Advances in Bioscience and Biotechnology
Vol.1 No.3(2010), Article ID:2359,7 pages DOI:10.4236/abb.2010.13020
Mitochondrial comparative proteomic analysis of sterile line and its maintain line of purple cytoplasmic rice (Oryza sativa)
![]()
1Food Crops Institute, Hubei Academy of Agricultural Sciences, Wuhan, China;
2Key Laboratory of MOE for Plant Developmental Biology, College of Life Sciences, Wuhan University, Wuhan, China.
Email: ricewei7@163.com
Received 23 May 2010; revised 7 June 2010; accepted 9 June 2010.
Keywords: ATP Synthase; Comparative Proteomic Analysis; Cytoplasmic Male Sterile (CMS); Ying xiang A; Ying xiang B
ABSTRACT
CMS/Rf systems in rice (Oryza sativa) have long been exploited for hybrid breeding to enhance productivity. Ying xiang CMS/Rf system is a new type. In this study, a mitochondrial comparative proteomic analysis of Ying xiang Sterile Line and its Maintain Line was started for a comprehensive investigation of the mitochondrial proteins’ functions in rice cytoplasmic male sterility. Mitochondria were prepared from rice shoots grown in the dark. Proteins were analyzed by two-dimensional electrophoresis and MALDI-TOF/ MS. Using Mascot, it was found that 7 proteins were not described previously for plant mitochondria, indicating novel mitochondrial functions. 3 of them were characterized.
1. INTRODUCTION
Cytoplasmic male sterility (CMS) is a maternally inherited phenotype characterized by the inability of a plant to produce functional pollen [1]. In rice (Oryza sativa), several CMS/Rf systems defined by the different CMS cytoplasm with distinct genetic features have been identified [2-4]. These systems have been widely used for hybrid rice breeding in China and other Asian countries as hybrid rice crops often produce higher yields than inbred varieties [5,6].
Cytoplasmic male sterility in plants is always associated with mitochondrial dysfunction. Molecular studies on CMS have revealed the existence of modifications in mitochondrial DNA and a correlation between the presence of chimaeric mitochondrial genes and the synthesis of new proteins [7-10], and some mitochondrial genes responsible for CMS are chimeric in structure [9,11]. Some of these genes encode a cytotoxic protein [12]. Proteomic analysis is a newly developed technique that has been demonstrated as a powerful approach in plant research [13-16].
Genetical resources play a key role in meeting global challenges in the fields of food security. For example, cytoplasmic diversity is very important for resistance to plant diseases and insect pests in agriculture. We constructed a new type of cytoplasmic male sterile (CMS) line ZidaoA. It can help us to reveal the mechanism of rice cytoplasmic male sterility because its microspores abort completely and earlier than the others, which is significant in hybrid breeding in rice [17,18]. Furthermore, it will enrich the diversity of the cytoplasmic genetic resource of rice, which can avoid the reduction of output caused by rice diseases and insect pests because of the genetic frangibility in rice. And Ying xiang CMS line Ying xiang A is a nucleus displacement product of ZidaoA of Purple rice CMS line and it has the identical cytoplasm and the same properties as the Purple-leaf rice CMS line, so research on Ying xiang CMS line and its maintenance can help us to elucidate the mechanism of purple-leaf rice cytoplasmic male sterility and be advantageous in hybrid breeding of rice. Our early study showed that RNA editing may play a role in the CMS. The translation of atp9 transcript might interfere with the construction of F0F1-ATPase, resulting in the decrease of ATPase activity and the abortion of pollen in Ying xiang A. We also studied the total proteins of leaves and young panicle by two-dimensional electrophoresis [19,20]. However, little is known about the defects of mitochondrial proteome in the Ying xiang CMS line so far. In the present study, we demonstrate and characterize the mitochondrial proteomics difference of CMS line and its maintain line using 2D-PAGE and MALDITOF/MS (matrix-assisted laser desorption/ionisationtime of flight mass spectrometry).
2. MATERIALS AND METHODS
2.1. Materials
Cytoplasmic male sterile (CMS) line and its maintain line of Zidao (Yunnan purple-leaf rice) type rice (Oryza sativa): Ying xiang A and Ying xiang B.
2.2. Rice Mitochondrial Isolation
The methods of rice mitochondrial isolation followed Mignouna et al. [21].
2.3. Two-Dimensional Gel Electrophoresis
Mitochondria protein samples (100 μg) were acetone extracted by addition of acetone to a final concentration of 80% (v/v) at –20℃. 1mg dried powder was used for two-dimensional electrophoresis. Proteins were visualized by Coomassie blue R350 staining for 10min at 100℃ and destained overnight with 10% (v/v) acetic acid. Mw standards were used to identify apparent molecular masses on second dimension separation.
