American Journal of Plant Sciences
Vol.06 No.18(2015), Article ID:60993,54 pages
10.4236/ajps.2015.618275
Nitrogen Nutrition, Its Regulation and Biotechnological Approaches to Improve Crop Productivity
Mettu Madhavi Reddy, Kandasamy Ulaganathan
Centre for Plant Molecular Biology, Osmania University, Hyderabad, India
Copyright © 2015 by authors and Scientific Research Publishing Inc.
This work is licensed under the Creative Commons Attribution International License (CC BY).


Received 10 September 2015; accepted 8 November 2015; published 11 November 2015

ABSTRACT
Nitrogen is the most important macronutrient needed for plant growth and development. The availability of nitrogen in the soil fluctuates greatly in both time and space. Crop plants, except leguminous plants, depend on supply of nitrogen as fertilizers. Large quantities of nitrogen fertilizers are applied to crop plants, but only 33% of it is utilized by the plant. Plants have developed efficient mechanisms to sense the varying levels of nitrogen forms and uptake them. They also have well developed mechanisms to assimilate the incoming nitrogen immediately or translocate to different parts of the plant wherever it is needed. Maintenance of nitrogen homeostasis is essential to avoid toxicity. Apart from translocation and assimilation, plants have developed different mechanisms, nitrogen efflux; vacuolar nitrogen storage and downward transport of nitrogen from aerial parts to roots, for maintaining nitrogen homeostasis. In crop plants the “grain yield per unit of available nitrogen in the soil” is referred as the nitrogen use efficiency (NUE) for which remobilization of nitrogen, mediated by various transporters plays a crucial role. All these processes are tightly regulated by proteins and microRNA in response to both external and internal nitrogen levels, carbon status of the plant and hormones. As most crop plants are non-leguminous and depend on soil nitrogen, more production could be achieved if crop plants can be made to utilize the available nitrogen efficiently. The recent explosion of research information and the mechanisms behind nitrogen sensing, signaling, transport and utilization enables biotechnological interventions for better nitrogen nutrition of crop plants. This review discusses such possibilities in the context of recent understanding of nitrogen nutrition and the genomic revolution sweeping the crop science.
Keywords:
Nitrogen, Nitrogen Use Efficiency, Genomics, Genetic Engineering, RNA-Seq, Nitrome, microRNA

1. Introduction
Plants require many mineral elements for their growth and development. Elements like nitrogen, potassium, calcium, magnesium, phosphorus, and sulphur are required in large quantities (macronutrients), while boron, chlorine, copper, iron, manganese, molybdenum and nickel are required in smaller quantities (micro nutrients) [1] [2] . Each mineral is equally important to the plant and deficiency of one or more of these mineral elements affect the growth, development and yield of crop plants. Soil is the source of mineral nutrients for plants and for crop plants mineral nutrients are supplied in the form of fertilizers to increase the yield. Among macro nutrients, nitrogen is next to carbon in importance to plants. It is an important constituent of amino acids, proteins, enzymes, vitamins, alkaloids and some growth hormones. Though, molecular nitrogen constitutes 78% of the atmosphere, only some microbes and leguminous plants can fix atmospheric nitrogen [3] . All other plants depend on external source for nitrogen and nitrogen is a prominent limiting factor in these plants [4] [5] . By supplying nitrogen as fertilizers along with genetic improvement of crop plants, green revolution achieved substantial increase in crop productivity [6] [7] .
Large quantities of nitrogen fertilizers are applied to crops worldwide each year. In 2010-2011, 57.5 million tons of nitrogen fertilizers (57.5% of global nitrogenous fertilizer consumption) were applied to cereals. Wheat was the main crop receiving nitrogen fertilizers, with 18.1% of global uses, followed by maize with 16.8% and rice with 15.4%. Other cereals accounted for 4.8% of the world total [8] . Though, nitrogen fertilizer application is the major factor responsible for huge increase in yield of crop plants, 2/3rds of the applied nitrogen is not utilized by the plants and is lost in the soil due to leaching, surface run-off, volatilization and de nitrification by microorganisms [9] - [16] . Huge quantities of energy (1.2% of total energy used globally) is used in the synthesis of nitrogen fertilizers, which significantly contributes to the production of greenhouse gases and the leached nitrogen fertilizers are the main contributors for eutrophication process [17] - [22] .
As most crop plants are non-leguminous and depend on soil nitrogen, more production can be achieved if crop plants can be made to utilize the available nitrogen efficiently [23] - [25] . Efficient utilization of nitrogen (nitrogen use efficiency) is measured in terms of “the grain yield per unit of available nitrogen in the soil” for crop plants and “the fresh or dry matter yield per unit of available nitrogen in soil” for biomass plants. Nitrogen use efficiency depends on nitrogen uptake, translocation, assimilation, vacuolar storage and remobilization [26] [27] .
Recently, an explosion of research information on the mechanisms behind nitrogen sensing, signalling, transport and utilization have occurred. Though a number of gaps still exist, the available information indicates that it is potentially possible to make biotechnological interventions for better nitrogen nutrition of crop plants. This paper aims at discussing such possibilities taking into consideration the voluminous information generated recently and the genomic revolution that is sweeping crop science.
2. Nitrogen in Soil
Plants predominantly acquire nitrogen from soil (rhizosphere) as inorganic nitrogen (nitrate, ammonium and urea) [28] [29] . But major part of soil nitrogen is present in organic form (organic matter) and plants cannot utilize organic nitrogen directly. Microorganisms present in the soil convert organic nitrogen (proteins and amino acids) into inorganic forms through the process of mineralization. Microbial utilization of inorganic nitrogen also depletes nitrogen available to plants. The balance among mineralization, nitrification and denitrification determines nitrogen availability to plants. The soil ammonium and nitrate concentration ranges from micro molar levels to hundreds of milli molar quantities [1] [30] . The diffusion coefficient of nitrate in soil is the consequence of nitrate available to plant roots and the nitrate lost through leaching (about 30% of inorganic nitrogen) [31] [32] .
Soil ammonium and nitrate concentrations in agricultural soils are in micro molar (less than 1 milli mole) and milli molar (1 - 5 milli moles) ranges, respectively [33] - [35] . When compared to nitrate, leaching of ammonium is less which is due to the relative immobility of ammonium in soil. Urea is the predominant type of nitrogenous fertilizer applied to crop plants [36] . Natural soils usually contain lower concentrations of urea (~3 micro moles) but in the fertilized soils it is present up to 70 μM level [37] . Microbial urease enzyme converts soil urea into ammonium and CO2. In addition to fertilizer application, arginine degradation (urea cycle) and ureide catabolism (catabolism of purines) are the sources of urea to plants [1] [36] [38] [39] .
