Journal of Water Resource and Protection
Vol.2 No.5(2010), Article ID:1778,7 pages DOI:10.4236/jwarp.2010.25045
Heterogeneity of Heterotrophic Bacteria Dominating in Brackish Lake Shira (Republic of Khakasia, Russia) Subjected to Anthropogenic Impact
1Krasnoyarsk Scientific Centre, Russian Academy of Sciences, Siberian Branch, Krasnoyarsk, Russia
2Institute of Biophysics, Russian Academy of Sciences, Siberian Branch, Krasnoyarsk, Russia
3Center of Hygiene and Epidemiology, Krasnoyarsk, Russia
E-mail: yanalobova@mail.ru
Received November 6, 2009; revised December 4, 2009; accepted February 10, 2010
Keywords: Anthropogenic Impact, Antibiotic Resistance, Halotolerance, Heterotrophic Bacteria, Heterogeneity
ABSTRACT
In this work data on heterogeneity in halotolerance and the resistance to antibiotics of heterotrophic bacteria dominating in Lake Shira (Republic of Khakasia, Russia) were presented. It was established that halotolerance of the bacteria is determined by genetic and epigenetic mechanisms. Influence of anthropogenic factor (entering the ecosystem allochthonous bacteria) on the appearance of multiple antibiotic resistance appearance by dominating bacterial species is discussed.
1. Introduction
Survival of bacteria in changing environments is frequently accompanied by acquiring new ecologically important properties as a result of adaptation of bacteria to the new environment. Adaptability of bacteria under the influence of one factor can differ not only among bacteria belonging to different species but also within one species. This is because, single cells in clonal population can differ in features of genomic structure from the main group of cells that is material for selection in changing environments [1], so heterogeneity is a very important ecological potential for survival of bacteria [1,2].
The water of Lake Shira (Republic of Khakasia, Russia) has unique properties and is used to treat diseases of the gastrointestinal and cardiovascular systems, locomotorium and various skin diseases [3]. Resorts for children and adults are situated at the shore of the lake. Annually this lake is visited by thousands of tourists from different regions of Russia. In our early works, we have shown that in the period of the resort season (June-August) lake is subjected to anthropogenic impact. In particular, plasmid-containing allochthonous bacteria with multiple antibiotic resistance enter the ecosystem with sewage from the resorts and village situated by Lake Shira and also at bathing of tourists [4-6], and as a result of this, resistance to antibiotics can be transferred from allochthonous to autochthonous bacteria by horizontal gene transfer. From literature it is well known about dissemination of drug resistance among bacteria in aquatic ecosystems under anthropogenic load [7-11]. To determine whether antibiotic resistance in cells of autochthonous bacteria of Lake Shira is maintained, the analysis of the structure of bacterial populations at the level of antibiotic resistance was carried out in this work. Another factor that has a key influence on the distribution of bacteria in the lake is salinity of the water [12]. Early we indicated that freshwater bacteria (i.e. that are unable to tolerate more than 3% NaCl) have predominated in the surface layers of the lake, and halophile bacteria and moderate halotolerant bacteria (surviving at 5% and 10% NaCl respectively) are predominated in the depth horizons of the ecosystem [4]. Since mineralization of the lake’s water varies through the basin, it was important to evaluate the potential of each species of autochthonous bacteria to survival at different salinity. These data might widen our understanding of the biological properties of autochthonous bacteria of Lake Shira and can be used for ecological monitoring of the ecosystem.
The aim of this work was to evaluate heterogeneity of heterotrophic bacteria dominating in brackish Lake Shira on their halotolerance and resistance to antibiotics.
2. Materials and Methods
2.1. Short Description of Lake Shira
Lake Shira is brackish and meromictic lake in the south of the Khakasia republic (Russia) (90°14’E, 54°30’N) (Figure 1). The lake is 9.35 km long and 5.3 km wide, the medium depth of the lake is 11.2 m, the maximal depth is 22 m. The lake has no outflow, and in the east, the Son River flows into the lake. The main anions of the lake’s water are sulphate at concentration from 7.3 to 10.8 g/L and chlorine-anion at 2 g/L. The basic cations of the water in Lake Shira are sodium (from 2.4 to 3.9 g/L) and magnesium (1-1.2 g/L). The water of Lake Shira is sulfate-chloride-sodium-magnetic. Mineralization of the lake’s water varies through the water basin. There is maximal salt concentration in the central part of the ecosystem. In the centre, salt concentration increases from 18 g/L in the surface layers of the lake to 30 g/L in the deep layers [12].
2.2. Sample Collection
Sample sites of Lake Shira differ in cation sodium concentration: 2.8 g/L in the central part (at the depth of 0.5 m), 3.7-4 g/L (at 10 m) and 4-5 g/L (at 21 m); 0.6 g/L in the south-east part of the lake by the Son River; 2.7 g/L in the resort part and 2.8 g/L in the north-east.
Samples of Lake Shira water were taken in different seasons: winter (February), spring (March, May), summer (June-August) and autumn (September, November) in 1997-2003, but the main part of the field works was carried out in summer period. Total amount of the samples analyzed was 196.
