Journal of Agricultural Chemistry and Environment
Vol. 2 No. 4 (2013) , Article ID: 39017 , 6 pages DOI:10.4236/jacen.2013.24011
Size fractionation and microbial community structure of soil aggregate
Jong-Shik Kim1,2*, David E. Crowley1 ![]() 1Department of Environmental Sciences, University of California, Riverside, USA;
1Department of Environmental Sciences, University of California, Riverside, USA;
*Corresponding Author: jskim@gimb.or.kr
2Gyeongbuk Institute for Marine Bioindustry, Uljin, Gyeongbuk, Republic of Korea
Copyright © 2013 Jong-Shik Kim, David E. Crowley.
This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received 28 May 2013; revised 1 July 2013; accepted 11 July 2013
ABSTRACT
The microbial community structure in various microaggregates in a loamy sand soil was investigated. The microaggregates were separated into outer and inner aggregates using a series of soil washes. Further physical fractionation of inner aggregates was achieved by separation into coarse and fine sand as macroaggregate fractions, coarse and fine silt as microaggregate fractions, and clay. Research on microbial communities and soil microaggregates can aid in our understanding of soil microhabitats and microorganisms in soil structures, with applications that may contribute to increasing crop production and maintaining sustainable agriculture. In order to study the microbial community structure of aggregates, polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) was performed using 16S rRNA genes. The PCR-DGGE of Bacteria, Actinomycetes and Archaea showed divergent results between the different aggregate fractions. The results showed that the bacterial community structure was highly similar between bulk soil and clay; the inner aggregate community structure of Actinomycetes was closely related between coarse and fine sand and coarse silt, and the Archaea community structure of outer and inner aggregates was more similar than that of total bacteria or Actinomycetes.
Keywords: Soil Aggregate; Microbial Community; Fractionation
1. INTRODUCTION
Microaggregates (diameter < 250 m) are very stable in water, whereas macroaggregates (diameter > 250 m) are easily broken down into microaggregates, coarse particles, and small pieces of organic debris in aquatic environments. Tisdall and Oades [1] demonstrated that roots and fungal hyphae are predominantly responsible for stabilizing macroaggregates. Whereas macroaggregation is also controlled by soil management practices (e.g., crop rotation and tillage), microaggregation is independent of management practices and depends on the persistence of organic binding agents. Organic binding agents include microbial polysaccharides that essentially glue fine mineral particles together [2].
Studies of soil aggregates have employed numerous separation methods, with varying degrees of success [3]. The aggregate hierarchy of Oxisol soils is drastically different from those of Alfisols and Mollisols, and organic matter (OM) content is one of the primary determinants of soil aggregation [4]. Therefore, the study of soil aggregates can be largely dependent on the nature and content of OM. Highly productive agricultural soils are significantly aided by stable soil structure and aggregate properties. The microbial metabolic products in soil have a greater impact on soil structure than does the sheer number of microbial species. Furthermore, fungi have been found to be involved in the binding of larger soil particles and bacteria in the stabilization of microaggregates and clay particles [5]. Bacteria play a major role in the formation and stabilization of microaggregates. The capsule surrounding many bacteria, especially Gramnegative bacteria, is composed of polysaccharides. This polysaccharide layer causes clay particles to physically adhere to the bacteria, and together with polyphenols attracted by ionic charges, protects the polysaccharide from microbial attack. This collection of clay particles and bacteria forms a microaggregate of about 20 μm in diameter. Additionally, saprotrophic fungal hyphae can grow between these microaggregates and continue the accumulation of material to produce larger aggregates, possibly by cation bridges between the hyphal polysaccharides and the clay particles [1,2].
The majority of microbial studies have been conducted on homogenized bulk soil samples, a practice which averages localized heterogeneity and does little to reveal the spatial origins of detected strains [6]. Many important microbial processes occur in spatially segregated microhabitats, and therefore improving our knowledge of the microbial community structure at the aggregate level deserves further investigation. Mendes and Bottomley [7] reported that microbial mineralization and enzyme activity vary among different sizes of soil aggregates, and several studies have explored the microbial community composition of aggregate fractions using cultivationbased and molecular techniques [6,8-13].
In general, the proportion of bacteria and fungi within a soil varies with aggregate size, as a greater proportion of bacteria are associated with microaggregates and a lesser proportion with macroaggregates [14]. Through the use of microarray techniques, Kim et al. [15] suggested that the ephemeral nature of soil aggregates in desert agricultural soils may reduce differences in the spatial distribution of bacterial populations when compared to those in soils with more stable aggregates. Ranjar et al. [10,11] utilized the genetic fingerprinting approach, based on ribosomal gene analysis, to compare the genetic structure of bacteria located in different size classes of aggregates. By using fractionated soil, a more accurate view of the bacterial composition of soil was obtained. Other researchers have used molecular methods, such as denaturing gradient gel electrophoresis (DGGE) [16], terminal restriction fragment length polymorphism (T-RFLP) [13,17], and cloning and sequence analysis of the 16S rRNA gene [6], to elucidate the relationship between aggregate fractions and microbial community structure in soils.