Plant roots receive nitrogen through transpiration dependent mass flow. Mass flow alone cannot meet the nitrogen demand of the roots and diffusion plays a complementary role in nitrogen uptake. Diffusion co-efficients of nitrate/ammonium and their concentration gradients determine their diffusion [1] [31] . Though the diffusion co-efficient of nitrate and ammonium in water is same, in soil it differs and is influenced by charge, ion size, soil moisture, temperature and water viscosity. Diffusion co-efficient of nitrate in soil is ca. 1 × 10−10 m2・s−4 and the diffusion co-efficient of ammonium is 10 - 100 folds less than that of nitrate [33] . The low diffusion coefficient of ammonium results in low leaching and less availability to roots for uptake [32] [40] [41] .
3. Sensing Nitrogen Levels in Soil
As soil nitrogen levels vary greatly, to cope with this versatile sensing and adaptive mechanisms were evolved in plants (Table 1) [42] - [50] . The very first thing in nitrogen uptake is to sense the levels of nitrogen in soil and then activation of appropriate transport systems for uptake of nitrogen from soil [51] .
3.1. Sensing of Soil Ammonium Levels
As ammonium concentration in the soil is highly variable, plants need to sense the level of ammonium and tightly regulate the
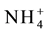 uptake and transport [52] - [54] . Plants have elaborate sensing and signaling mechanisms for ammonium in which ammonium transporting proteins AMT1; 1 and AMT1; 3 are involved [55] - [57] . The initial sensing process and the proteins involved in this are still not known. But it is clear that the unknown initial sensing mechanism changes the phosphorylation state of AMT1; 1 and AMT1; 3 by inducing the specific kinases involved in this [58] . They are expressed in the root hair rhizodermal cells [57] [59] . The trimeric form of these proteins has been shown to function as channels through which
uptake and transport [52] - [54] . Plants have elaborate sensing and signaling mechanisms for ammonium in which ammonium transporting proteins AMT1; 1 and AMT1; 3 are involved [55] - [57] . The initial sensing process and the proteins involved in this are still not known. But it is clear that the unknown initial sensing mechanism changes the phosphorylation state of AMT1; 1 and AMT1; 3 by inducing the specific kinases involved in this [58] . They are expressed in the root hair rhizodermal cells [57] [59] . The trimeric form of these proteins has been shown to function as channels through which
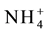 is transported [60] - [62] . Opening and closing of these channels is controlled by phosphorylation of conserved threonine residues [51] [56] [63] [64] . In AMT1; 1 protein Thr460 located in the conserved C-terminal region is the target for phosphorylation [55] [65] - [67] . In the un-phosphorylated state, the channel is open and amenable for transport of
is transported [60] - [62] . Opening and closing of these channels is controlled by phosphorylation of conserved threonine residues [51] [56] [63] [64] . In AMT1; 1 protein Thr460 located in the conserved C-terminal region is the target for phosphorylation [55] [65] - [67] . In the un-phosphorylated state, the channel is open and amenable for transport of
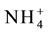 and when the concentration of ammonium is high in the soil it leads to phosphorylation of the AMT proteins which changes the conformation of these proteins leading to channel closure [57] [65] . Though the concentration dependent phosphorylation and closure of the channel has been proved, little is known about reversing this i.e. dephosphorylation and opening of the channel [68] .
and when the concentration of ammonium is high in the soil it leads to phosphorylation of the AMT proteins which changes the conformation of these proteins leading to channel closure [57] [65] . Though the concentration dependent phosphorylation and closure of the channel has been proved, little is known about reversing this i.e. dephosphorylation and opening of the channel [68] .
The involvement of AMT1; 3 in sensing ammonium and signaling is shown by its action on local ammonium concentration mediated lateral root growth. AMT1; 3 protein acts as a sensor but the signaling pathway is different [57] . Though a conserved threonine residue is present at 464th position, there is no evidence to support phosphorylation of this residue. High ammonium levels induce the oligomerization of AMT1; 3 and internalization of the oligomers [69] . This works as a shutoff mechanism to prevent ammonium toxicity. The internalization of AMT1; 3 oligomers is mediated by the clathrin-dependent and the membrane microdomain-associated endocytic pathways [70] .
3.2. Sensing of Soil Nitrate Levels
In plants, sensing of soil nitrogen concentration is mediated by the nitrate transporting NRT1.1 and NRT2.1 proteins [47] [71] - [77] . Wide variations in external nitrate levels are sensed by the dual affinity transporter NRT1.1. Changes in soil nitrate concentrations regulate the phosphorylation level of NRT1.1. It is phosphorylated (at Thr101) by the protein kinase CIPK23 when the nitrate concentration is low. Higher nitrate concentration prohibits phosphorylation of NRT1.1 and the dephosphorylated NRT1.1 switches from high to low affinity mode [73] [78] [79] . The calcium binding CBL9 protein activates CIPK23 kinase but the exact mechanism is
Table 1. Proteins involved in sensing soil nitrogen levels.
not known [80] [81] . Once activated, CIPK23 directly interacts with NRT1.1 in the plasma membrane [82] [73] . CIPK8 kinase mediates the high-level primary nitrate response under high nitrate conditions. Nitrate induced CIPK8 acts at low affinity phase. Like CIPK23, CIPK8 has been suggested to be activated by another CBL protein but no further evidence is available now [78] . Based on the ability of NRT2.1 to modulate the root system architecture according to the available nutrients, NRT2.1 has been predicted to sense nitrogen levels. Further investigations are needed to understand the mechanism behind this sensing [48] [83] .
4. Sensing Internal Nitrogen Levels
Plants possess multiple mechanisms for sensing the internal nitrogen levels. These include internal nitrogen sensing by nitrogen transporters, transcription factors, PII proteins, general amino acid control-based sensing, cytokinins and His-Asp phosphorelay-based sensing, inotropic glutamate receptors-based sensing and sensing by NIT2 proteins (Table 2) [84] .
4.1. Nitrogen Transporters as Internal Nitrogen Sensor
Nitrate transporting NRT1.1, NRT2.1and ammonium transporting AMT1; 3 proteins also acts as internal nitrogen sensors [47] [57] .
4.2. Sensing of Internal Nitrogen Levels by Transcription Factor Proteins
Nitrogen regulated transcription factors, MADS box transcription factor (ANR1), NLP7, LBD37/38/39 proteins, SPL9 and Dof1 are associated with internal nitrate sensing [85] - [87] . The localized stimulatory effect of nitrate on root growth requires ANR1 expression which is induced by nitrate. NRT1.1 acts upstream of ANR1 in this effect [48] [88] .
