Food and Nutrition Sciences
Vol.5 No.6(2014), Article ID:43561,7 pages DOI:10.4236/fns.2014.56057
Occurrence of Escherichia coli O157 in Retailed-Beef and Related Meat Products in Zaria, Nigeria
Salome Y. Tafida1, Jacob K. P. Kwaga2*, Mohammed Bello2, Junaidu Kabir2, Veronica J. Umoh3, Sabo E. Yakubu3, Andrew J. Nok4
1Department of Livestock and Pest Control Services, Federal Ministry of Agriculture, Gombe, Nigeria
2Department of Veterinary Public Health and Preventive Medicine, Faculty of Veterinary Medicine, Ahmadu Bello University, Zaria, Nigeria
3Department of Microbiology, Faculty of Science, Ahmadu Bello University, Zaria, Nigeria
4Centre for Biotechnology Research and Training, Ahmadu Bello University, Zaria, Nigeria
Email: *jacobkwaga@yahoo.com
Copyright © 2014 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).
http://creativecommons.org/licenses/by/4.0/


Received 20 July 2013; revised 20 August 2013; accepted 27 August 2013
ABSTRACT
This study evaluated retailed-beef and related meat products for E. coli O157 in Zaria, Nigeria. Sample types included raw meat, “suya” (roasted meat), “balangu” (barbequed meat), “kilishi” (spiced sun dried meat) and “dambu” (shredded fried meat). A total of 182 samples were analyzed for E. coli O157. Isolates were characterized using conventional biochemical methods and Microbact 12E test kit. Susceptibilities of the isolates to 18 commonly used antimicrobial agents were determined by the disk diffusion method. The carriage of stx1 and stx2 genes was determined by PCR. Microbact confirmed 4 E. coli isolates. All isolates exhibited multiple drug resistance to the antimicrobial agents tested. An overall prevalence of 2.2% was obtained for E. coli O157. All 4 isolates of E. coli O157 were isolated from raw meat; two of which harboured the stx1 gene.
Keywords:E. coli O157; Meat Products; Multiple Drug Resistance; Food Safety

1. Introduction
Escherichia coli O157:H7 is an enterohaemorrhagic strain of the bacterium Escherichia coli and a cause of food borne illness [1] . Escherichia coli O157:H7 strains are responsible for disease in animals and man, and have emerged to be important zoonotic agents [2] . While most strains of E. coli are harmless and normally found in the intestines of mammals, these strains may produce Shiga-like toxins, which cause severe illness, and are members of a class of pathogenic E. coli known as enterohaemorrhagic Escherichia coli or EHEC. They are also referred to by their toxin producing capabilities, verocytotoxin producing E. coli (VTEC) or Shiga-like toxin producing E. coli (STEC) [3] .
Escherichia coli O157:H7 have been implicated in severe human diseases, including bloody diarrhoea (haemorrhagic colitis) and haemolytic uremic syndrome (HUS) [4] which occasionally leads to kidney failure, especially in young children and elderly people. The growth of these strains in the human intestine is known to produce a large quantity of toxins, which can cause severe damage to the lining of the intestine and other organs of the body [2] . These toxins are similar to the toxins produced by Shigella dysenteriae [3] .
The pathogenicity of E. coli O157:H7 is mainly mediated by genes which are located either on the chromosome or on the transmissible 60 MDa plasmid [5] . The main virulence markers responsible for virulence of E. coli O157 are Shiga toxins Stx1 and Stx2 or Stx2 variants encoded by stx1 and stx2 genes respectively; and two factors are encoded by the pathogenicity island, LEE (locus of enterocyte effacement)—intimin (a product of the eaeA gene) and translocated intimin receptor (tir) [3] . Moreover, the plasmid-encoded E. coli enterohaemolysin (Ehly), which has been found in many O157 strains, has been suspected to have a role in pathogenicity of the organisms [6] . Although Stx toxins are considered the major virulence factors, it appears that combination of those and other markers are required for full virulence of E. coli O157 [4] . Cattle are the main reservoir of E. coli O157 strains which are transmitted to humans through foods contaminated with faecal material [7] .
