.S. (2001) The anti-inflammatory, analgesic and antipyretic activity of Nigella sativa. Journal of Ethnopharmacology, 76, 45-48. doi:10.1016/S0378-8741(01)00216-1
- Badary, O.A. and El-Din, A.M.G. (2001) Inhibitory effects of thymoquinone against 20-methylcholanthreneinduced fibrosarcoma tumorigenesis. Cancer Detection and Prevention, 25, 362-368.
- Houghton, P.J., Zarka, R., De las Heras, B. and Hoult, J.R.S. (2007) Fixed oil of Nigella sativa and derived thymoquinone inhibit eicosanoid generation in leukocytes and membrane lipid peroxidation. Planta medica, 61, 33-36. doi:10.1055/s-2006-957994
- Morsi, N.M. (2000) Antimicrobial effect of crude extracts of Nigella sativa on multiple antibiotics-resistant bacteria. Acta Microbiologica Polonica, 49, 63-74.
- Salem, M.L. and Hossain, M.S. (2000) Protective effect of black seed oil from Nigella sativa against murine cytomegalovirus infection. International Journal of Immunopharmacology, 22, 729-740. doi:10.1016/S0192-0561(00)00036-9
- Kamal, A., Arif, J.M. and Ahmad, I.Z. (2010) Potential of Nigella sativa L. seed during different phases of germination on inhibition of bacterial growth. E3 Journal of Biotechnology and Pharmaceutical Research, 1, 9-13.
- Akhtar, N., Alakloby, O.M., Aljabre, S.H.M., Alqurashi, A.R.M. and Randhawa, M.A. (2007) Comparison of antifungal activity of thymoquinone and amphotericin B against Fusarium solani in vitro. Scientific Journal of King Faisal University (Basic and Applied Sciences), 8, 1428H
- Halawani, E. (2009) Antibacterial activity of thymoquinone and thymohydroquinone of Nigella sativa L. and their interaction with some antibiotics. Advances in Biological Research, 3, 148-152.
- Randhawa, M.A. and Al-Ghamdi, M.J. (2002) A review of pharmaco therapeutic effect of Nigella sativa. Pakistan Journal of Medical Research, 41, 77-83.
NOTES
*Corresponding author.