Nitrate induction of many nitrate uptake and assimilatory genes is mediated by NLP7 protein which is highly expressed in vascular tissues, emerging secondary roots and root hairs. In the presence of nitrate NLP7 is retained or imported into the nucleus [89] . NLP7 is also expressed in stomata, and has a role in controlling stomata opening. The reduced expression or mutation in the NLP gene resulted in impaired nitrate uptake and assimilation. NLP7 protein has been found to negatively regulate nitrogen starvation inducible genes and positively regulate primary nitrate response (nitrate inducible) [78] [85] . Many genes associated with nitrate metabolism have NLP binding sites and co-coordinately regulated by NLP7 which helps the plant to adopt to different nitrogen levels [89] . Expression of LBD 37/38/39 genes was up regulated by nitrate, ammonium and glutamine while urea has no effect. LBD 37/38/39 transcriptionally regulates nitrate assimilatory genes, nitrate transporter
Table 2. Proteins involved in sensing internal nitrogen levels.
genes, and the internal N sensing PII protein genes [86] . Under low nitrogen concentrations, the growth and assimilation of plants is improved by over expression of DOF1 transcription factor [90] [91] . All these transcription factors act as nitrate signaling components and may have a role in internal nitrate sensing.
4.3. PII Protein-Based Sensing
Though PII proteins are known to regulate the nitrogen metabolism in bacteria, archaea and some of the eukaryotes, their function in plants is not understood and it has been suggested to play a role in sensing internal carbon/nitrogen ratio [92] - [96] . Plant PII proteins are nuclear encoded chloroplast proteins belonging to GlnB subfamily [97] . They are homotrimers of 12 - 13 KD and contain three conserved Loops (C loop, T loop and B loop). Though the trimeric PII protein has been found to regulate nitrogen metabolism at transcriptional level in bacteria and post-translational level in cyanobacteria, very little is known in plants [98] [99] . Both in bacteria and cyanobacteria PII proteins sense internal nitrogen status but the mechanism of sensing is different. Bacterial sensing of cellular nitrogen status involves covalent modification (uridylylation) of Try51 while cyanobacterial sensing of cellular nitrogen status involves phosphorylation of Ser-49 residue [100] - [102] . In plants, Tyr51 is replaced with Phenylalanine and the Ser52 residue is conserved [84] [103] . But, phosphorylation of the plant PII protein has not been proven yet [104] .
The following research findings suggest a role for PII proteins in internal nitrogen sensing: Over expression of plant PII protein impaired the ability to sense glutamine when grown under high sucrose and glutamine as nitrogen source. PII protein was found to interact with the arginine biosynthetic enzyme, N-acetyl glutamate kinase. No equivalent of the two component NRI/NRII system (bacterial) or the NtcA system (cyanobacterial) has been found in plants. The mechanism by which PII proteins convert the nitrogen depletion signal into transcription control in plants remains to be elucidated [84] [103] [105] - [109] .
4.4. General Amino Acid Control-Based Sensing
Yeast has a transcription regulation mechanism of amino acid biosynthesis under low nitrogen conditions (amino acid starvation) [110] . Uncharged tRNA, transcription factor GCN4, eIF2 and many other proteins are involved in this regulation [111] . Existence of a similar regulatory system has been implied in plants [112] - [115] . Although several homologs of the GAAC components have been found in plants, there is still no direct evidence linking up each component to verify their roles in the plant GAAC system [116] - [120] . No homolog to GCN4 protein, the main component of GAAC is found in plants [84] [103] [121] .
4.5. Cytokinins and His-Asp Phosphorelay-Based Sensing
Cytokinins mediate long-range nitrogen signaling in plants. There is a direct correlation between nitrogen supply and level of cytokinins [122] - [125] . The growth limitation caused by nitrogen limitation was partially overcome when supplied with exogenous cytokinins. The action of cytokinins as long distance nitrogen signal was exemplified by the increase in xylem sap cytokinin level after nitrogen treatment in Arabidopsis and maize [126] . Not only there was increase in cytokinin levels under nitrogen conditions but also there was decrease in levels of cytokinins under nitrogen limiting conditions. Cytokinins regulate genes associated with nitrogen uptake and assimilation at transcriptional level [127] [128] .
Expression of the cytokinin biosynthetic gene isopentenyl transferase 3 is induced by nitrate [129] . The root to shootnitrate translocation enhances xylem solute flux enabling root to leaf translocation of cytokinins, which in turn triggers His-Asp multi-step phosphorelay mediated cellular signals [125] [130] [131] . The His-Asp multi-step phosphorelay consists of histidine protein kinase, response regulator and histidine-phospho transfer protein [132] - [134] . Histidine protein kinase contains the conserved cytokinin receptor, the CHASE domain [135] . Both cytokinins and nitrate induce the expression of response regulator genes. Cytokinins His-Asp phosphorelay signal is induced upon sufficient supply of nitrogen that signifies a proper time for growth and development. The role of cytokinin as a systemic relay that integrates nitrogen status and the regulation of plant growth are not proven yet. Correlation between nitrogen signals and responses of cytokinins His-Asp phosphorelay has not yet been proven and functions of many response regulator proteins are not known. The mechanismthat distinguishesoverlapping signals from cytokinin and nitrate pathways is not clear but microarray studies have revealed that only a portion of genes responsive to cytokinin are nitrate inducible [136] .
4.6. Ionotropic Glutamate Receptors-Based Sensing
Homologs of synaptic transmission associated animal inotropic glutamate receptorsare found in plants [137] - [139] . Plant ionotropic glutamate receptors regulate carbon and nitrogen metabolisms, using amino acids as the signaling molecules [140] . Transgenic A. thaliana under-expressing ionotropic glutamate receptor alters the C/N sensitivity of the plant during seed germination and alters accumulation of several enzymes/transporters associated with C/N metabolism [95] . Abscisic acid regulates nitrate mediated root growth in plants. Nitrogen signals ( or amino acids) act as the positive signals stimulating glutamate receptors to inhibit the ABA biosynthesis and promoting root growth. Carbon signals (sucrose) act negatively toward ionotropic glutamate receptors to reduce the expressions of carbon and nitrogen metabolism genes and elevate ABA biosynthesis [140] [141] . Ionotropic glutamate receptors from A. thaliana are root specific and highly expressed in roots and they may take part in sensing the change of amino acid composition in soil, leading to the adjustment of root architecture for colonizing the nutrient-rich patch of soil [141] [142] .
or amino acids) act as the positive signals stimulating glutamate receptors to inhibit the ABA biosynthesis and promoting root growth. Carbon signals (sucrose) act negatively toward ionotropic glutamate receptors to reduce the expressions of carbon and nitrogen metabolism genes and elevate ABA biosynthesis [140] [141] . Ionotropic glutamate receptors from A. thaliana are root specific and highly expressed in roots and they may take part in sensing the change of amino acid composition in soil, leading to the adjustment of root architecture for colonizing the nutrient-rich patch of soil [141] [142] .