2.3. Media
Water samples were plated on agar medium M9 with peptone that contained per liter of distilled water: 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl, 5 g peptone and 20 g agar. After autoclaving, 1 mL of 20%
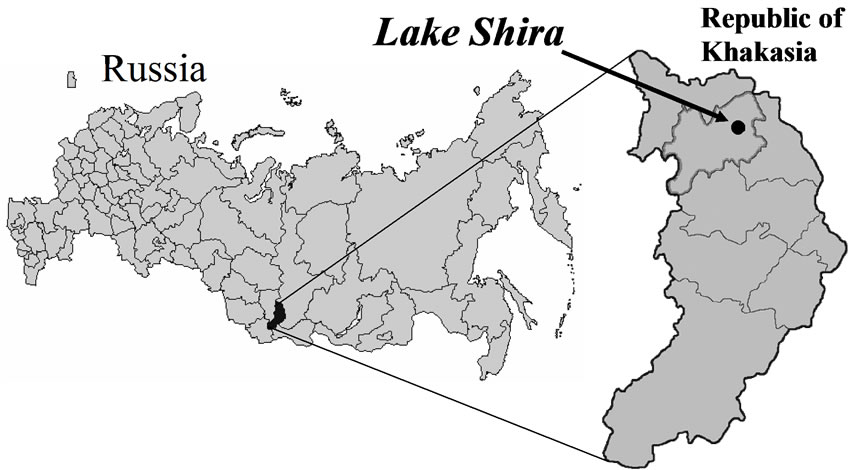
Figure 1. Location of Lake Shira.
MgSO4 and 1 mL of 0.5% CaCl2 were added [13].
2.4. Isolation of Bacteria
To isolate heterotrophic bacteria, water samples were plated on agar medium M9 containing peptone and cultivated thermostatically at 25℃. To isolate populations of halotolerant bacteria, sodium chloride was used at different concentrations 5.0, 50.0 and 100.0 g/L, at presence of which, survival of bacteria indicates of belonging of them to groups characterizing by different halotolerance. We analyzed the 1) freshwater bacteria (tolerate not more than 3% NaCl), 2) halophilic bacteria (able to survive at 0-5% NaCl) and 3) moderate halotolerant bacteria (tolerate not more than 10% NaCl). Cultivation of bacteria was carried out thermostatically at 25℃ [14].
2.5. Test for Sensitivity to Antibiotics
Resistance of the isolated bacteria to antibiotics was determined by the replica plating [13]. The antibiotic resistance of bacteria was tested on solid media amended with two different antibiotics: ampicillin (an inhibitor of the cell wall synthesis) and kanamycin (an inhibitor of protein synthesis). Antibiotics were added at concentration of 50 µg/mL. The antibiotic resistance of heterotrophic bacteria was estimated by the number of 1) bacteria resistant to ampicillin; 2) bacteria resistant to kanamycin; 3) bacteria resistant to both ampicillin and kanamycin (multiple antibiotic resistance); and 4) bacteria sensitive to the antibiotics. The number of bacteria sensitive to ampicillin and kanamycin was calculated from the number of bacteria unable to grow on the antibiotic-containing medium.
2.6. Identification of the Bacteria
Morphological and biochemical properties of the bacteria were investigated at the Krasnoyarsk Centre for State Sanitary-Epidemiological Inspection. The bacteria were identified according to Bergey’s manual of determinative bacteriology [15].
2.7. Quantitative Assessment of Heterogeneity
Quantitative assessment of heterogeneity of populations of isolated bacteria was carried out on the base of index of variation of qualitative feature—Vp [16] which was calculated according to the follow formula:
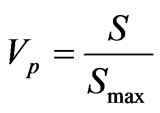 where S is variability of qualitative feature and it is determined from the formula
where S is variability of qualitative feature and it is determined from the formula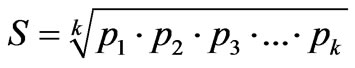 , where
, where 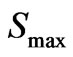 is maximal possible index for S at the same
is maximal possible index for S at the same 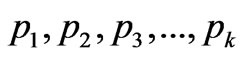 (parts of features in a sample). This index can varies from 1 (maximal heterogeneity of feature) to 0 (at absence of heterogeneity of a feature).
(parts of features in a sample). This index can varies from 1 (maximal heterogeneity of feature) to 0 (at absence of heterogeneity of a feature).
3. Results
3.1. Isolation of Heterotrophic Bacteria Dominating in Lake Shira
The analysis of the morphological diversity of colonies and cells of heterotrophic bacteria isolated from Lake Shira in different seasons in 1997-2003 was carried out. It was shown that during of the period of the ecosystem investigation heterotrophic bacteria were presented by the twenty morphotypes. The analysis of appearance of the morphotypes of the heterotrophic bacteria in the water samples showed that nine morphotypes were dominating and each of them held certain morphological features of the colonies and the cells. Taxonomic composition of dominating in the lake morphotypes of heterotrophic bacteria was determined (Table 1). Bacteria isolated were shown to belong to the following genuses of Acinetobacter, Alcaligenes, Flavobacterium, Micrococcus and Pseudomonas that are typical inhabitants of aquatic environments.
The next stage of our work was to investigate heterogeneity of the populations of bacteria dominating in Lake Shira on halotolerance and antibiotic resistance that are indicating the influence on the ecosystem of natural factor (salinity of the water) and anthropogenic factor (the resistance to antibiotics) correspondingly.