Much has been reported on this organism all over the world, but there is insufficient information about the organisms in Nigeria, especially the northern parts of the country. In the light of the foregoing, this study therefore aimed at determining the prevalence of E. coli O157 in meat and meat products, through identification and characterization of isolates using conventional biochemical methods, Microbact (Oxoid, UK) 12E Identification System, evaluating the in vitro susceptibilities of the isolates to commonly use antimicrobial agents and to determine the habourage of stx1 and stx2 genes using the polymerase chain reaction (PCR).
2. Materials and Methods
2.1. Study Area
The study was conducted in Zaria, which is situated in the centre of Northern Nigeria, located on a plateau at a height of 2200 feet above sea level [8] . It is positioned between Latitude 11˚3'N and 7˚42'E. Its climate is tropical continental characterized by cool, humid wet seasons and cold or hot dry seasons [8] .
2.2. Samples
Convenience sampling was carried out from March 2009 to March 2010 from 4 major markets in Zaria metropolis and from a local abattoir. Samples were bought in wraps as they would normally be sold to the consumer. All samples were appropriately labeled, placed in a Coleman flask on ice, transported to the laboratory and analysed within 2 h of collection. Raw meat was collected in the morning hours while the meat products were sampled in the evening based on availability.
A total of 182 samples were screened for E. coli O157 consisting of 52 raw muscle meat, 18 intestine, 15 liver, 11 rumen, 10 omasum, 10 lung, 5 heart, 3 spleen, 1 reticulum, 5 abomasum, 23 “Suya” (roasted meat), 21 “Balangu” (barbequed meat), 5 “Kilishi” (spiced sun dried meat) and 3 “Dambu” (shredded fried meat).
2.3. Isolation and Identification of E. coli O157 in Meat and Meat Products
2.3.1. Enrichment and Plating
Ten (10) g of each sample was weighed and placed in a stomacher bag followed by addition of 90 ml of modified tryptone soya broth (Oxoid, UK) supplemented with novobiocin which served as the enrichment broth. This was then homogenised in a laboratory stomacher and incubated at 37˚C for 24 h. Cefexime Rhamnose Sorbitol MacConkey agar (CR-SMAC) (Oxoid, UK) was used as the selective plating medium. A loopful of the enrichment homogenate was streaked onto the CR-SMAC and incubated at 37˚C for 24 h.
Colonies that appeared straw coloured on CR-SMAC indicative of non-sorbitol fermenting organisms were picked and stored on nutrient agar at 4˚C, pending further characterization.
2.3.2. Biochemical Characterization of E. coli Isolates
Prior to testing of isolates stored on nutrient agar slants, they were purified on CR-SMAC. All presumptive cultures were characterized based on standard techniques [9] . Organisms showing typical characteristics in the substrates were identified as E. coli. Typical reactions of E. coli were indole positive, methyl red positive, Voges Proskauer negative, citrate negative, H2S negative, oxidase negative, urease negative, positive for fermentation of mannitol, lactose, arabinose, rhamnose, but negative for inositol, adonitol and sorbitol.
2.4. Microbact 12E Gram-Negative Bacillus (GNB) Rapid Identification System
A 24 h culture of presumptive E. coli colonies on selective media was obtained and the test was carried out and interpreted as recommended by the manufacturer (Oxoid, UK).
A 4 digit code was then obtained which was fed into the computer identification software; which gave the probable identity of the organism tested in percentage score. The Microbact software recommends a 75% cut-off point for a probable identification. All tests that gave less than 75% were not accepted as E. coli.
2.5. Evaluation of the in Vitro Susceptibility of the Isolates to Antimicrobial Agents
All the isolates identified as E. coli O157 were tested for susceptibility to eighteen (18) antimicrobial agents with the following disc contents; tetracycline, TE (30 µg), streptomycin, S (10 µg), amoxicillin/clavulanic acid, AMC (30 µg), kanamycin, K (30 µg), ampicillin, AMP (10 µg), chloramphenicol, C (30 µg), erythromycin, E (15 µg), penicillin, P (10 IU), trimethoprim, W (5 µg), sulphamethoxazole/trimethoprim, SXT (25 µg), gentamicin, CN (10 µg), lincomycin, MY (10 µg), ciprofloxacin, CIP (5 µg), nitrofurantoin, F (50 µg), neomycin, N (10 µg), polymyxin B, PB(300 µg), methicillin, MET (10 µg), oxacillin, OX (5 µg), by the disk diffusion method described by [10] based on recommendations of CLSI [11] .