4.7. NIT-2 Protein-Based Sensing
The GATA transcription factor NIT-2 regulates many nitrogen metabolism genes in fungi, the homologs of which are found in plants [143] - [146] . In Arabidopsis, though a nitrate inducible GATA transcription factor has been identified, its role in nitrate regulation of gene expression is not clear [147] . Plant NIT2-like protein (NLT1) and NIT2 binding site (GATA element) on the promoter of plant genes have been identified. A single zinc finger domain is found in plant NIT2 proteins that showed high homology with fungal NIT2 proteins [144] . The NIT2- like regulatory system has been suggested to be involved in the regulation of nitrate and nitrite reductases. The exact N sensing mechanism of NIT2 proteins in plants is still unclear.
5. Nitrogen Transporters
Plants have two transport systems for nitrogen based on the nitrogen concentration (nitrate, ammonium and urea). The low affinity nitrogen transport system functions at high nitrogen levels (millimolar concentrations >1 mM) and the high affinity nitrogen transport is mediated by transporter proteins which function at sub-millimo- lar nitrogen concentrations (Table 3) [61] [148] -[156] . Further, these transporters are classified into constitutive and inducible transport systems. In constitutive transport system, the expression of transporter genes is independent of nitrogen while in the inducible transport system the expression of the transporter genes is induced by nitrogen. The transporters of all three nitrogen forms fall under the above classifications schemes [157] - [162] . The membrane localized nitrogen transporters, using the free energy provided by the proton motive force, rapidly depolarize the membrane. Ammonium transporters act as uniporters i.e. transport of ammonium along the electrochemical gradient or as NH3/H+ co-transporters while the nitrate transporters acts as symporters, transporting nitrate along with protons via a 2H+:
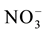 symport [61] [152] [163] [164] . Urea transporting proteins act as urea/H+ symporters [165] [166] .
symport [61] [152] [163] [164] . Urea transporting proteins act as urea/H+ symporters [165] [166] .
Different gene families coding for the transporters of nitrate, ammonium and urea exist in plants. Plant ammonium transport is mediated by transporter gene families, AMT1, AMT2, AMT3, AMT4 and AMT5. AMT1 proteins contribute to high affinity transport of ammonium into the plant roots; AMT2 and other AMT proteins function as low affinity transporters [167] - [171] . For transport of nitrate, five families of transporters, NRT1, NRT2, CLC, SLAC1/SLAH and ALMT1 (aluminium activated malate transporter1), are found in crop plants [153] [164] [172] - [176] . Members of NRT1 proteins, except NRT1.1 and NRT1.3 are low affinity transporters. Both NRT1.1 and NRT1.3 functions as a dual-affinity tranceptors. NRT2 proteins act as high affinity transporters and require an additional protein NAR2.1 and function as a 2 × 2 tetramer [177] - [179] . The tonoplast localized CLC (chloride channel family) proteins (CLCa and CLCb), mediate vacuolar nitrate accumulation by proton-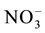 exchange mechanism [180] [181] . SLAC1 (slow anion channel 1) and SLAH3 (SLAC1 homologue 3) proteins showed nitrate transport activity. ALMT12 is a novel class of anion transporter involved in stomatal closure and it is permeable more to nitrate than to chloride [175] [182] [183] . Like other Nitrogen transporters, Urea transporters are of two types, low affinity transporters (Major intrinsic proteins) and high affinity transporters (DUR3 proteins). The low affinity major intrinsic proteins are channel-like, allowing passive urea transport down the concentration gradient while the high affinity DUR3 transporters transport urea against concentration gradient actively using energy [36] [158] .
exchange mechanism [180] [181] . SLAC1 (slow anion channel 1) and SLAH3 (SLAC1 homologue 3) proteins showed nitrate transport activity. ALMT12 is a novel class of anion transporter involved in stomatal closure and it is permeable more to nitrate than to chloride [175] [182] [183] . Like other Nitrogen transporters, Urea transporters are of two types, low affinity transporters (Major intrinsic proteins) and high affinity transporters (DUR3 proteins). The low affinity major intrinsic proteins are channel-like, allowing passive urea transport down the concentration gradient while the high affinity DUR3 transporters transport urea against concentration gradient actively using energy [36] [158] .
Table 3. Nitrogen transport proteins.
5.1. Ammonium Transporters
The ammonium transporting AMT proteins belong to ammonium transporter/methyl-ammonium permease/ rhesus family (AMT/MEP/Rh). These proteins are highly hydrophobic membrane proteins of 45 - 55 kDa size. They are trimers with substrate conduction channel within each monomer [60] [184] [185] . Ammonia channel proteins have either 11 or 12 transmembrane helices and divided into structurally similar two halves with a N- out, C-in topology (long cytoplasmic C-terminal) but organized with pseudo-two-fold symmetry [186] [187] . Ammonium is transported along a pore in between these two halves of the protein [188] [189] . A cytosolic loop connects the two halves (5th & 6th helices) and the residues of the helices are involved in recruitment and transport of ammonium, but the mechanism involved is not known [184] . Oligomerization is a general feature of Amt/Mep/Rh proteins and plant ammonium transporters have been found to form oligomers (homo or hetero) in the plasma membrane of roots. A functional pore is formed by each subunit of the oligomer [65] . The cytosolic C terminus (CCT) containing three conserved Ser/Thr residues regulate the function of AMT oligomers. When the external ammonium concentration is high, the AMT proteins are phosphorylated at the C-terminus (at Thr-460) leading to rapid shut down of ammonium transport into the cell [68] . Thr-460 is conserved among all AMT1 homologs except in AMT1; 5 [190] . Mutations or truncation of the C-terminus resulted in the inactivation of the AMT proteins [55] [56] . The interactions among CCT, the loop between 5 - 6 transmembrane helices and adjacent loops, serve as an allosteric switch that changes the conformational state of the monomers in the complex thereby controlling access to substrate or transport of ammonium through the pore [163] . This kind of allosteric regulation efficiently monitors ammonium transport capacities of AMT oligomer complexes in plant roots [69] . Vacuolar ammonium loading is mediated by tonoplastic intrinsic proteins (TIPs) TIP1; 2, TIP2; 1 and TIP2; 3 [191] - [193] .