2.4. Mass Spectrometry Analysis and Protein Identification
Protein spots were manually excised from stained gels and characterized after in-gel trypsin digestion by Voyager-DE STR (Applied Biosystems, CA) according to Zhang et al. [22]. Then the peptide mixture was loaded onto a MALDI plate for MS analysis using a Reflex III type Maldi-Tof-MS (BRUKER) analyses. For protein identification purposes, mass spectrometric data were searched using MASCOT software (www.matrixscience. com). The search parameters used with this software were as follows: one missed cleavage; 0.1 Da mass accuracy; six peptides allowed. For hits that did not fit with the tolerated mass accuracy, namely the occurrence of six peptides per protein and a coverage greater than 20%, cross-matching was carried out by comparing the molecular weight and pI of the predicted protein with the observed molecular mass and pI of the excision site on the two-dimensional gel. Database searches of MALDITOF/MS were performed using the NCBI database (O. sativa specified databases and the reversed databases as a control), and positive identifications were made when the scores were above the significance threshold value (P < 0.05). The proteomics tools at http://www.expasy.org/ tools/ were used in computing the pI and Mw and functional analysis.
3. RESULTS AND ANALYSIS
3.1. Identification of Proteins via 2DE, Mass Spectrometry and Bioinformatics
In order to get the differentially accumulated mitochondrial proteins in sterile Ying xiang A and maintain lines Ying xiang B, the proteome maps visualized by Coomassie blue R350 of the mitochondia were compared, and protein spots with distinctly difference (Figure 1) were analysed using adobe Photoshop software. Only 13 protein spots were chosen (Figure 1). The 13 protein spots were excised manually and digested in-gel with trypsin, and analyzed using MALDI-TOF/MS and 12 protein spots were produced peptide mass fingerprint
(PMF) data. The 13 spots were numbered, as indicated in Figure 1 and Table 1. Database searches using these PMF data at http://www.matrixscience.com/cgi/ using MASCOT revealed the identities of 12 of the protein
 (a)
(a)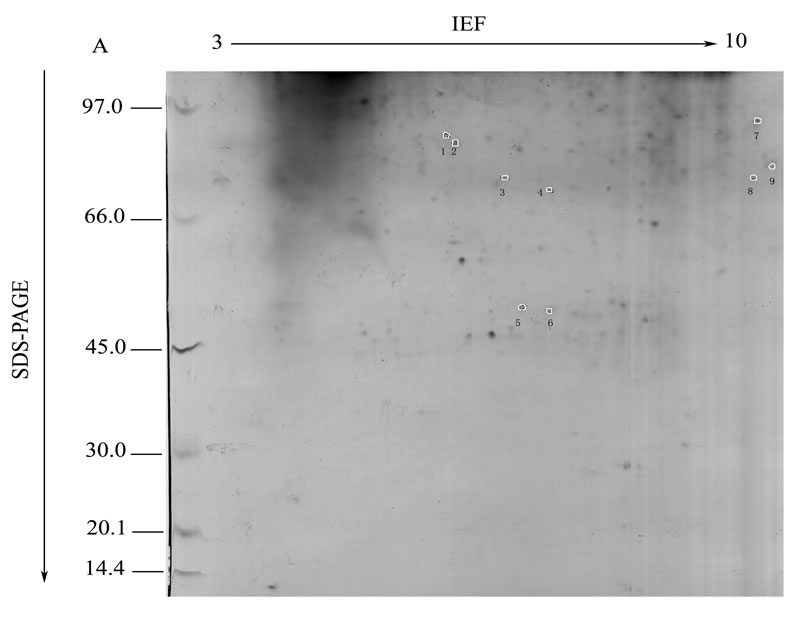 (b)
(b)
Figure 1. Representative 2-DE maps of the total mitochondrial protein from CMS rice Ying xiang A (a) and its maintainer Ying xiang B (b). IEF in the first dimension was carried out on linear pH 3-10, 18 cm IPG strips loaded with 1 mg of protein. In the second dimension a 13% SDS-PAGE gel was used and proteins were visualised by Coomassie Brilliant Blue R-350 staining. The spots analysed by MALDI-TOF/MS are circled and the arbitrary identifiers correspond to those listed in Table 1.
spots and most of the 12 identified protein spots contained only one protein, while 4 spots (spots 5, 6, 12 and 13) contained two or more proteins with the sequences highly conserved to the best matched proteins. But only 3 spots (spots 3, 4 and 10) were significant after being compared with the observed molecular mass and pI of the excision site on the two-dimensional gel.
3.2. Functional Analysis of Identified Proteins
All 3 identified proteins were analyzed using ClustalX at home and BLAST at http://www.ncbi.nlm.nih.gov/. Furthermore, to get the function and other characters of the proteins proteomics, tools at http://www.expasy.org/ tools/ were also used. To our knowledge, spot 3 is identifiable as r40c1 protein, whose function is not characterized and that belongs to family PD683597. Spot 4 is identifiable as hypothetical protein OsJ_002947, which has a domain of ATP synthase F1, beta subunit (Figure 2). Spots 10 is identifiable as OSJNBb0017I01.1 protein (GenBank: CAE05721.1), which function seem to be Phototransformation of protochlorophyllide (Pchlide) to chlorophyllide (Chlide) (Table 2).