In this study, we performed soil fractionation to evaluate the community structure of Bacteria and Archaea in various microenvironments. We measured the structure of Bacterial, Actinomycetes and Archaea communities associated with the aggregate size fractions of a desert agricultural soil.
2. MATERIALS AND METHODS
2.1. Size Fractionation of Soil Aggregates
Nine 10-cm soil cores were systematically collected from fields planted with lettuce in the Imperial Valley, California, as explained previously [15]. The samples were immediately transferred to our laboratory in ice coolers and then stored at 4˚C for 72 h before processing. The Imperial soil is a loamy sand (fine, smectitic, calcareous, hyperthermic Vertic Torrifluvents) containing 79.5% sand, 6.5% silt, and 14.0% clay.
Prior to aggregate fractionation, the soil samples were passed through a 2-mm sieve using a Tyler Ro-Tap Sieve Shaker (Mentor, OH, USA) and then pooled. The soil microenvironments were fractionated as described previously [11]. The entire fractionation procedure was performed twice on 30 g of soil. The method involved 15 successive soil washes in a sterile 0.8% NaCl solution using a rotary shaker (100 rotation∙min−1) for 1 min. The supernatants were then pooled and centrifuged at 9800 × g for 20 min. The pellet (outer fraction) was resuspended in 50 mL of 0.8% NaCl solution, and the remaining washed soil (inner fraction) was further fractionated into five separate particle size fractions as described