5.2. Nitrate Transporters
The low affinity NRT1 transporters belong to NPF family (previously called PTR/PTO family [194] . NRT1 transporters show sequence similarity to plant and animal peptide transporters (PTR). The earlier under standing off unctional distinction of NRT1 (nitrate transport) and PTR (peptide transport) members need to be revised asrecent investigations have shown that NRT1 proteins, in addition to transporting nitrate, also transporthormones, dipeptides and glucosinolates. Plant NRT1 proteins contain “12 transmembrane (TM) domains and a plant specific long hydrophilic loop between TM6 and TM7 domains”. NRT1 transporters contain one or two MFS domains [47] [195] - [198] .
Dual affinity nitrate transporters form dimer at the plasma membrane, this dimerization is depends on phosphorylation at Thr101 of the NRT1.1 transporter. Thr101 is closed by a pocket formed by transmembrane helices 2, 3 and 4, which is located at the homodimer interface. The substrate binding site is formed by Leu 49, Val 53, Leu 78 and Phe 511. The NRT1.1 contains His 356 at TMH7 which has a role in stabilization of nitrate in the binding site. The mechanism of dual affinity mode of action is mediated by oligomerization. In the un-phosphorylated state NRT1.1 transporters form a homodimer and function as low affinity nitrate transporter. When phosphorylated NRT1.1 homodimers are disrupted, each monomer functions as independent high affinity transporters. The long N-terminal structure of NRT1.1 forms a cleft between the NRT1.1 proteins in the dimer which provides the binding site for kinases and other enzymes [199] .
The high affinity NRT2 transporters belong to Nitrate-Nitrite Porter family (NNP) under major facilitator super family [152] [200] . Like NRT1 transporters, NRT2 proteins also contain twelve transmembrane spanning domains and a cytosolic oriented loop in between TM6 and TM7 domains. Members of NRT2 have not been shown to transport anything else other than nitrate. Although both NRT1 and NRT2 proteins contained MFS domains, the NRT2 transporters contained an additional “nitrate motif” (A-G-W/L-G-N-M-G) located in the fifth transmembrane domain. The C terminal domain is long and is cytosolic. R87 and R459 located in transmembrane domains 2 and 8 respectively were identified as essential residues for substrate binding by mutation analysis.
CLC proteins, the vacuolar nitrate transporters, belong to secondary active transporters which is a subfamily of chloride channel proteins [201] - [203] . Plant CLCa proteins show selectivity in transporting nitrate than chloride. This transport is dependent on proton coupling which is a unique feature in plants when compared to animal and bacterial CLC proteins where it is not dependent on proton coupling [180] . The nitrate selectivity of CLC proteins is based on the presence of conserved amino acids [204] [205] . A serine residue (Ser107) is conserved in human and bacterial CLC proteins while in plants a proline residue (Pro160) is conserved in the conserved motif which selects nitrate over chloride [201] [206] . Along with proline there are 2 other conserved amino acids called gating glutamate and proton glutamate essential for the transport activity of CLCa protein. Seven and four members of CLC protein genes were found in Arabidopsis and in rice, respectively. Four of the Arabidopsis CLC proteins contained proline as the conserved residue while others contained serine as the conserved residue in the selectivity filter motif. Except for CLCa, b the physiological functions of other proteins are not known [180] [207] - [209] .
SLAC1 are slow anion channel proteins located in guard cells capable of nitrate transport [182] . Homologues of SLAC1 located in other tissues are called SLAH proteins which are activated by the CPK21 kinase in a nitrate induced manner. The plasma membrane localized SLAC1 and SLAH member proteins contain 10 transmembrane helices. The aluminum activated malate transporter12 (ALMT12) is another protein found to transport nitrate to the guard cells. This guard cell plasma membrane localized protein contains six transmembrane α-helices with an N-out and C-out topology. The structural features determining the selectivity of these proteins is yet to be determined [210] .
5.3. Urea Transporters
Plant urea transport proteins come under sodium solute symporter super family [36] [166] . Among the five urea transport proteins known in plants, DUR3 is the lone high affinity transporter while others are low affinity transporters. The membrane localized DUR3 contains 14 - 15 transmarine domains and N out and C in topology [211] . Domains 1 - 6 and 7 - 12 are conserved in most plants, but the length of C-terminal is variable. In between 8th and 9th transmembrane domains, an outside-oriented loop is located which contains the conserved Walker A motif (P-loop) [212] . The low affinity passive urea transport is carried out by aquaporins which are major intrinsic proteins (MIPs). They are of four types: Nodulin 26-like membrane intrinsic proteins, plasma membrane intrinsic proteins, small basic membrane intrinsic proteins and tonoplast intrinsic proteins [213] .
Major intrinsic proteins contain “six transmembrane spanning helices, five interconnecting loops (A-E) with N out and C out topology” [214] . A, C and E loops are apoplastic oriented in plasma membrane intrinsic proteins and Nodulin 26-like membrane intrinsic proteins and vacuolar space oriented in tonoplast intrinsic proteins. The conserved NPA motif (asp-pro-ala sequence) is located in the partly membrane embedded small loops B and E. The NPA motif acts as the selectivity filter. The B and E loops bring together two highly conserved NPA-motifs (asparagine-proline-alanine) in the centre of the membrane and contribute to pore formation [215] [216] . The substrate specificity of these proteins can be due to size exclusion at two main pore constrictions, H-bonding mediated stereo specific recognition of the substrate and hydrophobic interactions within the pore. Along with urea, aquaporins which are able to transport ammonia were also found and they belong to the tonoplast intrinsic proteins family. The selectivity between urea and ammonia (NH3) is due to the presence of the histidine instead of a hydrophobic aromatic amino acid in the pore constriction region [193] .