As for spot 4, this predicted molecular mass is slightly larger than the apparent molecular mass of the mature peptide as estimated by 2D-PAGE. We think it might be a defective protein. Marilyn and Robert [23] proposed that because the ATPase subunit 2 polypeptide was synthesized in the cytoplasm, it likely was synthesized as a larger precursor with a pre-sequence for mitochondrial targeting and localization. But it is not the case here. Because the protein spot is almost not detectable in the corresponding fertile line’s proteomic map, if it is true, the fertile lines can not survive.
4. DISCUSSION
CMS was defined as maternally inherited male sterility resulting from a specific (mitochondrial) gene whose expression impairs the production of viable pollen without otherwise affecting the plant. That means that cytoplasmic male sterility in plants is associated with mitochondrial dysfunction. Up to now, 12 mitochondrion DNA regions associated with CMS have been identified, and most of them are involved in the genes encoding F0F1-ATPase subunits [11]. The N-terminal region of atp9 in petunia is present in the CMS-associated pcf gene. atp6 provides the 5’ regulatory sequences for the CMS associated urf13-T gene in maize [24]. The CMSassociated gene orf522 found in sunflowers, co-transcribes with atpA and a tissue-specific increase in the level of polyadenylated atpA-orf522 transcripts correlates with the tissue-specific instability of atpA-orf522 mRNA in male florets of the restored hybrid plants [25]. Rice (Oryza sativa L.) CMS-Boro II is associated with an abnormal copy of the mitochondrial gene atp6 [12,26] that transcribes aberrant mRNAs containing an additional orf named orf79 [27]. In Honglian (HL) rice orfH79 is a mitochondria chimeric gene being responsible for the CMS trait [28,29]. Furthermore, RNA editing plays a role in the CMS [10].
Mitochondrion is the site of both the tricarboxylic acid cycle and oxidative phosphorylation pathway and plays a crucial role in energy and carbon metabolism in eukaryotic cells. The oxidative phosphorylation pathwayconsists of the electron transfer chain (ETC), including complex I–IV, and F0F1-ATPase (complex V). The F1- ATP synthase beta subunit (β-subunit) is a highly con-
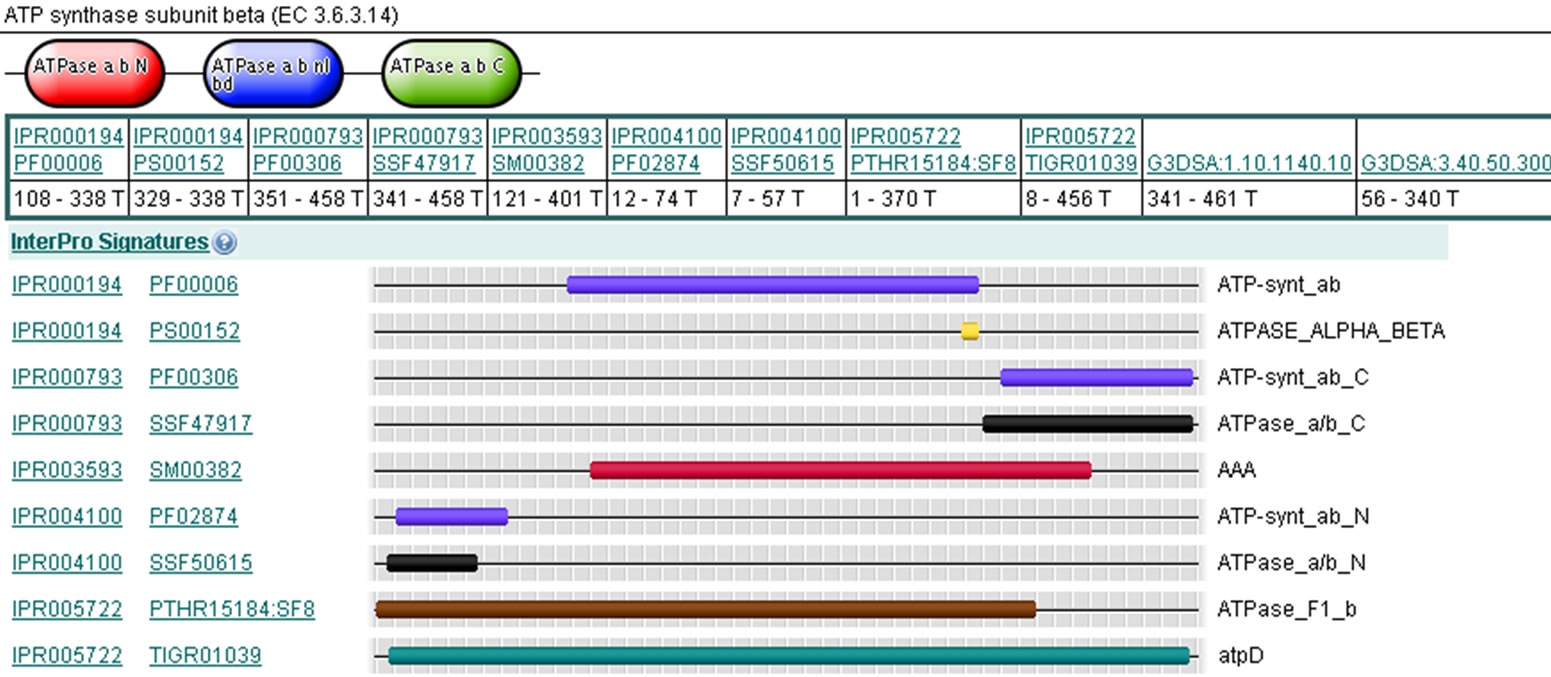
Figure 2. Graphical view of domain structure at http://www.ebi.ac.uk/interpro/of spot 4.