previously [9]. The inner fraction was divided into coarse and fine sand fractions (2000 - 250 µm and 250 - 53 µm, respectively), coarse and fine silt fractions (53 - 20 µm and 20 - 2 µm, respectively), and dispersible clay fractions (<2 µm). The selected chemical properties of the aggregate fractions are shown in Table 1.
To quantify the fraction of various aggregate sizes, an AMRAY 3200 (AMRAY Inc., Bedford, MA) scanning electron microscope using a backscatter electron detector was used to obtain images of the fractionation of soil aggregates. Scanning electron microscopy (SEM) indicated that the aggregate separation was well fractionated. The sample left after being washed was fractionated as the inner soil portion, which consisted of coarse and fine sand fractions for the macroaggregates, and coarse and fine silt fractions for the microaggregates (Figure 1). After 15 consecutive washes, the outer aggregate portions were collected and the outer part was dried and observed as a photograph because direct observations were insufficient for describing the true characteristics of the outer portion.
2.2. Denaturing Gradient Gel Electrophoresis
The DNA of soil aggregate fractions was extracted using the Fast DNA Spin Kit for Soil (Bio 101, QBio
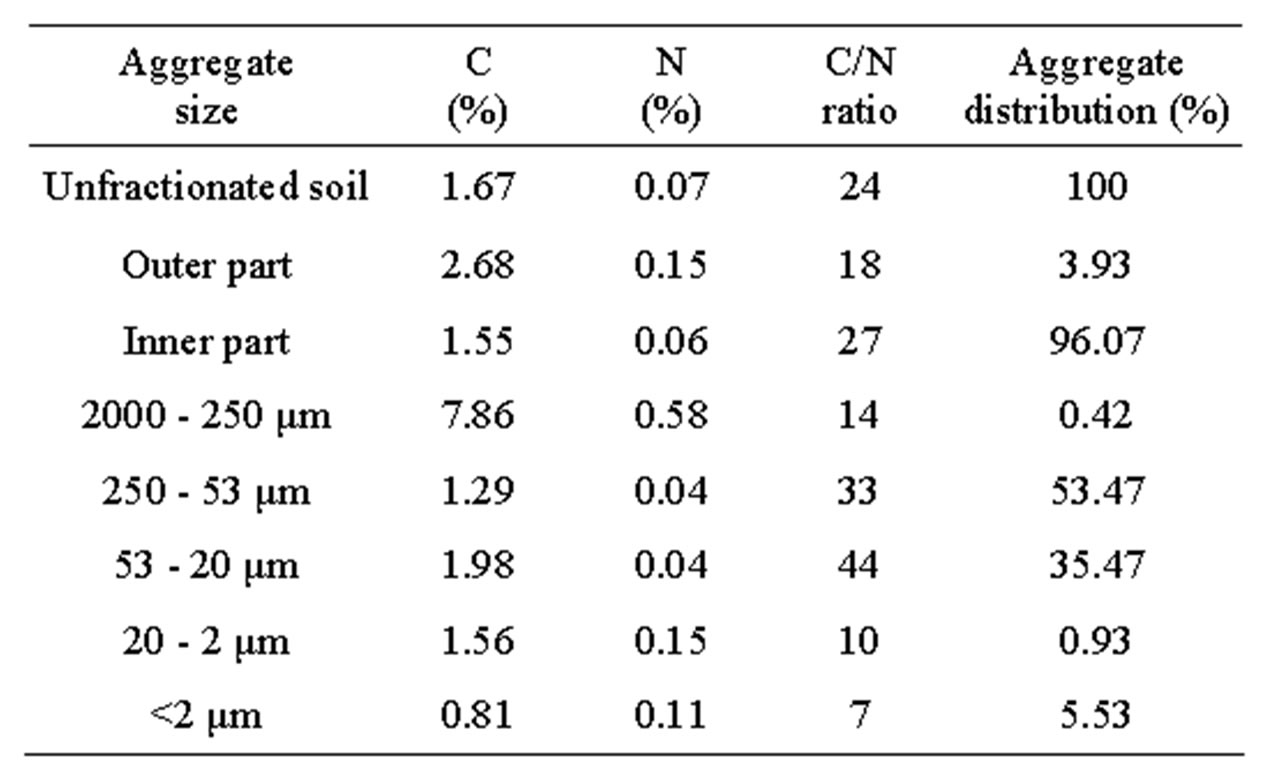
Table 1. Representative properties of unfractionated soil and aggregate fractions.
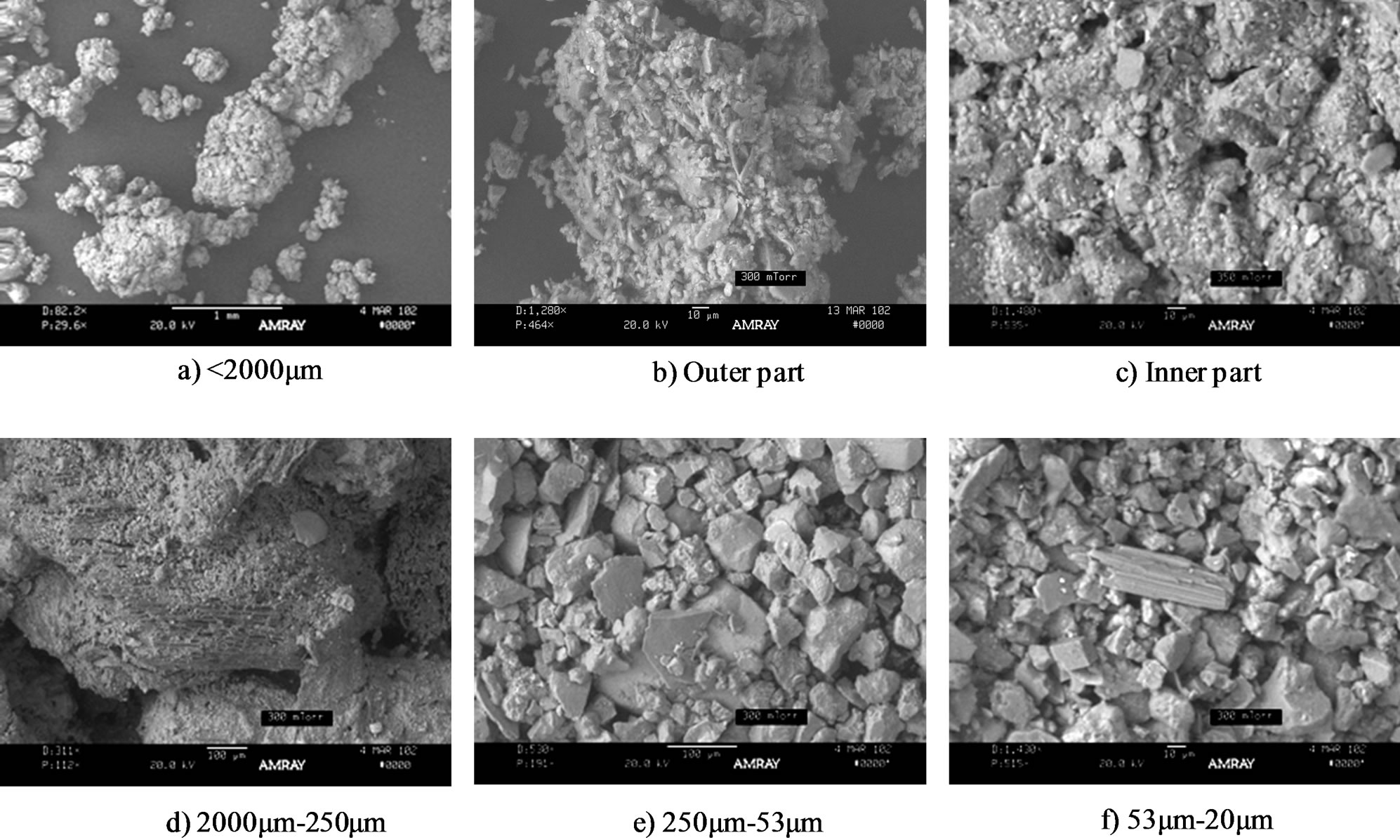
Figure 1. Scanning electron micrograph of soil aggregate fractions.