6. Nitrogen Uptake
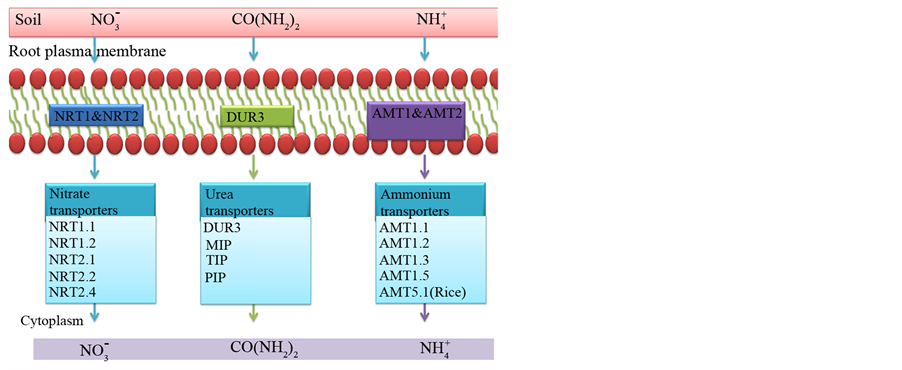
The most important function of nitrate, ammonium and urea transporters is the acquisition of different forms of nitrogen from the soil into the plant roots. Though many nitrogen transporters have been identified in plants, only few of them are found to be associated with soil nitrogen uptake (Figure 1). AMT1; 1, AMT1; 2, AMT1; 3, AMT1; 5, AMT2; 1 and AMT5; 1 proteins mediate ammonium uptake. Nitrate uptake is mediated by NRT1.1, NRT1.2, NRT2.1, NRT2.2 and NRT2.4. Urea uptake is mediated by DUR3 and MIP transporters (Table 4).
6.1. Ammonium Uptake
Ammonium uptake is carried out by AMT1:1, AMT1; 2, AMT1; 3 and AMT1; 5 proteins in Arabidopsis and other plants. However, in rice two additional proteins, AMT2; 1 and AMT5.1 were found to uptake ammonium [59] [163] [167] [171] [217] [218] . AMT1; 1, AMT1; 2, AMT1; 3 and AMT1; 5 are high affinity ammonium transporters while AMT2; 1 and AMT5; 1 are low affinity ammonium transporters. All these proteins show root specific expression. AMT1; 1 and AMT2; 1 are constitutively expressed but ammonium increases expression of these genes in roots. AMT1; 2 is an ammonium inducible protein while AMT1; 3, and AMT1; 5 are repressed by nitrogen [162] [218] . In Arabidopsis all AMT proteins are constitutively expressed but in maize, AMT1; 1 and AMT1; 3 are ammonium inducible [219] . AMT1; 1, AMT1; 3 and AMT1; 5 are expressed mainly in the
Figure 1. Nitrogen transporters involved in N uptake from the soil. NRT1.1, NRT1.2, NRT2.1, NRT2.2 and NRT2.4 are nitrate transporters, while AMT1.1; AMT1.2; AMT1.3; AMT1.5 & AMT5.1 are the ammonium transporters involved in uptake of nitrogen. Urea transporters DUR3, MIP, TIP and PIP proteins are involved in nitrogen uptake. All these proteins are plasma membrane localized and highly expressed in the roots.
Table 4. Proteins associated with nitrogen uptake.
root epidermal cells and root hair regions [59] [67] [218] . AMT1; 2 is expressed in outer cortical cells, central root cylinder, endodermis cells, sclerenchyma cells, root hairs and pericycle cells of primary root tip [67] [220] [221] .
Among all the AMT proteins AMT1; 1 has the highest substrate affinity and responsible for efficient ammonium uptake from the soil even under very low concentrations (nano molar). Under high ammonium conditions over-expression of OsAMT1; 1, increased the ammonium uptake and reduced the total biomass leading to impaired growth and development. In nitrogen deficient roots AMT1; 1 and AMT1; 3 carryout bulk of high affinity transport (1/3rd of) When the availability of
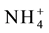 in the soil is low, AMT1; 3 acts in an additive manner with AMT1; 1 in taking up
in the soil is low, AMT1; 3 acts in an additive manner with AMT1; 1 in taking up
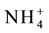 ions About 90% of root ammonium uptake is mediated by AMT1; 1, AMT1; 2 and AMT1; 3 transporters [59] [160] [162] [217] [222] [223] . AMT transporters act either as
ions About 90% of root ammonium uptake is mediated by AMT1; 1, AMT1; 2 and AMT1; 3 transporters [59] [160] [162] [217] [222] [223] . AMT transporters act either as
 uniporters or NH3/H+ co-transporters. Structural studies with AMT1; 1 have shown that ammonium is taken up from the soil and it is recruited at the external pore by a coordinated interplay of highly conserved Trp178 and tyr133 residues. Then ammonium passes through the pore residues during which it is de-protonated (NH3) by transfer of the protons to the two histidine residues (His168 and His318) within the pore. The NH3 moves down the pore and again re-protonated before release into the cytoplasm using the proton transferred from histidine residues [61] [62] [150] [163] .
uniporters or NH3/H+ co-transporters. Structural studies with AMT1; 1 have shown that ammonium is taken up from the soil and it is recruited at the external pore by a coordinated interplay of highly conserved Trp178 and tyr133 residues. Then ammonium passes through the pore residues during which it is de-protonated (NH3) by transfer of the protons to the two histidine residues (His168 and His318) within the pore. The NH3 moves down the pore and again re-protonated before release into the cytoplasm using the proton transferred from histidine residues [61] [62] [150] [163] .
6.2. Nitrate Uptake
Nitrate uptake is mediated by NRT1.1, NRT1.2, NRT2.1, NRT2.2 and NRT2.4 proteins. The dual affinity NRT1.1 transporter acts as an inducible low affinity transporter (except in rice and tomato where it was found to be constitutive) [161] [222] [224] . NRT1.2 is a low affinity constitutive nitrate transporter (except in tomato where it is inducible). While NRT2.1 and NRT2.2 proteins are inducible high affinity nitrate transporters [159] [225] [226] . The constitutive NRT2.4 transporter gene is induced by N limiting conditions [227] .
All nitrate uptake transporters show expression in the root tissue. NRT1.1 isexpressed in primary and secondary root tips, pericycle cells, epidermal cells, root hairs and root meristem quiescent centre [72] [157] . NRT1.1 uptakes nitrate both at low and a high soil nitrate condition which is made possible by the phosphorylation based switching (see nitrogen sensing for details about this switching). The low affinity nitrate uptake by NRT1.1 is pH dependent [73] [79] . NRT1.2 is a root dominant gene, expressed in rhizodermis and this expression is independent of root age [228] . In tomato it has been found to express in root hairs [222] . Antisense lines of NRT1.2 in Arabidopsis showed decreased nitrate uptake indicating the functional role of NRT1.2 [157] . NRT2.1 gene showed a root specific expression [178] [229] - [231] .