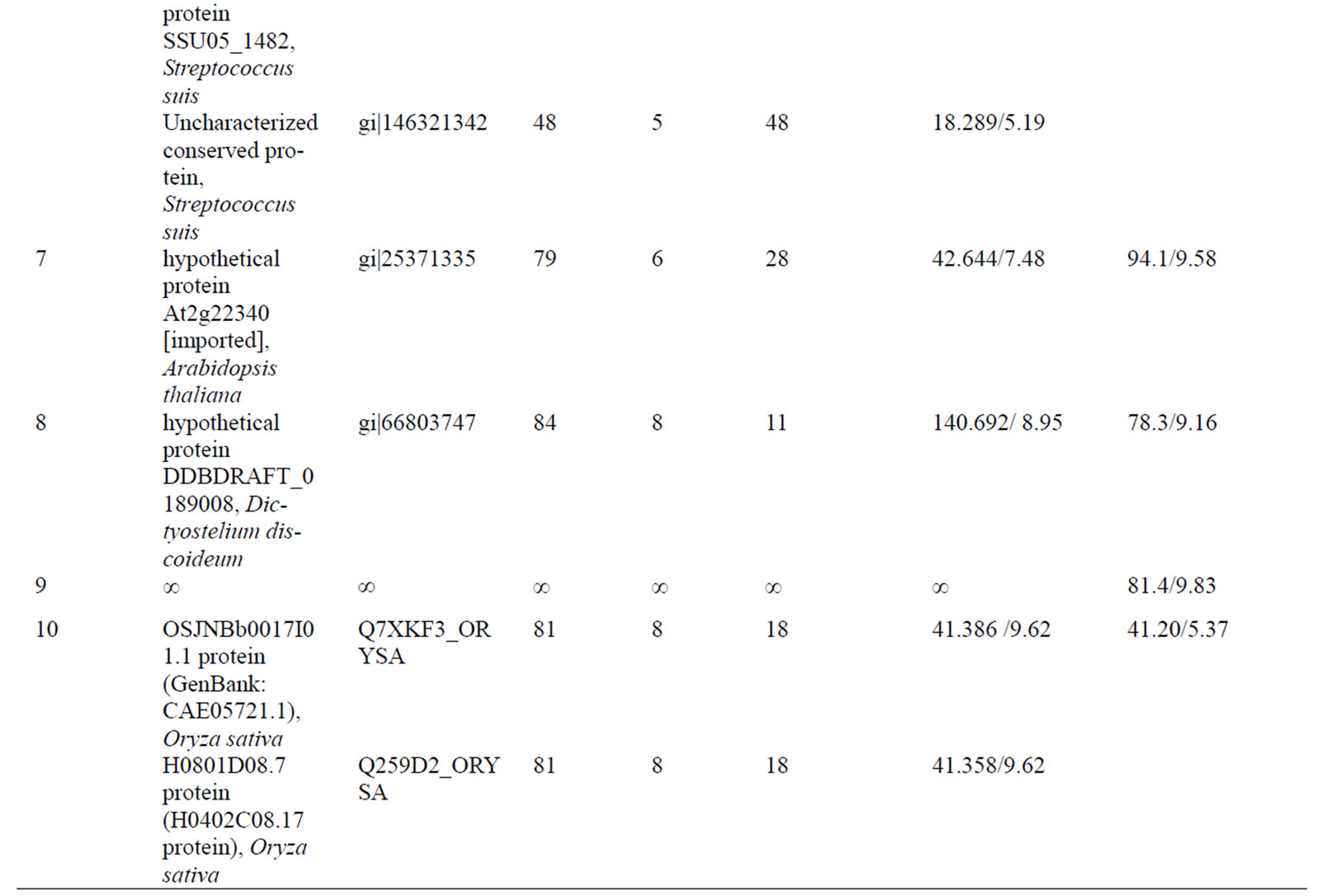
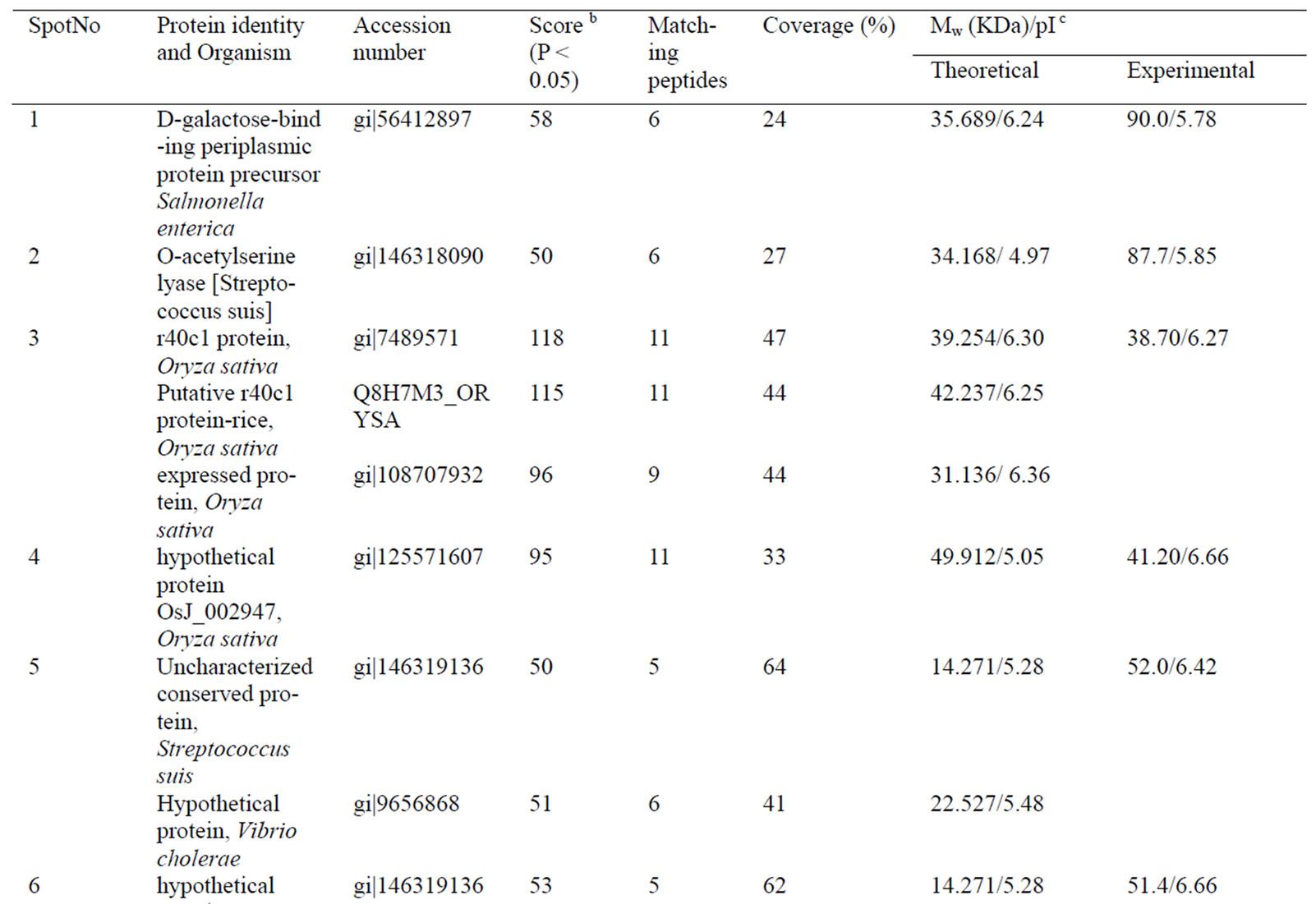
Table 1. Differentially accumulated proteins identified by mass spectrometry analysis and database searching.