Briefly, two to three (2 - 3) colonies of the appropriate cultures were inoculated into 5 ml tryptone soy broth and incubated at 37˚C until the turbidity approximated 0.5 McFarland’s standard.
Mueller-Hinton agar plates were prepared and used according to manufacturers’ instructions. Sterile swabs were dipped into the broth culture with the excess broth drained by pressing on the inner side of the tube; and used to streak the Mueller-Hinton agar in three directions at 180˚ until the entire surface was covered. A known Staphylococcus aureus methicillin resistant strain (ATCC 33591) was used as a control strain. The plates were allowed to dry at room temperature for 10 minutes and the antimicrobial discs were dispensed into the plates using the multiple disc dispenser (Oxoid, UK). The disks were further pressed with sterile forceps to ensure complete contact with medium.
The Petri dishes were then inverted and incubated at 37˚C for 18 h. After incubation, the zones of incubation were measured to the nearest millimetre and interpreted based on interpretation of zone diameter of test culture provided by CLSI [11] .
2.6. Detection of Shiga Toxin-Encoding Genes among E. coli O157 Isolates by Multiplex Polymerase Chain Reaction
DNA Extraction
All isolates were inoculated into 5 ml tryptone soya broth (TSB) and incubated for 24 hrs at 37˚C. DNA extraction was carried out using the ZR Fungal/Bacterial DNA MiniPrep™ ZRD6005 (Zymo research, CA, USA). All protocols were followed and ultra pure DNA was eluted into 50 µl DNA elution buffer. All isolates that were identified as E. coli O157 were subjected to the PCR reaction, with the stx1 and stx2 genes being targeted. Primers used were obtained from Fermentas™, Germany and were designed based on the sequences of the stx1 and stx2 genes [12] .
The primer sequences for stx1 were forward primer 5’GAAGAGTCCGTGGGATTACG-3’ and reverse primer 5’AGCGATGCAGCTATTAATAA-3’ with an expected amplicon size of 130 bp, while the primer sequences for stx2 were forward primer 5’TTAACCACACCCACGGCAGT-3’ and reverse primer 5’GCTCTGGATGCATCTCTGGT-3’, with an expected amplicon of 346 bp.
PCR was carried out in a total volume of 25 µl containing 3 µl template DNA, 0.5 µl of the forward and reverse primers (5 mM), 2.5 µl of 10 × buffer, 0.5 µl dNTPs (5 mM), 0.5 µl (2.5 units) Taq polymerase and 1.5 µl Mgcl2; 16 µl of nuclease free water was also added. PCR was performed in a DNA thermal cycler (Applied Bio systems, Gene Amp PCR system 9700). After initial denaturation step of 5 min at 95˚C, 40 cycles of amplification were performed. Each cycle consisted of the following steps; 1 min at 95˚C (denaturation), 1 min at 55˚C (primer annealing) and 1 min at 72˚C (extension) and 72˚C for 10 min for final extension. Ten micro litres of the reaction mixture was mixed with gel loading buffer and then resolved by electrophoresis on 2% agarose gels with the 100 bp DNA ladder (Fermentas™, Germany). Negative control consisted of all contents of reaction mixture excluding template DNAwhich was substituted with 3 µl sterile water. The reaction products were visualized by staining with ethidium bromide. Image documentation was carried out with a Gene snap ultra violet transluminator machine and viewed on a computer.
3. Results
3.1. Frequency of Isolation of E. coli O157 from Retailed-Meat and Meat Products
Out of eight (8) suspected E. coli identified by the conventional biochemical methods, the Microbact Identification System confirmed 4 to be E. coli and the remaining 4 isolates as Proteus vulgaris, Acinetobacter baumanii, Citrobacter freundii and Serratia liquefaciens.
3.2. In Vitro Susceptibilities of the Isolates to 18 Antimicrobial Agents
All 4 E. coli O157 isolates tested were found to be susceptible to kanamycin, gentamicin, ciprofloxacin, nitrofurantoin and polymyxin B and resistant to ampicillin, penicillin G, trimethoprim, lincomycin, methicillin and oxacillin. One isolate was resistant to erythromycin and neomycin. All isolates exhibited multiple drug resistance (MDR) and each gave a unique antibiogram as follows: AMP, P, MET, OX, MY, AMC, N, S, TE, C, W, SXT; AMP, P, MET, OX, MY, AMC, S, TE, C, E, W, SXT; AMP, P, MET, OX, MY, AMC, S, TE, C, W, SXT and AMP, P, MET, OX, MY, SXT, with resistance to 6, 11, 12 and 12 drugs respectively.