3.2. Heterogeneity of Dominating Bacteria on Halotolerance
Since Lake Shira is a brackish lake and its mineralization varies in both horizontal and vertical (at increasing depth of the lake) [3] the heterogeneity of each heterotrophic bacterial species dominating in the lake was investigated.
It was shown that each of bacterial species under the study was presented by the freshwater bacteria, halophilic bacteria and moderate halotolerant bacteria (Figure 2). The analysis of indexes of varying of the qualitative feature on the halotolerance of bacteria dominating in the lake showed that the main part of them exhibited a high level of heterogeneity (Table 2). In particular, Acinetobacter sp.1, Acinetobacter sp.2, Alcaligenes sp.1, Flavobacterium sp.1, Pseudomonas sp.2 and Pseudomonas sp.3 were characterized by the most expressed of heterogeneity on halotolerance (Figures 2(a)-2(d) and 2(h)-2(i), Table 2). This indicates the ability of the bacterias to survive at different salinity. Pseudomonas pseudoalcaligenes and Pseudomonas sp.1 vise versa in main were presented by populations of freshwater bacteria
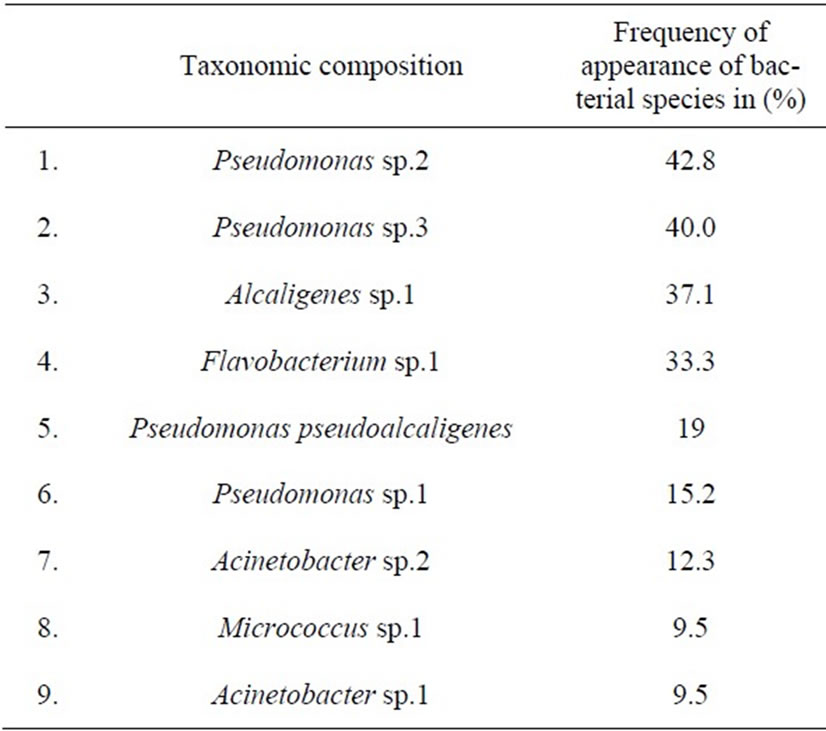
Table 1. Frequency of appearance of bacterial species dominating in the water samples of Lake Shira in 1997-2003.
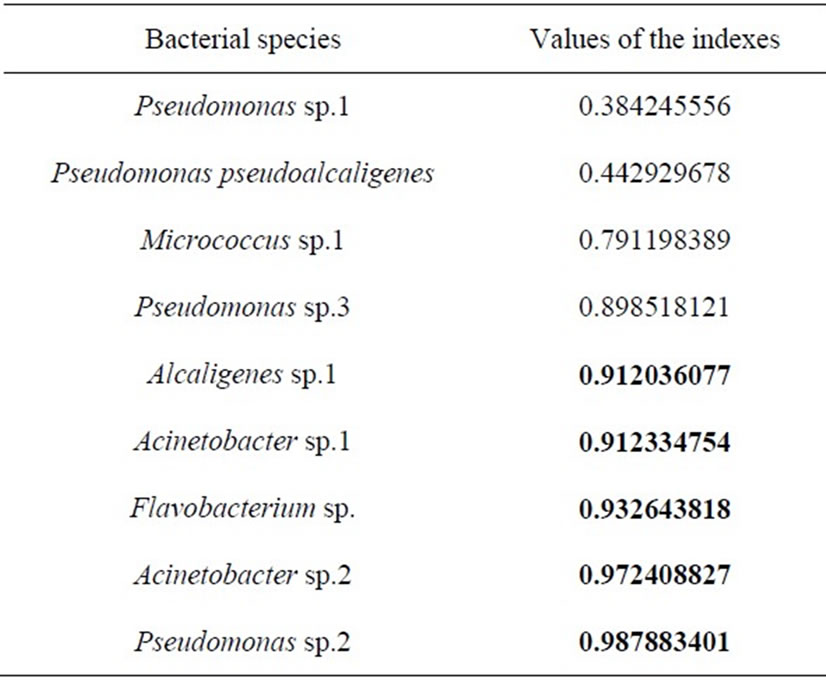
Table 2. Index of varying of qualitative feature on halotolerance of heterotrophic bacteria dominating in Lake Shira.