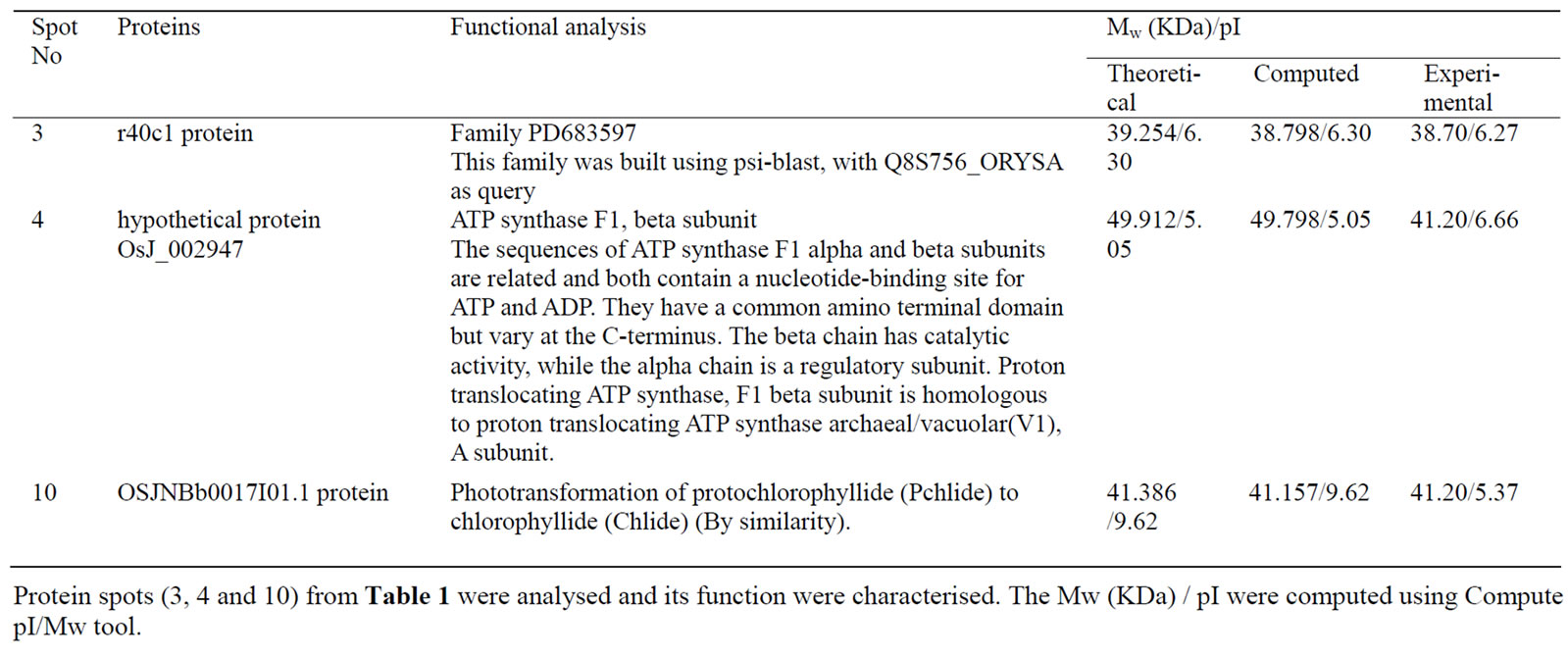
Table 2. Functional analysis of identified proteins by ClustalX, BLAST and proteomics tools.
served single copy gene encoding a 480 amino acid protein which is a catalytic site for the ATPase reaction [30]. It plays an important role in the ATPase activity with the alpha subunit. If the β-subunit is defective, it will cause the dysfunction of F0F1-ATPase, which may impact the energy output of mitochondria, resulting in abnormal anther development with non-functional pollens. It has been reported that there are very highly energy demands during pollen development in higher plants [31]. The dysfunction of F0F1-ATPase in CMS line Ying xiang A may impact the energy output of mitochondria, resulting in abnormal anther development with non-functional pollens.
A widely accepted hypothesis on the mechanism of CMS is that the increased demand for respiratory function and cellular energy in the form of ATP during anther development may be compromised by expression of the aberrant mitochondria genes. The mitochondrial enzyme F0F1-ATP synthase synthesizes adenosine triphosphate (ATP), the universal currency of chemical energy in the cell. The ATPase is installed in the inner membrane of the mitochondrion, with the F1-sector protruding into the matrix compartment and the F0-sector embedded in the inner membrane where it forms a proton-translocating channel. Using the pH-gradient between the cytosolic site and the matrix, the membrane embedded F0-part drives the synthesis of ATP in the F1-sector. The latter contains the three nucleotide binding pockets of the enzyme, which are formed mainly by the residues of the three β-subunits, which is thought to be the most conserved in its amino acid sequence [32].
In mitochondria, the majority of ATP is synthesized by F0F1-ATPase and driven by the electron transport on the inner membrane. If there are defects in the F0F1-ATPase, a further disturbance of the electron transport will magnify the disruptions of ATP synthesis, and consequently, the metabolism process in cells will be interfered with. For testing the link between F0F1-ATPase defects and CMS in HL (Hong-lian) rice, Zhang et al. [33] used inhibitor of ETC to interfere the respiration of seedlings and observed their growth. The results revealed that seedling growth delayed in the sterile line due to the disruption of phosphorylation, suggesting that F0F1- ATPase activity was suppressed in the CMS plants. Peng et al. [29] found that the content of reactive oxygen species (ROS) in the transformants that expressed ORFH79 was increased by 31%, and ATP was decreased by 41% compared with the control. Higher ROS content caused a more swift decrease of F0F1-ATP activity and ATP contents in YtA than those in YtB [34]. Sun et al. [35] deduced that reduction of the proteins associated with energy production and lesser ATP equivalents detected in CMS anther indicated that the low level of energy production played an important role in inducing CMS-HL.