gene, Vista, CA, USA) as described in detail [15]. We performed PCR of DNA using 16S rRNA genes and DGGE primers from Invitrogen (Carlsbad, CA, USA) (Table 2). The PCR conditions were 5 min at 95˚C, 30 cycles of 30 s at 94˚C, 30 s at 50 or 55˚C, 1 min at 72˚C, and a final extension for 10 min at 72˚C. The PCR products were examined using DGGE. All 16S rRNA gene PCR forward primers were utilized with the universal reverse primer r518 (with GC-clamp). The gels were prepared as described [18]. Electrophoresis was performed for 13 hours at a constant voltage of 50 V. Staining was performed with ethidium bromide and the gels were scanned using Gel-doc and then analyzed using Quantity One (Bio-Rad, Hercules, CA, USA). On the basis of the DGGE profile, community similarities were analyzed using cluster analysis (Minitab, State College, PA, USA). Similarities in community structure quantified by cluster analysis were determined using the single linkage method with Euclidean distance measurements for the determination of differences between clusters.
3. RESULTS AND DISCUSSION
The inner fraction was a combination of macroaggregates, microaggregates, and smaller sizes. Previously, Tisdall and Oades [1] utilized SEM in the analysis of 53 - 250 μm, 90 - 250 μm, and 20 - 90 μm fractions of an Alfisol. The fine silt and clay fractions were difficult to observe in detail due to the limited resolution of SEM. The three-dimensional arrangement of particles identified by SEM indicated the following: 1) macroaggregates, coarse sand (0.42% for distribution, 7.86 for total carbon; 2) microaggregates, fine sand (53.47% for distribution, 1.29 for total carbon); 3) microaggregates, coarse silt
(35.47% for distribution, 1.98 for total carbon); 4) fine silt (1.56 for total carbon); and 5) clay (0.81 for total carbon). The microaggregates in our soil (Entisol) contained 89.94% fine sand and silt, which explained the carbon content of this size fraction. The coarse silt fraction contained predominantly quartz grains (Figure 1).
While the distribution of fine sand and coarse silt was very widespread (found in 88.9% of the aggregates), the cation exchange capacity (CEC) was not low (33.19 cmolc/kg). The carbon content of clay (%) was very low (0.81%). Christensen (1996) reported that clay generally accounts for >50% of the carbon in some soils, with clay and silt (<20 μm) together accounting for >90%. The proportion of carbon in carbohydrates generally increases with decreasing particle size and a greater microbial input to clay than to silt which is indicated by hexose to pentose ratios. Cultivation has been found to induce shifts in soil carbon composition from coarser to finer size fractions [19]. Soil fractionation methods vary widely, making a direct comparison of results difficult. The loss of carbon is due to the mineralization and transfer of soil organic matter (SOM) to siltand clay-sized particles. As indicated in the review [20], soil aggregation increased as much as 92% in an Entisol under notillage cultivation.
The microbial community structure of different soil aggregate sizes was assessed by PCR-DGGE using 16S rRNA gene primers (Figure 2). Bacteria and Actinomycetes populations were not highly related between the outer and inner aggregate regions, whereas Archaea populations had a high degree of homology with 96.3% similarity (Figure 2).