The role of NRT2 proteins in high affinity nitrate transport was identified from null mutant studies of NRT2:1 and NRT2:2. About 80% reduction in nitrate uptake was observed when genes coding for these two proteins are disrupted [148] [159] [232] - [234] . These two proteins need NAR2 protein for their functional activity. However, NRT2.4 does not need this additional protein for nitrogen uptake. The expression of NAR2 protein is induced by nitrate and T-DNA insertion of NAR2 also decreased the high affinity nitrate uptake. NAR2 directly interacts with the NRT2 proteins to form a hetero oligomer and are localized to the plasma membrane [179] [226] . NRT2.4 gene is expressed in the root primordium and epidermis of young lateral roots [152] . Mutational analysis of Arabidopsis NRT2.4 gene proved its role in nitrate uptake under N limiting conditions [227] .
Nitrate uptake into the plant roots, depolarizes the plasma membrane and coupled with proton gradient. The depolarization is counteracted by the nitrate inducible plasma membrane H+-ATPase [235] [236] . Both high and low affinity transporters mediate nitrate transport as 1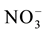 /2H+ and the energy is provided by proton motive force. The internal nitrogen status and the root age determine the contribution of each transporter in nitrate uptake. Genes coding for NRT1.1 and NRT2.4 are expressed in the young roots while NRT2.1 is specific for mature roots. The nitrate binding site is closely bound both extra cellular (TM1-TM2 and TM7-TM8) and intracellular gates (TM4-TM5 and TM10-TM11). Proton coupled nitrate uptake is mediated by, protonation of His 356 followed by nitrate binding. Nitrate binding closes the extracellular gate and conformational changes occur in the protein, and opens up intra cellular gate to transport nitrate and protons in to the cell [237] .
/2H+ and the energy is provided by proton motive force. The internal nitrogen status and the root age determine the contribution of each transporter in nitrate uptake. Genes coding for NRT1.1 and NRT2.4 are expressed in the young roots while NRT2.1 is specific for mature roots. The nitrate binding site is closely bound both extra cellular (TM1-TM2 and TM7-TM8) and intracellular gates (TM4-TM5 and TM10-TM11). Proton coupled nitrate uptake is mediated by, protonation of His 356 followed by nitrate binding. Nitrate binding closes the extracellular gate and conformational changes occur in the protein, and opens up intra cellular gate to transport nitrate and protons in to the cell [237] .
6.3. Urea Uptake
Urea uptake in plants is mediated by the high affinity DUR3 and low affinity NIP proteins [238] . It is expressed mainly in the root hairs, rhizodermis, root cortex and root xylem and localized to the plasma membrane. Nitrgen starvation and urea up regulates DUR3 gene expression while ammonium and nitrate repress the gene [158] [165] [239] [240] . DUR3 mediated urea uptake involves proton and membrane potential dependent 1 urea/1H+ symport the details of it is yet to be understood [158] [165] . Arabidopsis DUR3 T-DNA insertion mutants showed impaired growth. Low affinity urea transporters NIP2; 1, NIP2.4 and NIP6.1 are associated with uptake of urea [241] - [243] . NIP2; 1 and NIP2.4 are expressed constitutively in the root epidermal cells. NIPs function as channels through which urea can pass and is independent of pH [191] . NIP6; 1 contained alanine instead of tryptophan in helix 2 and this creates a much wider pore at the ar/R region which is permeable to urea [244] .
7. Nitrogen Efflux
For some unknown reasons, plants excrete nitrate outside and is induced by acidic conditions. The excretion may be passive or mediated by transporters NAXT1 and NRT1.5 (Table 5) [245] [246] . NAXT1 gene is specifically expressed in the mature root cortical cells and this protein is localized to the plasma membrane [246] .
Table 5. Proteins associated with nitrogen efflux.
NAXT1 is excretes
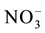
Ammonium accumulation in plants leads to toxicity and reduced nitrogen uptake efficiency [247] - [249] . For maintaining ammonium homeostasis, plants efflux ammonium into soil. Plants respond differently towards ammonium levels and based on this plants are classified as ammonium sensitive and tolerant species [250] . Under high ammonium conditions ammonium efflux occurs mainly at root elongation zone and depends on root respiration leading to repression of root growth and plant development [248] . The proteins associated with ammonium efflux are not yet identified. Efflux may be a mechanism in plants to protect from toxicity and to maintain root growth under excess ammonium conditions.
8. Nitrogen Translocation and Assimilation
Nitrogen (nitrate, ammonium and urea) taken up into the roots can either be transported to other parts of root and the shoot through xylem/phloem or stored in the vacuoles or assimilated (Figure 2 & Figure 3) (Table 6). Nitrogen assimilation depends on nutrient availability, diurnal cycle and carbon availability [95] . Among the three nitrogen sources, ammonium is toxic, so assimilated immediately after uptake into the roots and transported to shoots in an organic form. The non-toxic nitrate and urea is either assimilated immediately in the roots or assimilated after translocation to shoots.
8.1. Translocation and Assimilation of Ammonium
Sources of ammonium includes direct ammonium uptake from the soil by AMT1 and AMT2 transporters, nitrate reduction, protein degradation and photo respiration [251] - [253] . Photorespiration produces large amounts (20 fold higher than nitrate reduction) of ammonium. As high ammonium is toxic, it is assimilated immediately into organic nitrogen forms [254] . Assimilation of ammonium into glutamine is mediated by glutamine synthetase enzyme. Glutamate synthase converts glutamine into two glutamate molecules (Glutamine and glutamates are used for synthesis of other amino acids [255] [256] .
Glutamine synthetase exists as two isoforms, GS1 and GS2.GS1 isoform found in leaf cells and root phloem is associated with the root primary nitrogen assimilation. The GS2 isoform occurring in plastids is involved in re assimilation of ammonium released via photorespiration. Photorespiration plays an important role in maintaining the nitrogen status of the plant [257] [258] . The GS/GOGAT cycle incorporates photo respiratory and non-res- piratory ammonium and provides nitrogen for transport and maintaining nitrogen status in the plant [259] [260] . Assimilation of one
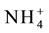
Though ammonium is assimilated immediately after uptake or generation, there are specific instances in plants where ammonium is translocated. Transport of ammonium to pollen is essential for its development, germination and growth of pollen tube. This transport is mediated by the constitutively expressed pollen grains and pollen tube localized AMT1; 4 ammonium transporter [264] . The expression pattern of AMT2; 2 and AMT2; 3 in shoots and roots indicate their role in translocation of ammonium but the exact mechanism of the translocation process is not known [171] [220] [265] - [267] .