3.3. stx1 and stx2 Detection by Polymerase Chain Reaction
Two of the 4 E. coli O157 isolates were positive for the stx1 gene (Figure 1) and none was positive for the stx2.
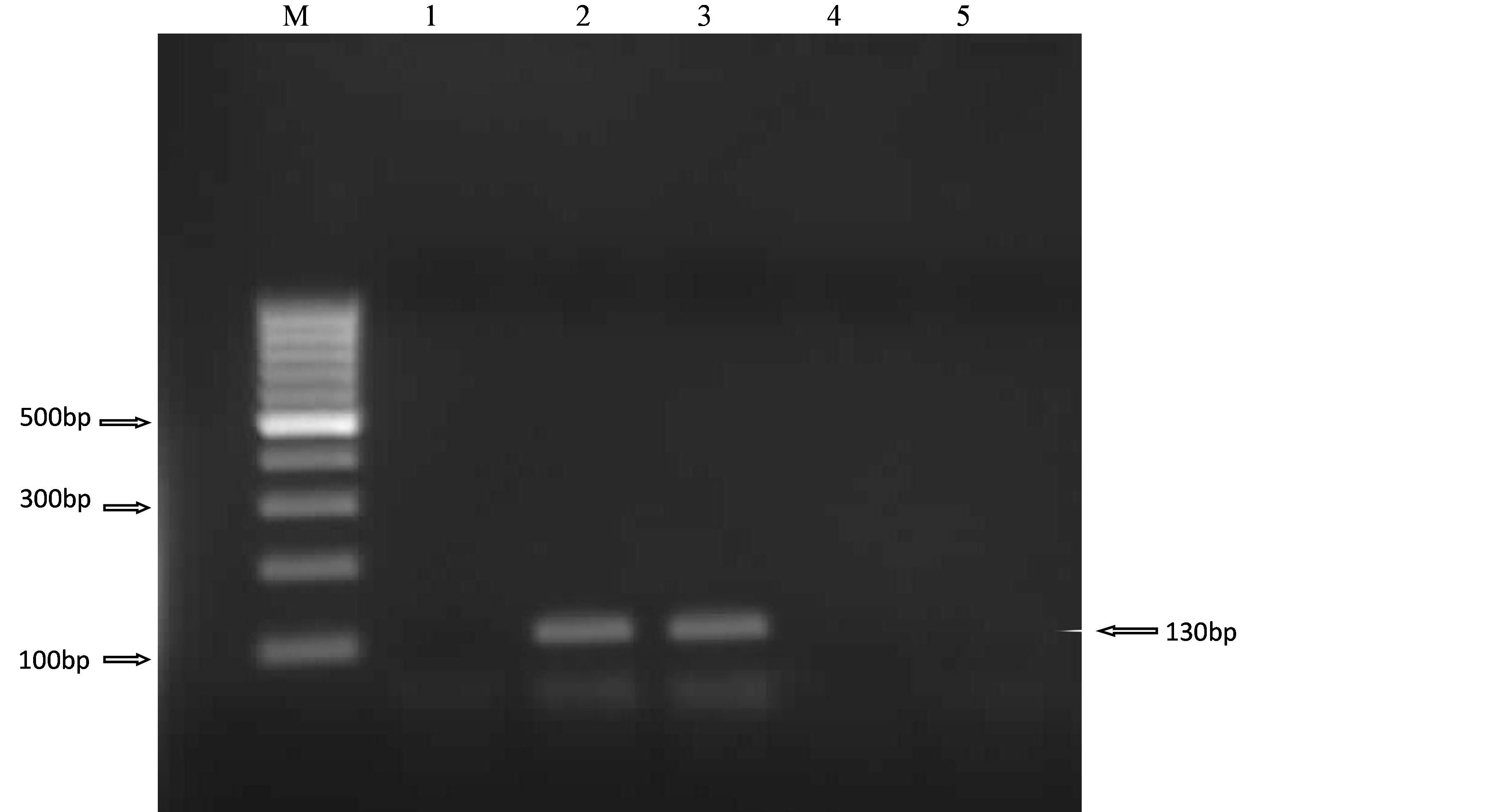
Figure 1. PCR amplification of the stx1 gene in 4 E. coli O157 isolates. Lane 1, negative control, Lanes 2 - 5, E. coli O157 isolates (L2-SMM18, L3-SMM15, L4-TW13, L5-ZNLU3) and M 100 bp ladder (Fermentas™, Germany).