(Figures 2(f) and (g)). Micrococcus sp.1 (Figure 2(e)) was also characterized by the low level of heterogeneity on halotolerance, but in comparison with Pseudomonas pseudoalcaligenes and Pseudomonas sp.1 (Figures 2(f) and (g)) was presented by the populations of the halophilic bacteria.
3.3. Heterogeneity of Dominating Bacteria on Resistance to Antibiotics
Another feature appearing by bacteria from Lake Shira is resistance to antibiotics. A test for sensitivity to antibiotics of the bacteria was carried out with drugs widely used in medical practice and belonging to different classes of antibiotics: 1) ß-lactam antibiotics (ampicillin) and 2)
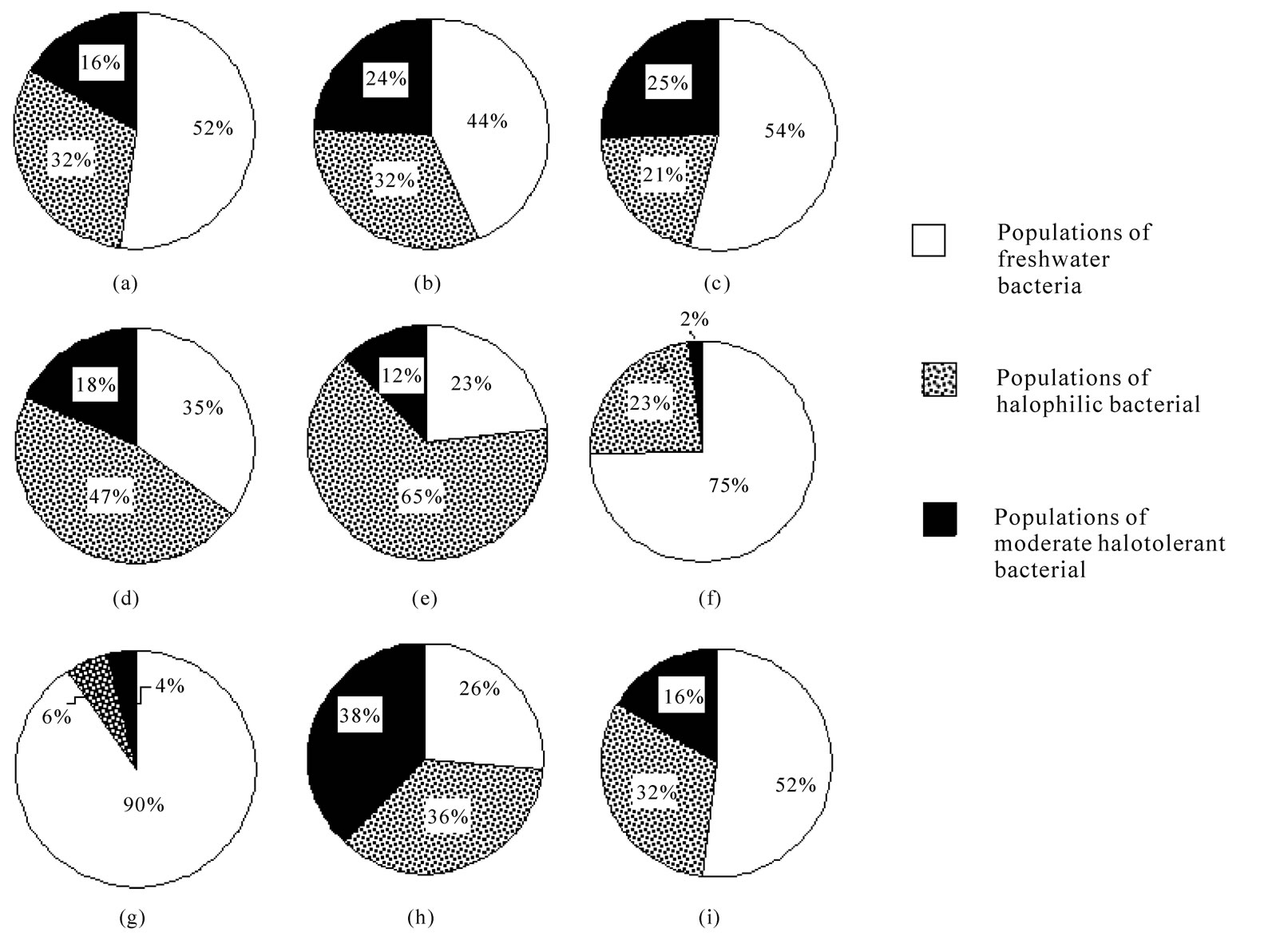
Figure 2. Heterogeneity of heterotrophic bacteria dominating in Lake Shira in their halotolerance: (a) Acinetobacter sp.1; (b) Acinetobacter sp.2; (c) Alcaligenes sp.1; (d) Flavobacterium sp.1; (e) Micrococcus sp.1; (f) Pseudomonas pseudoalcaligenes; (g) Pseudomonas sp.1; (h) Pseudomonas sp.2; (i) Pseudomonas sp.3. Bacteria were isolated from the ecosystem in 1997-2003.
aminoglycosed antibiotics (kanamycin). Bacterias under the study were found to have the low level of heterogeneity on antibiotic resistance and the maximal value of index of varying of this property was just 0.648 for Pseudomonas sp.3, and for other isolated species the values of indexes were the same and lower (Figure 3, Table 3).