In our earlier study, we reported the RNA editing of transcripts of the mitochondrial atp9 gene from Ying xiang A and Ying xiang B of the purple rice cytoplasm and discussed the different editing in them [36]. The atp9 transcript of Ying xiang A was shown to have no editing sites and the transcript of Ying xiang B was shown to have 2 editing sites with changes affecting the amino acid sequence of the protein product. The editing of the atp9 transcript from Ying xiang B was found to change an arginine codon into a stop codon, shortening the protein of Ying xiang B to the “standard” size. And the Ying xiang A transcript, which has no stop codon, cannot be translated to a normal protein. The results demonstrate the important role of RNA editing in the production of the functional ATP9 subunit and RNA editing is associated with cytoplasmic male sterility. In the present study, protein spot 4 was identified to be beta subunit of ATP synthase F1 sector and it is deduced to be a defective protein. Its incorporation into the ATP synthase complex may lead to the dysfunction of F0F1- ATPase. An abnormal ATP synthase may impact the energy output of mitochondria, resulting in abnormal anther development with non-functional pollens in Ying xiang A. Our earlier study also proved that the mitochondrial activity of fertile lines is much higher than that of the sterile lines [17].
In conclusion, CMS of Ying xiang A in rice may have multiple causes. The abnormal protein complex of ATP synthase may cause the dysfunction of mitochondrion. We can propose that the cooperation of abnormal beta subunit and ATP9 protein may cause the cytoplamic male strility of Ying xiang A. Further research should be carried out to characterise the function of other different proteins.
5. ACKNOWLEDGEMENTS
We thank Dr. Josephine Richardson for her critical reading of the manuscript. This study was supported by the National Science Foundation of China (no. 30571143).
REFERENCES
- Pring, D.R., Van, T.H. and Schertz, K.F. (1995) Cytoplasmic male sterility and organelle DNAs of sorghum. In: Levings, C.S., III and Vasil, I.K., Eds, Advances in Cellular and Molecular Biology of Plants, Kluwer Academic, Dordrecht, 461-495.
- Shinjyo, C. (1969) Cytoplasmic genetic male sterility in cultivated rice, Oryza sativa L. II. The inheritance of male sterility. Japanese Journal of Genetics, 44(3), 149-156.
- Lin, S. and Yuan, L. (1980) Hybrid rice breeding in China. Innovative Approaches to Rice Breeding, International Rice Research Institute, Los Banos, Philippines, 35-51.
- Rao, Y. (1988) Cytohistology of cytoplasmic male sterile lines in hybrid rice. In: Smith, W.H., Bostian, L.R. and Cervantes, E.P., Eds., Hybrid Rice, International Rice Research Institute, Manila, Philippines, 115-128.
- Li, J. and Yuan, L. (2000) Hybrid rice, genetics, breeding, and seed production. In: Janick, J., Ed, Plant Breeding Reviews, John Wiley & Sons, New York, 17, 15-158.
- Virmani, S.S. (2003) Advances in hybrid rice research and development in the tropics. In: Hanoi, Vietnam, S., Virmani, S., Mao, C. and Hardy, B., Eds, Hybrid Rice for Food Security, Poverty Alleviation, and Environmental Protection, International Rice Research Institute, Los Baňos, Philippines, 7-20.
- Dewey, R.E., Levings, C.S., III and Timothy, D.H. (1986) Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas malesterile cytoplasm. Cell, 44(3), 439-449.
- Levings, C.S., III (1990) The Texas cytoplasm of maize: Cytoplasmic male sterility and disease susceptibility. Science, 250(4983), 942-947.
- Hanson, M.R. (1991) Plant mitochondrial mutations and male sterility. Annual Review of Genetics, 25, 461-486.
- Hernould, M., Suharsono, S., Zabaleta, E., Carde, J.P., Litvak, S., Araya, A. and Mouras, A. (1998) Impairment of tapetum and mitochondria in engineered male-sterile tobacco plants. Plant Molecular Biology, 36(4), 499-508.
- Hanson, M.R. and Bentolila, S. (2004) Interaction of mitochondria and nuclear genes that affect male gametophyte development. The Plant Cell, 16(Suppl 1), S154-S169.
- Wang, Z., Zou, Y., Li, X., Zhang, Q., Chen, L., Wu, H., Su, D., Chen, Y., Guo, J., Luo, D., Long, Y., Zhong, Y. and Liu, Y. ( 2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. The Plant Cell, 18(3), 676-687.
- Rakwal, R., Agrawal, G.K. (2003) Rice proteomics: Current status and future perspectives. Electrophoresis, 24(19-20), 3378-3389.
- Tanaka, N., Mitsui, S., Nobori, H., Yanagi, K. and Komatsu, S. (2005) Expression and function of proteins during development of the basal region in rice seedlings. Molecular & Cellular Proteomics, 4(6), 796-808.
- Kim, D.S., Cho, D.S., Park, W.M., Na ,H.J. and Nam, H.G. (2006) Proteomic pattern-based analyses of light responses in Arabidopsis thaliana wild-type and photoreceptor mutants. Proteomics, 6(10), 3040-3049.