4. Discussion
E. coli O157 is one of the most common agents of food borne illnesses in humans and have been isolated from beef and dairy cattle at all stages of production. The shedding of the organisms is intermittent and can be difficult to detect, although they appear to be fairly widespread throughout the bovine population [13] .
E. coli O157 was isolated from 4 (2.2%) of 130 raw meat samples analysed. This closely agrees with a previous study of [14] in which E. coli O157 was isolated from raw meat and “suya” with an isolation rate of 2.3% in Benin, Nigeria. This study also had similar results with a study in Botswana [15] , which reported a prevalence of 2.3% from fresh beef sausage. In that study, they also reported a prevalence of 5.2% in meat cubes and 3.8% from raw ground beef. A prevalence of 2% in sausages was also reported in Egypt [13] . The presence of E. coli O157 in sausage was attributed to contamination from faeces of infected animals as well as the unsatisfactory hygienic measures during manufacturing and handling [13] .
Antimicrobial resistance (AMR) has clearly reached alarming levels in human pathogens, but some fears have also been raised with regards to pathogens and commensals from animals [16] . Multiple drug resistance was observed in all the isolates of E. coli O157 tested in this study, with the E. coli O157 isolates exhibiting resistance to 6 or more antimicrobial agents. Two isolates were resistant to 12 antimicrobial agents including tetracycline, augmentin, chloramphenicol, trimethoprim, sulphamethoxazole, streptomycin and the β-lactam group.
The worldwide overuse or misuse of antimicrobials in different fields, including human medicine, veterinary medicine and agriculture, and as prophylactic supplements or growth-promoting agents in the feed of food animals, has created enormous pressure for the selection of antimicrobial resistance among bacterial pathogens and endogenous microflora [17] . However, antibiotic therapy in food animal production is increasingly coming under close scrutiny, largely because of the fear of increased levels of resistance in food-borne human pathogens, such as Salmonella, Campylobacter and Escherichia coli [18] .
The determination of antimicrobial susceptibility of clinical isolates and surveillance for resistant pathogens is often crucial for the optimal antimicrobial therapy of infected patients. This need is becoming more urgent with increasing resistance and the emergence of multidrug-resistant microorganisms [19] .
The extensive multi drug resistance observed in this study, even though among few isolates, may be suggestive of the role of an integron system operative in these microorganisms because they have been reported in gram negative genera including Salmonella and E. coli [20] . Further tests may be needed to confirm the suspicion that integrons played a role in the exhibition of the multiple drug resistance observed.
The findings of the present study ascertain that these organisms have developed resistance for routinely prescribed antimicrobial drugs and pose considerable health hazards to the consumers, unless prudent control measures are instituted.
Shiga toxin 1 (stx1) gene was detected in 2 out of the 4 E. coli O157 isolates tested by PCR; but stx 2 was not detected in any of the isolates tested. Similar occurrences were observed in studies conducted where STEC isolates carried either stx1 or stx2 genes [21] [22] . Conversely, [23] reported the detection of both stx1 and stx2 genes in majority of the isolates tested, but also detected either stx1 or stx2 genes among some isolates tested. The absence of stx1 gene in the remaining isolates may have been caused by instability of the phages carrying stx genes. Loss of stx genes in serial cultures is seen after long storage/culturing of the organism [24] .
Detection of stx1genes suggests that the strains are potentially pathogenic, thus expansion of this study to include genes encoding accessory virulence factors, such as intimin or the plasmid-encoded haemolysin [25] , may be necessary to further evaluate the significance of STEC strains in Zaria.
The ability to accurately detect and identify microorganisms that are capable of causing infectious disease has become increasingly important in environmental surveillance and clinical medicine. Rapid tests for bacterial identification might contribute to, but not replace, bacteriological culture techniques. The isolation of organisms is still needed for serotyping and determination of resistance profiles, and also for epidemiological studies. However, in a routine diagnosis, it should be considered that a large number of samples may be processed in a relatively short period of time using the PCR assay [26] .