In bacterial composition, the outer fraction was clustered with the 20 - 2 μm fraction (94.4% similarity). The
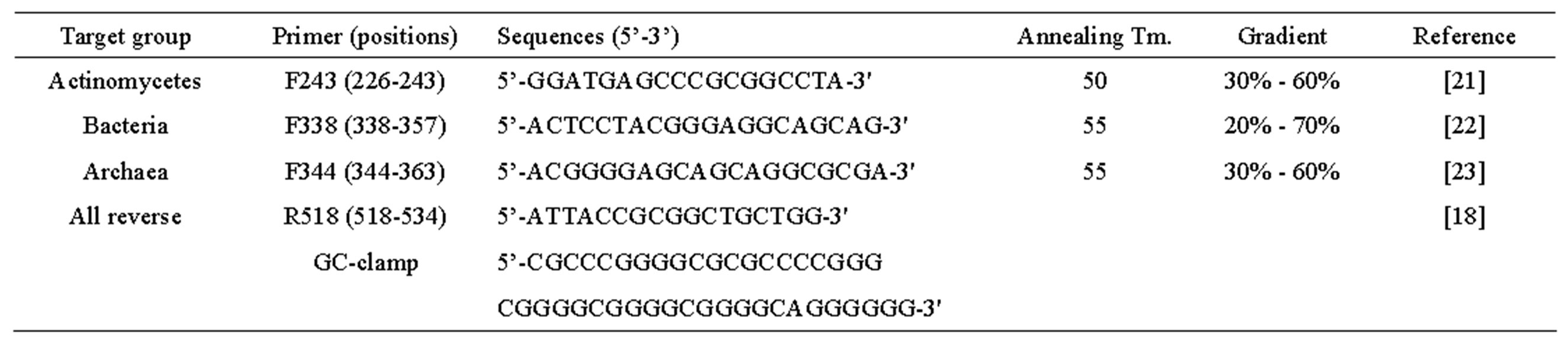
Table 2. Primers and conditions used in PCR-DGGE experiments.
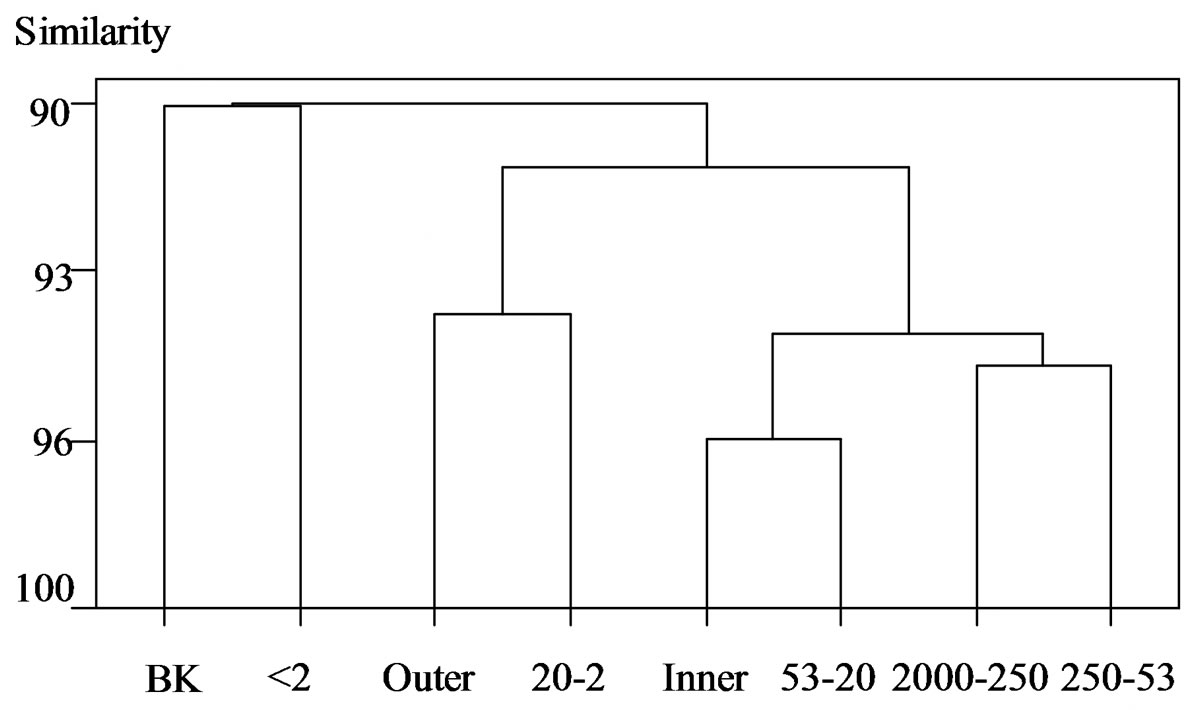
Figure 2.(a) Bacteria.