- Plomion, C., Lalanne, C., Claverol, S., Meddour, H., Kohler, A., Bogeat-Triboulot, M.B., Barre, A., Provost, G., Dumazet, H., Jacob, D., Bastien, C., Dreyer, E., Daruvar, A., Guehl, J.M., Schmitter, J.M., Martin, F. and Bonneu, M. (2006) Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics, 6(24), 6509-6527.
- Wei, L., Ding, Y., Liu, Y. and Yu, J. (2002) Microcalorimetric analysis of anthers of male sterile rice sterile line. Journal of Wuhan Botanical Research, 20(4),308-310.
- Hu, Y., Wu, Q., Liu, S., Wei, L., Chen, X., Yan, Z., Yu, J., Zeng, L. and Ding, Y. (2005) Study of rice pollen grains by multispectral imaging microscopy. Microscope Research and Technique, 68(6), 335-346.
- Wei, L., Ding, Y., Hu, Y. and Yu, J. (2002) Analysis of leaf proteins of zidao male sterile line by 2D-PAGE. Acta Genetica Sinica, 29(8), 696-699.
- Hu, Y., Wei, L., Liu, S., Yu, J. and Ding, Y. (2005) Analysis of male sterility-related protein of young panicle in rice (Oryza sativa L.) by two-Dimensional electrophoresis. Wuhan University Journal of Natural Sciences, 10(3), 597-601.
- Mignouna, H., Vermin, S.S. and Briquet, M. (1987) Mitochondrial DNA modification associated with cytoplasmic male sterility in rice. Theoretical and Applied Genetics, 74(5), 666-669.
- Zhang, Y., Fan, X., Chen, R., Xiao, Z., Feng, X., Tian, X. and Chen, Z. (2005) Comparative proteome analysis of untreated and Helicobacter pylori-treated HepG2. World Journal of Gastroenterology, 11(22), 3485-3489.
- Marilyn, E. and Robert, B. (1990) Respiration and mitochondrial biogenesis in germinating embryos of maize. Plant Physiology, 93(1), 295-304.
- Hanson, M.R., Pruitt, K.D. and Nivison, H.T. (1989) Male sterility loci in plant mitochondrial genes. Oxford Surveys in Plant Molecular and Cellular Biology, 6, 61-85.
- Gagliardi, D. and Leaver, C.J. (1999) Polyadenylation accelerates the degradation of the mitochondria mRAN associated with cytoplasmic male sterility in sunflower. The EMBO Journal, 18(13), 3757-3766.
- Iwabuchi, M., Kyozuka, J. and Shimamoto, K. (1993) Processing followed by complete editing of an altered mitochondrial apt6 RNA restores fertility of cytoplasmic male sterile rice. The EMBO Journal, 12(4),1437-1446.
- Akagi, H., Sakamoto, M., Shinjyo, C., Shimada, H. and Fujimura, T. (1994) A unique sequence located downstream from the rice mitochondrial apt6 may cause male sterility. Current Genetics, 25(1), 52-58.
- Yi, P., Wang, L., Sun, Q. and Zhu, Y. (2002) Discovery of mitochondria chimeric gene associated with male sterility of Honglian-rice. Chinese Science Bulletin, 47(9), 744-747.
- Peng, X., Li, F., Li, S. and Zhu, Y. (2009) Expression of a mitochondrial gene orfH79 from the CMS-HongLian rice inhibits Saccharomyces cerevisiae growth and causes excessive ROS accumulation and decrease in ATP. Biotechnology Letters, 31(3), 409-414.
- Pedersen, P.L. and Amzel, L.M. (1993) ATP synthases. Structure, reaction center, mechanism, and regulation of one of nature’s most unique machines. Journal of Biological Chemistry, 268(14), 9937-9940.
- Tedege, M. and Kuhlemeier, C. (1997) Aerobic fermentation during tobacco pollen development. Plant Molecular Biology, 35(3), 343-354.
- Runswick, M. and Walker, J. (1983) The amino acid sequence of the β-subunit of the ATP synthase from bovine heart mitochondria. Journal of Biological Chemistry, 258(5), 3081-3089.
- Zhang, H., Li, S., Yi, P., Wan, C., Chen, Z. and Zhu, Y. (2007) A Honglian CMS line of rice displays aberrant F0 of F0F1-ATPase. Plant Cell Reports, 26(7), 1065-1071.
- Hu, C., Sun, Q., Peng, X., Huang, Q., Wang, M., Li, S. and Zhu, Y. (2010) Flow cytometric analysis of mitochondrial populations in HL-CMS systems of rice under H2O2 stress. Protoplasma, 241(1-4), 91-98.
- Sun, Q., Hu, C., Hu, J., Li, S. and Zhu, Y. (2009) Quantitative proteomic analysis of CMS-related changes in Honglian CMS rice anther. The Protein Journal, 28(7-8), 341-348.
- Wei, L., Yan, Z. and Ding, Y. (2008) Mitochondrial RNA editing of F0-ATPase subunit 9 gene (atp9) transcripts of Yunnan purple rice cytoplasmic male sterile line and its maintainer line. Acta Physiologiae Plantarum, 30(5), 657-662.