The analysis of the antibiotic resistance of bacterial species dominating in Lake Shira showed that these bacteria can be divided into the three groups. Acinetobacter sp.1, Micrococcus sp.1 and Pseudomonas sp.1 belonged to the first group; they were presented generally by the populations of bacteria which were sensitive to the antibiotics (Figures 3(a), 3(e) and 3(g)). Flavobacterium sp.1, Pseudomonas sp.2 and Pseudomonas sp.3 belonged to the second group, in the populations of which vise versa, bacteria with multiple antibiotic resistance (resistant to both ampicillin and kanamycin) dominated (Figures 3(d), 3(h) and 3(i)). Acinetobacter sp.2 and Pseudomonas pseudoalcaligenes belonged to the third group; that were presented by the populations of bacteria characterizing in main by the multiple antibiotic resistance and resistance to ampicillin (Figures 3(b) and 3(f)). Alcaligenes sp.1, in comparison with other bacterial species under the study, was presented by two populations, the one of them was resistant to ampicillin (39%), and the other was sensitive to antibiotics (41%) (Figure 3(c)).
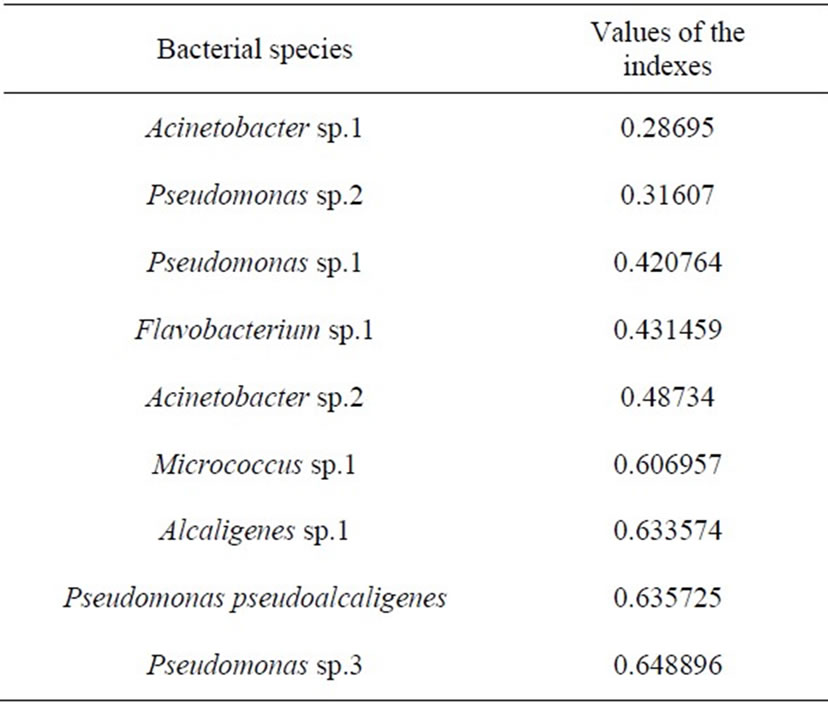
Table 3. Index of varying of qualitative feature on the resistance to antibiotics of heterotrophic bacteria dominating in Lake Shira.
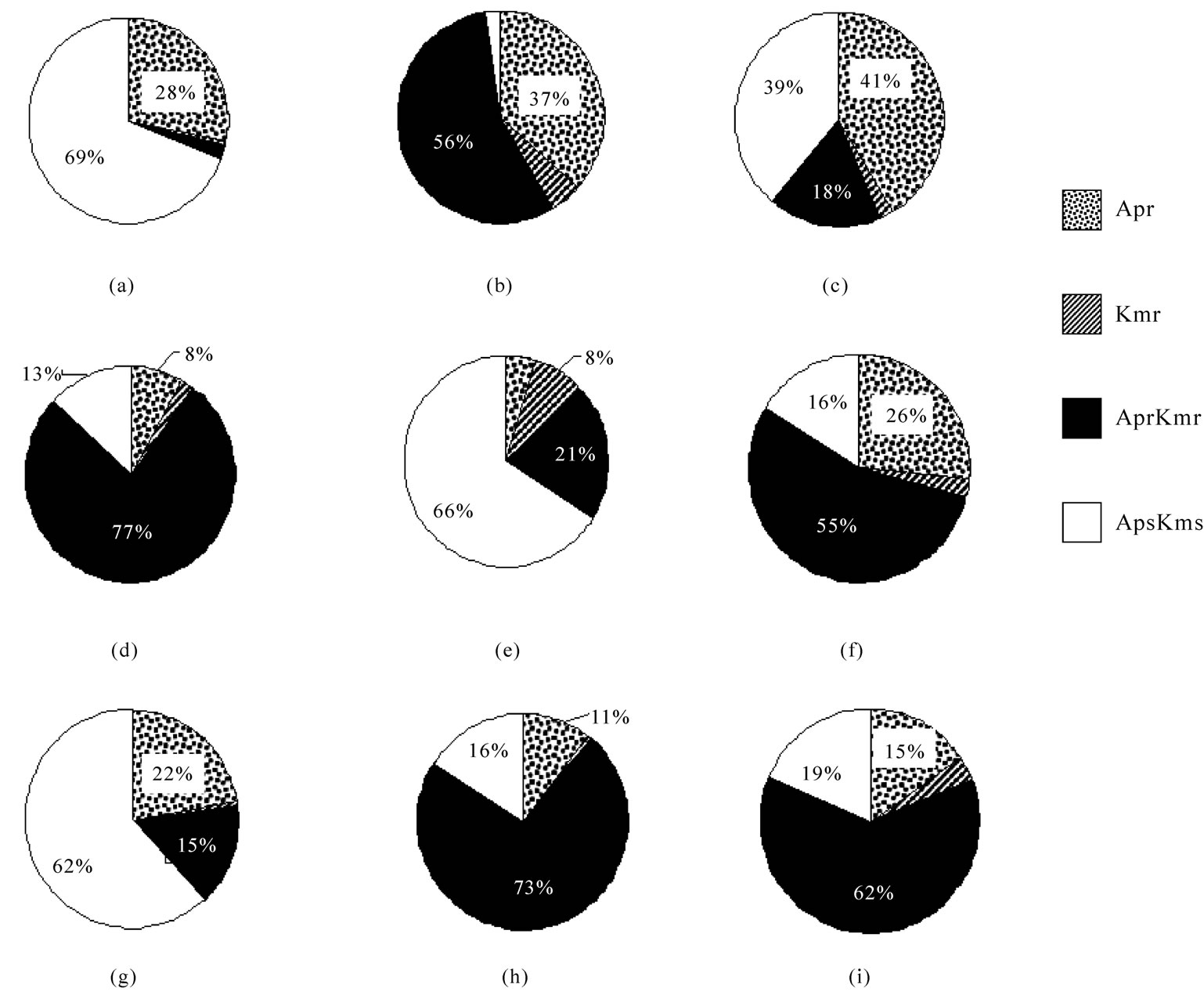