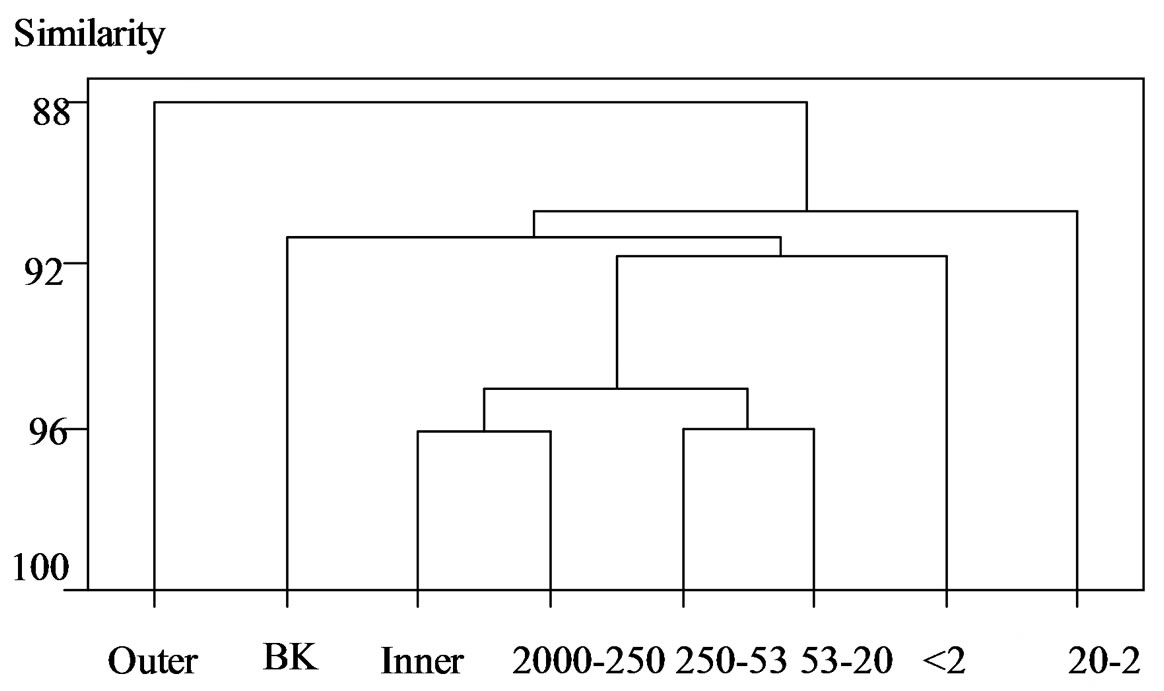
Figure 2 .(b) Actinomycetes
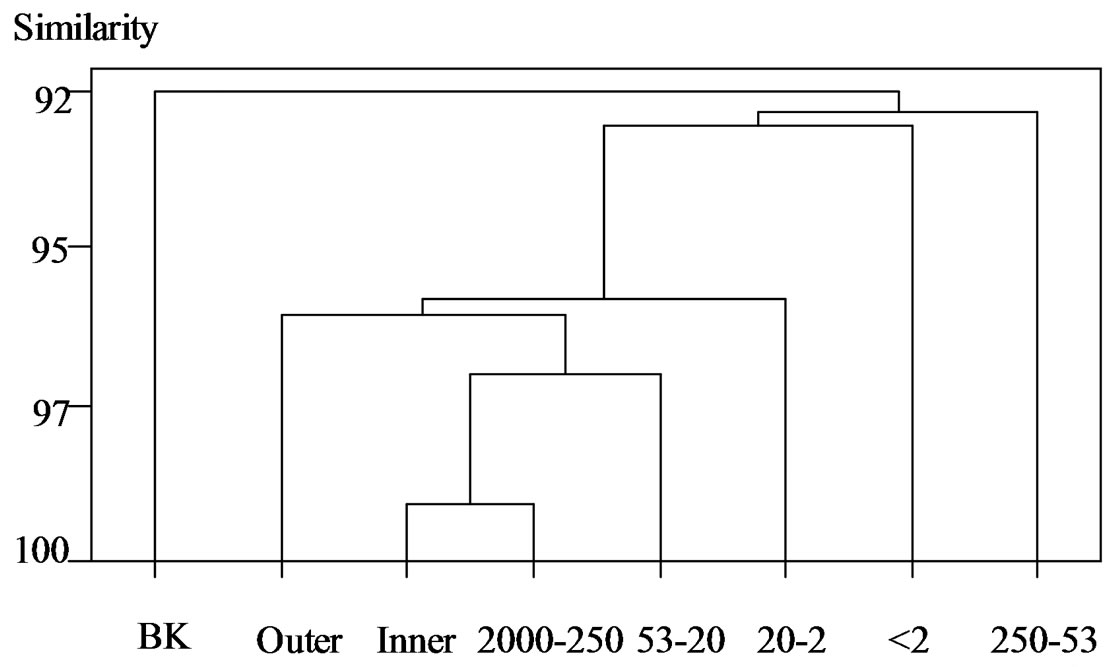
Figure 2 .(c) Archaea
Figure 2. Cluster analysis of DGGE banding patterns for (a) Bacteria, (b) Actinomycetes, and (c) Archaea within the inner and outer aggregate fractions. BK, unfractionated soil; Outer, outer fraction; Inner, inner fraction; 2000 - 250, 2000 - 250 μm; 250 - 53, 250 - 53 μm; 53 - 20, 53 - 20 μm; 20 - 2, 20 - 2 μm; <2, <2 μm.