The findings of this study have implications on the public health burden of E. coli O157 in Zaria and possibly Nigeria as a whole. The isolation of this pathogen suggests that contaminated meat and meat products are sold to consumers and thus exposing them to food borne hazards. Furthermore, it shows that food hygiene and handling practices remain a great challenge in a developing country such as Nigeria.
The findings in this study may also be an indication of the poor sanitary environment under which animals are slaughtered, transported, processed and sold. This is in agreement with earlier work [27] that reported poor sanitary conditions of slaughter environment in the study area and contamination of carcasses due to use of nonpotable water.
It is important to realize that management of meat safety risks should be based on an integrated effort and approach that applies to all sectors, from the producer through the processor, distributor, packer, retailer, food monitoring authorities and consumer. The report of the presence of E. coli O157 in this study should prompt relevant authorities to bear in mind that most food borne illnesses may be due to mishandling of foods, while animal-borne pathogens introduced into the environment lead to illness associated with consumption of contaminated meat. Thus, consumer education and environmental pollution issues should be major targets in efforts to improve meat and food safety.
This study was funded by the University Board of Research, Ahmadu Bello University Zaria, Nigeria.
References
- Karch, H., Tarr, P. and Bielazewska, M. (2005) Enterohaemorrhagic Escherichia coli in Human Medicine. International Journal of Medical Microbiology, 295, 405-418. http://dx.doi.org/10.1016/j.ijmm.2005.06.009
- Nataro, J.P. and Kaper, J.B. (1998) Diarrheagenic Escherichia coli. Clinical Microbiology Reviews, 11, 142-201.
- Paton, J.C. and Paton, A.W. (1998) Pathogenesis and Diagnosis of Shigatoxin Producing Escherichia coli Infections. Clinical Microbiology Reviews, 11, 450-479.
- Coia, J.E. (1998) Clinical, Microbiological and Epidemiological Aspects of Escherichia coli O157 Infection. FEMS Immunology and Medical Microbiology, 20, 1-9. http://dx.doi.org/10.1016/S0928-8244(97)00105-3
- Schmidt, H., Beutin, L. and Karch, H. (1995) Molecular Analysis of the Plasmid-Encoded Hemolysin of Escherichia coli O157:H7 Strain EDL933. Infection and Immunity, 63, 1055-1061.
- Beutin, L., Montenegro, M.A., Orskov, I., Orskov, F., Prada, J., Zimmermann, S. and Stephan, R. (1995) Close Association of Verotoxin (Shiga-Like Toxin) Production with Enterohemolysin Production in Strains of Escherichia coli. Journal of Clinical Microbiology, 27, 2259-2264.
- Armstrong, G.L., Hollingsworth, J. and Morris Jr., J.G. (1996) Emerging Food Borne Pathogens: Escherichia coli O157:H7 as a Model of Entry of a New Pathogen into the Food Supply of the Developing World. Epidemiology Reviews, 18, 29-51. http://dx.doi.org/10.1093/oxfordjournals.epirev.a017914
- Mortimore, M.J. (1970) Zaria and Its Region. Annals of the Association of American Geographers, 60, 73-80.
- Barrow, G.I. and Feltham, R.K.A. (1995) Cowan and Steel’s Manual for the Identification of Medical Bacteria. Cambridge University Press, 94-163.
- Bauer, A.W., Kirby, W.M., Sherris, J.C. and Turck, M. (1966) Antibiotic Susceptibility Testing by a Standardized Single Disk Method. American Journal of Clinical Pathology, 45, 493-496.
- Clinical Laboratory Standards Institute (CLSI) (2006) Performance Standards for Antimicrobial Susceptibility Testing. Sixth Informational Supplement CLSI Document M100-516, Wayne.
- Al-Gallas, N., Ben Aissa, R., Attia Annabi, Th., Bahri, O. and Boudabous, A. (2002) Isolation and Characterization of Shiga Toxin-Producing Escherichia coli from Meat and Dairy Products. Food Microbiology, 19, 389-398. http://dx.doi.org/10.1006/fmic.2002.0488
- Abd El-Atty, N.S. and Meshref, A.M.S. (2007) Prevalence of Salmonella and E. coli O157 in Some Foods. Beni-Suef Vet. Medicine Journal, 5th. Scientific Conference, 73-78.