Figure 3. Heterogeneity of heterotrophic bacteria dominating in Lake Shira in their resistance to antibiotics: (a) Acinetobacter sp.1; (b) Acinetobacter sp.2; (c) Alcaligenes sp.1; (d) Flavobacterium sp.1; (e) Micrococcus sp.1; (f) Pseudomonas pseudoalcaligenes; (g) Pseudomonas sp.1; (h) Pseudomonas sp.2; (i) Pseudomonas sp.3. Bacteria were isolated from the ecosystem in 1997-2003. Apr-resistance to ampicillin, Kmr-resistance to kanamycin, AprKmr-resistance to ampicillin and kanamycin, ApsKms-sensitivity to antibiotics.
The population of Alcaligenes sp.1 appearing the multiple antibiotic resistance was not predominated and not exceeded 18% (Figure 3(c)).
Analyzing the antibiotic resistance of heterotrophic bacteria dominating in Lake Shira it was noted that resistance to kanamycin was revealed by bacteria at the low level, and such bacterial features as the resistance to ampicillin and also the multiple antibiotic resistances were appeared by all bacterial species under the study.
4. Discussion
Survival of bacteria in changing environment can be determined as modification changes that do not modify of cell’s genotype (on level of a phenotype) [17] and genetically mechanisms as well via an acquiring of new properties by bacteria as a result of horizontal gene transfer. On the sample of heterotrophic bacteria dominating in Lake Shira the analysis of heterogeneity of their populations on account of influence on the bacteria of natural factor (salinity of the lake’s water) and anthropogenic factor (entrance of allochthonous bacteria the ecosystem) and the attempts to find out possible mechanisms contributing to adaptation of the bacteria to the factors were made.