inner fraction was clustered with the coarse silt fraction. There was a 95.4% similarity between the coarse and fine sand fractions; whereas the clay fraction showed little similarity between the outer and inner fractions. Ranjard et al. [11] showed similarities among the coarse and fine sand fractions and the coarse and fine silt fractions. More specifically Ranjard et al. [11] found that the outer fraction was similar to clay, whereas the present data show a similarity to the fine silt fraction. Ranjard et al. [11] also found the inner fraction and bulk soil to be located in the same cluster, whereas the present data show the clay and bulk soil in the same cluster, similar to the findings [16]. Ranjard et al. [11] analyzed their data using rRNA Intergenic Spacer Analysis (RISA) and DGGE analysis. Kandeler et al. [16] showed that bulk soil was clustered more closely with the clay than with other fractions, and silt (63 - 2 μm) and fine sand were closer than coarse sand. In our data, bulk soil was clustered closer to coarse sand and fine sand than to coarse silt and fine silt. These differences may be due to different separation methods of soil aggregates and different soil types [9,17].
In Actinomycetes, there was a difference in clustering between the outer and inner fractions. The composition of coarse and fine sand and coarse silt fractions was close to the inner fraction, while small particles (fine silt and clay) were far from the outer and inner fractions. The inner fraction cluster was related to coarse sand (macroaggregates), followed by fine sand and coarse silt fractions. Therefore, the Actinomycetes composition of the inner fraction was more similar to macroaggregates (coarse sand) than to microaggregates (fine silt and clay). In the inner fraction, the distribution of Actinomycetes overlapped with coarse and fine sand and coarse silt.
In Archaea, there was 95% similarity between the outer and inner fractions. The cluster of the inner fraction and coarse sand had similarities of 99% and 97% with coarse silt, 96% with the outer fraction, and 95.5% with fine silt. However there was not a high similarity with either fine sand or clay. Nonetheless, the main cluster and the two particle sizes did not have a low similarity. That is, the outer and inner fractions were very similar in terms of species composition and they may have a similar species composition to those of coarse sand and coarse silt. To our knowledge, this work is the first DGGE analysis of Archaea in soil aggregates. Clay and fine sand may have specific compositions. That is, Archaea may be in between microaggregates, or Archaea may use the aggregates as habitat including microaggregates, which are composed of a specific composition of clay and fine sand.
In conclusion, in Bacteria, the DGGE separation of 16S rRNA gene fragments revealed that the microbial diversity was predominantly within the clay fraction. Actinomycetes was predominantly in the outer fraction. In the inner fraction Actinomycetes was clustered with coarse and fine sand and coarse silt, and Archaea was clustered with coarse and fine sands.
4. ACKNOWLEDGEMENTS
The authors are grateful to Inmaculada Lebron for the SEM analysis.
REFERENCES
- Tisdall, J.M. and Oades, J.M. (1982) Organic matter and water-stable aggregates in soils. Journal of Soil Science, 33, 141-163. http://dx.doi.org/10.1111/j.1365-2389.1982.tb01755.x
- Tisdall, J.M. (1996) Formation of soil aggregates and accumulation of soil organic matter. In: Carter, M.R. and Stewart, B.A., Eds., Structure and Organic Matter Storage in Agricultural Soils, Boca Raton, FL Lewis.
- Ranjard, L. and Richaume, A. (2001) Quantitative and qualitative microscale distribution of bacteria in soil. Research in Microbiology, 152, 707-716. http://dx.doi.org/10.1016/S0923-2508(01)01251-7
- Oades, J.M. and Waters, A.G. (1991) Aggregate hierarchy in soils. Australian Journal of Soil Research, 29, 815- 828. http://dx.doi.org/10.1071/SR9910815
- Lynch, J.M. and Bragg, E. (1985) Microorganisms and soil aggregate stability. Advances in Soil Science, 2, 133- 171. http://dx.doi.org/10.1007/978-1-4612-5088-3_3
- Mummey, D., Holben, W., Six, J. and Stahl, P. (2006) Spatial stratification of soil bacterial populations in aggregates of diverse soils. Microbial Ecology, 51, 404-411. http://dx.doi.org/10.1007/s00248-006-9020-5
- Mendes, I. and Bottomley, P. J. (1999) Distribution of a population of Rhizobium leguminosarum bv. trifolii among different size classes of soil aggregates. Applied and Environmental Microbiology, 64, 970-975.