- Enabulele, S. and Uraih, N. (2009) Enterohaemorrhagic Escherichia coli O157:H7. Prevalence in Meat and Vegetables sold in Benin City, Nigeria. African Journal of Microbiological Research, 5, 276-279.
- Magwira, C.A., Gashe, B.A. and Collison, E.K. (2005) Prevalence and Antibiotic Resistance Profiles of Escherichia coli O157:H7 in Beef Products from Retail Outlets in Gaborone, Botswana. Journal of Food Protection, 68, 403-406.
- Catry, B., Laevens, H., Devriese, L.A., Opsomer, G. and De Kruif, A. (2003) Antimicrobial Resistance in Livestock. Journal of Veterinary Pharmacology and Therapeutics, 26, 81-93. http://dx.doi.org/10.1046/j.1365-2885.2003.00463.x
- World Health Organization (WHO) (2000) WHO Global Principles for the Containment of Antimicrobial Resistance in Animals Intended for Food. Report of a WHO Consultation, WHO Department of Communicable Disease Surveillance and Response, Geneva, 1-7.
- Threlfall, E.J., Angulo, F.J. and Wall, P.G. (1998) Ciprofloxacin-Resistant Salmonella Typhimurium DT104. Veterinary Record, 142, 255.
- Fluit, A.C., Jones, M.E., Schmitz, F.J., Acar, J., Gupta, R. and Verhoef, J. (2000) For the SENTRY Participants Group. Bacteremia in European Hospitals, Incidence and Antimicrobial Susceptibility. Clinical Infectious Diseases, 30, 454- 460. http://dx.doi.org/10.1086/313710
- Fluit, A.C. and Schmitz, F.J. (1999) Class 1 Integrons, Gene Cassettes, Mobility and Epidemiology. European Journal of Clinical Microbiology and Infectious Diseases, 18, 761-770. http://dx.doi.org/10.1007/s100960050398
- Schmidt, H., Geitz, C., Phillips, I.T., Matthias, F. and Karch, H. (1999) Non-O157 Pathogenic Shiga Toxin Producing Escherichia coli: Phenotypic and Genetic Profiling of Virulence Traits and Evidence for Clonality. Journal of Infectious Diseases, 179, 115-123. http://dx.doi.org/10.1086/314537
- Pradel, N., Boukhors, K., Bertin, Y., Forestier, C., Martin, C. and Livrelli, V. (2001) Heterogeneity of Shiga ToxinProducing Escherichia coli Strains Isolated from Hemolytic-Uremic Syndrome Patients, Cattle and Food Samples in Central France. Applied and Environmental Microbiology, 67, 2460-2468. http://dx.doi.org/10.1128/AEM.67.6.2460-2468.2001
- Adwan, M.G. and Adwan, M.K. (2004) Isolation of Shiga Toxigenic Escherichia coli from Raw Beef in Palestine. International Journal of Food Microbiology, 97, 81-84. http://dx.doi.org/10.1016/j.ijfoodmicro.2004.03.032
- Karch, H., Meyer, T., Rüssmann, H. and Heesemann, J. (1992) Frequent Loss of Shiga-Like Toxin Genes in Clinical Isolates of Escherichia coli upon Subcultivation. Infection and Immunity, 60, 3464-3467.
- Arthur, T.M., Barkocy-Gallagher, G.A., Rivera-Betancourt, M. and Koohmaraie, M. (2002) Prevalence and Characterization of Non-O157 Shiga Toxin-Producing Escherichia coli on Carcasses in Commercial Beef Cattle Processing Plants. Applied and Environmental Microbiology, 68, 4847-4852. http://dx.doi.org/10.1128/AEM.68.10.4847-4852.2002
- Cortez, A.L.L., Carvalho, C.F.B., Ikuno, A.A., Bu¨rger, K.P. and Vidal-Martins, A.M.C. (2006) Identification of Salmonella spp. Isolates from Chicken Abattoirs by Multiplex-PCR. Research in Veteterinary Science, 81, 40-344. http://dx.doi.org/10.1016/j.rvsc.2006.03.006
NOTES

*Corresponding author.