As it was early mentioned in the work, the mineralization of the water in Lake Shira is varying, there are approximately 3% NaCl in the surface water layers of the lake and 5% NaCl in the depth. Thus, bacterias from the lake can be able to survive at different salinity. As shown in Figure 2 and Table 2, all bacterias studied are halotolerant i.e., they survive at presence both the low concentration of sodium chloride (3%) and the high of it concentrations (5% and 10%). The high level of heterogeneity of the bacteria on this property determines of their appearance through the lake. It should be noted that all bacteria under the study hold a potential to survive at the higher concentration of sodium chloride (10%) that it is not found in the lake, so resistance of the bacteria to 10% NaCl is not necessary for their survival. Additionally, all bacteria presented in this work, except for bacteria of genus of Micrococcus, have no taxonomical feature of halotolerance. The high potential of bacteria belonging to genuses of Acinetobacter, Alcaligenes, Flavobacterium and Pseudomonas to survive in environments with high salinity is well known. This property is expressed more frequently by the following bacterial genuses of Acinetobacter, Flavobacterium and Pseudomonas that are able to grow at presence of 0.05%-20% NaCl and higher [18,19]. The main osmoprotector mechanisms of halotolerant bacteria in comparison with halophilic bacteria (growing only at presence of high concentration of NaCl) are either part of anion lipids increasing in contain of cell’s membrane (more often phosphatidylclycerol and/or glycolipids) [18,20] or intracellular concentration increasing of such aminoacids as asparagine, glutamine and proline [18]. Genetic mechanisms of haloadaptation, in particular by means of mobile genetic elements such as plasmids, are known for halophilic bacteria. For example, bacterial strain of Halomonas elongata at loosing of its single plasmid was not able to grow at presence of 19.7% NaCl [21]. On the sample of cyanobacterial strain of Synechococcus sp. NKDG 042902, it was shown that copy number of the plasmid pSY10 with size of 2.7 kb increased by five orders after cultivation of the strain in medium with 3% NaCl [22]. The plasmid of the same size (2.7 kb) was found in the cells of four halotolerant bacterial strains isolated from the central part of Lake Shira. Two strains from them are belonging to genius of Micrococcus, and two other strains to Pseudomonas. It was indicated that at increasing of sodium chloride concentration from 0.05 to 10% in medium of the strains cultivation, the copy number of the plasmid increases too and in consequence of this, the limit of the strains halotolerance increases [23]. The plasmid with size of 2.7 kb isolated from the four halotolerant bacterial strains is of interest for investigation in future with the aim to get the information concerning of genetic mechanism of bacterial adaptation to high salinity. Investigation of other plasmids obtained from bacteria of Lake Shira has not shown dependence (decreasing or increasing) of their copy numbers on the salt concentration in medium of the bacteria cultivation [4]. As result of this, we concluded that presence of those plasmids in the bacterial cells does not contribute to their halotolerance. In our early work it was shown that resistance to ampicillin is marker property of autochthonous bacteria and its appearance by bacteria does not depend on presence of plasmids in their cells [24]. The multiple antibiotic resistance is a marker property of allochthonous bacteria entering the ecosystem with sewage [4,6]. It should be noted that allochthonous bacteria contain simultaneously in their cells from two till four plasmids differing in size and some of these plasmids are found in cells of autochthonous bacteria. However, the comparative analysis of the plasmid profile of allochthonous and autochthonous bacteria appearing the same antibiotic resistance pattern showed that there is no correlation between phenotypes of the bacteria and their plasmid profile. Moreover, some of autochthonous plasmids-bearing bacteria are sensitive to antibiotics [4]. Thus, the analysis of the plasmid DNA emerging in bacteria from Lake Shira and their antibiotic resistance does not allow making a conclusion that this connection exists. Investigation of functional significance of the plasmids genes for autochthonous bacteria of the lake is the task for our ongoing studies. The special attention will be paid to such species as Acinetobacter sp.2 Flavobacterium sp.1, Pseudomonas sp.2, Pseudomonas sp.3 and Pseudomonas pseudoalcaligenes that were presented in main by populations of bacteria appearing the multiple antibiotic resistance. Although pseudomonads are naturally multiresistant bacteria [25] the analysis of origin of their resistance to the antibiotics will be investigated. The presence of plasmids DNA having the same sizes in cells of autochthonous and allochthonous bacteria isolated from the parts of the ecosystem the most subjected to anthropogenic impact might be in consequence of lateral gene transfer from allochthonous plasmid-bearing bacteria entering the ecosystem from sewage of the resorts and the village to autochthonous bacteria.
5. Conclusions
For the first time, data on the heterogeneity on the halotolerance and the antibiotic resistance of heterotrophic bacteria dominating in Lake Shira were obtained. Increase either in the level of drug resistance of autochthonous bacteria or plasmid diversity of them can be used as an indicator of anthropogenic impact onto the ecosystem.
6. Acknowledgements
This work was supported by the Netherlands Organization for Scientific Research (NWO) and Russian Foundation for Basic Research (RFBR) Grant no. 047.017.012 and by the Russian Science Support Foundation (Award to Tatiana Lobova) and by the Federal Targeted Programme “Scientific and Scientific-Pedagogical Personnel of the Innovative Russia in 2009-2013”.
REFERENCES
- I. R. Booth, “Stress and the Single Cell: Intrapopulation Diversity is a Mechanism to Ensure Survival Upon Exposure to Stress,” International Journal of Food Microbiology, Vol. 78, No. 1-2, September 2002, pp. 19-30.
- N. Dhar and J. D. McKinney, “Microbial Phenotypic Heterogeneity and Antibiotic Tolerance,” Current Opinion Microbiology, Vol. 10, No. 1, February 2007, pp. 30-38.
- V. P. Parnachev and A. G. Degermendzhy, “Geographical, Geological and Hydrochemical Distribution of Saline Lakes in Khakasia, Southern Siberia,” Aquatic Ecology, Vol. 36, No. 2, April 2002, pp. 107-122.
- T. I. Lobova, E. Y. Maksimova, L. Yu. Popova and N. S. Pechurkin, “Geographical and Seasonal Distribution of Multiple Antibiotic Resistance of Heterotrophic Bacteria of Lake Shira,” Aquatic Ecology, Vol. 36, No. 2, April 2002, pp. 299-307.
- T. I. Lobova, L. V. Listova and L. Yu. Popova, “Distribution of Heterotrophic Bacteria in Lake Shira,” Microbiology, Vol. 73, No. 1, January 2004, pp. 89-93.