- Hattori, T. and Hattori, R. (1976) The physical environment in soil microbiology an attempt to extend principles of microbiology to soil microorganisms. Critical Reviews in Microbiology, 4, 423-461. http://dx.doi.org/10.3109/10408417609102305
- Kabir, M., Chotte, J.L., Rahman, M., Bally, R. and Jocteur, M.L. (1994) Distribution of soil fractions and location of soil bacteria in a vertisol under cultivation and perennial ryegrass. Plant Soil, 163, 243-255. http://dx.doi.org/10.1007/BF00007974
- Ranjard, L., Nazaret, S., Gourbiere, F., Thioulouse, J., Linet, P. and Richaume, A. (2000a) A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints. FEMS Microbiology Ecology, 31, 107-115. http://dx.doi.org/10.1111/j.1574-6941.2000.tb00676.x
- Ranjard, L., Poly, F., Combrisson, J., Richaume, A., Gourbiere, F., Thioulouse, J. and Nazaret, S. (2000b) Heterogeneous cell density and genetic structure of bacterial pools associated with various soil microenvironments as determined by enumeration and DNA fingerprinting approach (RISA). Microbial Ecology, 39, 263-272.
- Schutter, M.E. and Dick, R.P. (2002) Microbial community profiles and activities among aggregates of winter fallow and cover-cropped soil. Soil Science Society of America Journal, 66, 142-153. http://dx.doi.org/10.2136/sssaj2002.0142
- Blackwood, C.B., Dell, C.J., Smucker, A.J.M. and Paul, E.A. (2006) Eubacterial communities in different soil macroaggregate environments and cropping systems. Soil Biology and Biochemistry, 38, 720-728. http://dx.doi.org/10.1016/j.soilbio.2005.07.006
- Monreal, C.M., Schulten, H.-R. and Kodama, H. (1997) Age, turnover and molecular diversity of soil organic matter in aggregates of a Gleysol. Canadian Journal of Soil Science, 77, 379-388. http://dx.doi.org/10.4141/S95-064
- Kim, J.S, Dungan, R.S. and Crowley, D. (2008) Microarray analysis of bacterial diversity and distribution in aggregates from a desert agricultural soil. Biology and Fertility of Soils, 44, 1003-1011. http://dx.doi.org/10.1007/s00374-008-0291-5
- Muyzer, G, De Waal, E.D. and Uitterlinden, A.G. (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Applied and Environmental Microbiology, 59, 695-700.
- Kandeler, E., Tscherko, D., Bruce, K.D., Stemmer, M., Hobbs, P.J., Bardgett, R.D. and Amelung, W. (2000) Structure and function of the soil microbial community in microhabitats of a heavy metal polluted soil. Biology and Fertility of Soils, 32, 390-400. http://dx.doi.org/10.1007/s003740000268
- Sessitsch, A., Weilharter, A., Gerzabek, M.H., Kirchmann, H. and Kandeler, E. (2001) Microbial population—In soil particle size fractions of a long-term fertilizer field experiment. Applied and Environmental Microbiology, 67, 4215-4224. http://dx.doi.org/10.1128/AEM.67.9.4215-4224.2001
- Christensen, B.T. (1996) Carbon in primary and seconddary organomineral complexes. In: Carter, M.R. and Stewart, B.A., Eds., Structure and Organic Matter Storage in Agricultural Soils, Boca Raton, FL Lewis.
- Dalal, R.C. and Bridge, B.J. (1996) Aggregation and organic matter storage in subhumid and semi-arid soils. In: Carter, M.R. and Stewart, B.A., Eds., Structure and Organic Matter Storage in Agricultural Soils, Boca Raton, FL Lewis.
- Heuer, H., Krsek, M., Baker, P., Smalla, K. and Wellington, E.M.H. (1997) Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied and Environmental Microbiology, 63, 3233-3241.
- Lane, D.J. (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E. and Goodfellow, M. Eds., Nucleic Acid Techniques in Bacterial Systematics, Wiley, New York, 115- 175.
- Raskin, L., Stromley, J.M., Rittmann, B.E. and Stahl, D.A. (1994) Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Applied and Environmental Microbiology, 60, 1232-1240.