- T. I. Lobova, Y. V. Barkhatov, O. V. Salamatina and L. Y. Popova, “Multiple Antibiotic Resistance in the Littoral Zone of Lake Shira as an Indicator of Human Impact on the Ecosystem,” Microbiological Research, Vol. 163, No. 2, March 2008, pp. 152-160.
- R. J. Ash, B. Mauck and M. Morgan, “Antibiotic Resistance of Gram-Negative Bacteria in Rivers, United States,” Emerging Infectious Diseases, Vol. 8, No. 7, July 2002, pp. 713-716.
- N. Esiobu, L. Armenta and J. Ike, “Antibiotic Resistance in Soil and Water Environments,” International Journal of Environmental Health Research, Vol. 12, 2002, pp. 133-144.
- P. T. Biyela, J. Lin and C. C. Bezuidenhout, “The Role of Aquatic Ecosystems as Reservoirs of Antibiotic Resistant Bacteria and Antibiotic Resistance Gene,” Water Science and Technology, Vol. 50, No. 1, 2004, pp. 45-50.
- S. Mukherjee, B. Bhadra, R. Chakraboty, A. Gurung, S. Some and R. Chakraboty, “Unregulated Use of Antibiotics in Siliguri City Vis-A-Vis Occurrence of MAR Bacteria in Community Waste Water and River Mahananda, and their Potential for Resistance Gene Transfer,” Journal of Environmental Biology, Vol. 26, No. 2, 2005, pp. 229-238.
- R. Zhang, Y. Wang and J.-D. Gu, “Identification of Environmental Plasmid-Bearing Vibrio Species Isolated from Polluted and Pristine Marine Serves of Hong Kong and Resistance to Antibiotics and Mercury,” Antoine van Leeuwenhoek, Vol. 89, No. 3-4, May 2006, pp. 307-315.
- G. S. Kalacheva, V. G. Gubanov, I. V. Gribovskaya, I. A. Gladchenko, G. K. Zinenko and S. V. Savitsky “Chemical Analysis of Lake Shira Water (1997-2000),” Aquatic Ecology, Vol. 36, No. 2, April 2002, pp. 123-141.
- J. H. Miller, “Experiments in Molecular Genetics,” Cold Spring Harbor Laboratory, Gold Spring Harbor, New York, 1972.
- D. J. Kushner, “Microbial Life in Extreme Environments,” Academic Press, London, New York, San Francisco, 1978.
- D. H. Bergey and J. G. Holt, “Bergey’s Manual of Determinative Bacteriology,” 9th Edition, Williams & Wilkins, Philadelphia, 1994.
- A. V. Smiryaev, S. P. Martinov and A. V. Kilchevskiy “Biometrics in Genetic and Selection of Plants” Michael Schaap, Moscow, 1992.
- E. Kussell, R. Kishony, N. Q. Balaban and S. Leibler, “Bacterial Persistence: A Model of Survival in Changing Environments,” Genetics, Vol. 169, April 2005, pp. 1807- 1814.
- A. Ventosa, J. J. Nieto and A. Oren “Biology of Moderately Halophilic Aerobic Bacteria,” Microbiology and Molecular Biology Reviews, Vol. 62, No. 2, June 1998, pp. 504-544.
- S. C. De la Rosa-Garsía, A. A. Muñoz-García, L. F. Barahona-Pérez and M. M. Gamboa-Angulo, “Antimicrobial Properties of Moderately Halotolerant Bacteria from Cenotes of the Yucatan Peninsula,” Letters Applied Microbiology, Vol. 45, No. 3, June 2007, pp. 289-294.
- N. J. Russell, “Adaptive Modifications in Membranes of Halotolerant and Halophilic Microorganisms,” Journal of Bioenergetics and Biomembranes, Vol. 21, No. 1, February 1989, pp. 93-113.
- R. H. Vreeland, “Mechanisms of Halotolerance in Microorganisms,” Critical Reviews in Microbiology, Vol. 14, No. 4, 1987, pp. 311-356.
- H. Takeyama, J. G. Burgess, S. Hiroaki, K. Sode and T. Matsunaga, “Salinity-Dependent Copy Number Increase of Marine Cyanobacterial Endogenous Plasmid,” FEMS Microbiology Letters, Vol. 90, No. 1, December 1991, pp. 95-98.
- T. I. Lobova, S. N. Zagrebelny and L. Yu. Popova “The Influence of Salt Concentrations on the Copy Number of Plasmid of pSH1 Replicating in Micrococcus sp.9,” Microbiol, Vol. 74, No. 3, May 2005, pp. 349-356.
- T. I. Lobova, Y. V. Barchatov and L. Y. Popova “Antibiotic Resistance of Heterotrophic Bacteria in Shira Lake: Natural and Anthropogenic Impacts,” Aquatic Microbial Ecology, Vol. 30, No. 1, 2002, pp. 11-18.
- M. Goñi-Urriza, M. Capdepuy, C. Arpin, N. Raymond, P. Caumette and C. Quentin, “Impact of an Urban Effluent on Antibiotic Resistance of Riverine Enterobacteriaceae and Aeromonas spp,” Applied and Environmental Microbiology, Vol. 66, No. 1, January 2000, pp. 125-132.