AMT3; 1, weakly expressed in root but more in the stem and the new leaves is involved in translocation of ammonium from roots to the leaves [171] [267] . N starvation induces the expression of AMT3; 1 in roots, AMT3; 2 is expressed in roots and shoots, is an inducible ammonium transporter involved intrans location of ammonium from the roots through the xylem. AMT3; 3 is an inducible ammonium transporter expressed in old leaves and in the stems and is associated with translocation of ammonium [169] [220] [267] .
Figure 2. Role of nitrogen transporters in nitrogen assimilation in plants. Once taken inside nitrate, ammonia and urea are assimilated into different forms. Nitrate is assimilated to nitrite then to ammonia finally to amino acids. Urea is immediately converted to ammonium and then to amino acids. Nitrate transporter NRT1.1 and Ammonium transporters AMT1.2, AMT1.3 are involved in the N assimilation.
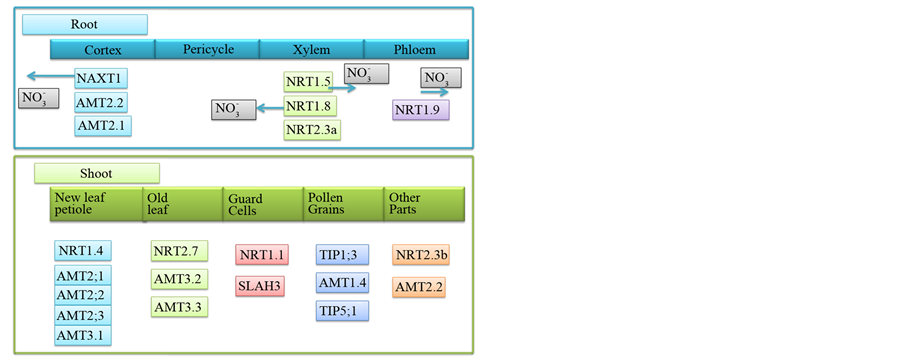
Figure 3. Nitrogen translocation. Nitrogen once taken inside the root is distributed to different parts of the plant and involves different N transporters. Different nitrogen transporters are involved in different organs of the plant.
Table 6. Proteins involved in nitrogen translocation and assimilation.
An ammonium transporter AMT1; 2 is involved in ammonium retrieval (Figure 3). AMT1; 2 is induced by both ammonium and nitrate [222] [268] [269] . AMT1; 2 contains serine rich plastid transit peptide. Two T-DNA insertions of AMT1; 2 mutant analysis showed that, the accumulation of ammonium is not altered showing that it has major function in the intracellular ammonium transport than uptake. AMT1; 2 is involved intransfer of ammonium to the vascular tissues [56] . Nitrate assimilation produces ammonium in both roots and shoots and AMT1; 2 is required to maintain these ammonium pools in the plastids. It is involved in efflux ammonium from the plastids to cytosol for further assimilation or transport into the chloroplast for further re assimilation [30] . AMT1; 2 is expressed root apoplast, endodermal and cortical cells and in the leaves and expression is regulated by whole plant nitrogen status. Further work required for the functional characterization of AMT1; 2 in nitrogen assimilation [162] [218] [270] .
8.2. Translocation and Assimilation of Nitrate
The extent to which nitrate is assimilated in the roots and how much nitrate is transported to shoots is not clear and is influenced by various factors like the species of plant, developmental stage of the plant, light intensity and temperature [95] [271] - [273] . In the root, highest assimilation is found behind the root tip. Nitrate reductase reduces the incoming nitrate to nitrite immediately in a NAD (P) H-dependent manner in the cytosol and it is a rate limiting step in assimilation of nitrate [274] [275] .
Then nitrite reductase reduces the nitrite transported into the chloroplast (by diffusion or by the nitrite transporter (Nitr)) into ammonium and is finally converted to amino acids by GS/GOGAT pathway [276] . Nitrate assimilation responds to internal and external signals (nitrate, light, sucrose, circadian rhythms, glutamine, auxin and pentose phosphate pathway metabolites) integrating nitrate assimilation with carbon and energy metabolism, ensuring availability of sufficient carbon and energy for the assimilation of inorganic nitrogen. They also influence nitrogen uptake by regulating root development. The nitrogen status of the plant regulates genes coding for nitrate transporters and N-assimilatory enzymes at transcriptional, translational and post-translational levels [276] [277] . Expression of nitrogen assimilatory genes nitrate reductase, nitrite reductase, glutamate synthase and carbon metabolism genes are induced by nitrate [136] [278] - [283] . Assimilation of one molecule of
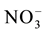
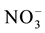
Translocation of nitrate from roots to shoots is mediated by transporters which load and unload nitrate into xylem and phloem by which nitrate is redistributed to different parts of the plant. Xylem is the major route of nitrate transport in plants. The low NRT1.5 and the high affinity NRT2.3 a load nitrate into xylem. Unloading of nitrate from xylem is mediated by NRT1.4 and NRT1.8. NRT1.5 is a nitrate inducible bi-directional transporter regulated by pH. It transports nitrate from root through xylem [173] [245] . Abiotic stresses down regulate expression of NRT1.5 by which the root nitrate is retained during stress contributing to stress [285] [286] . NRT2.3 a is another transporter that loads nitrate into xylem for transport to shoots. It is a vascular specific transporter expressed in the root stele and the scutellum tissue of rice [287] [288] .
Unloading of nitrate from xylem is mediated by both NRT1.4 and NRT1.8 proteins. NRT1.4 is localized to petiole and it retrieves nitrate from the xylem and translocate to other parts of the leaf [228] [289] . It is mainly responsible for accumulation of nitrate in the petiole by which it regulates leaf nitrate homeostasis [164] . NRT1.8 is the main xylem nitrate unloaded and is expressed mostly in parenchyma cells of xylem [286] . Nitrate is also transported through phloem and the level of nitrate in phloem was found to be 1.9 to 8.1 mM under N non-limiting conditions [290] [291] . NRT1.9 and NRT2.4 load nitrate into phloem. NRT1.9 is a phloem loader, predominantly expressed in the companion cells of root phloem. It facilitates downward movement of nitrate which is considered to be a mechanism regulating shoot nitrate transport [292] .
8.3. Translocation and Assimilation of Urea
Like nitrate, urea assimilation also occurs both in roots and shoots, but the mechanism is not known for this partition [293] . Urea is assimilated to NH3 and CO2 by the enzyme urease [157] . The GS/GOGAT pathway enzymes assimilate the NH3 generated into amino acids. Urease is a nickel metalloenzyme requiring accessory proteins, UreD, UreF, and UreG, for activation [294] . Sources of urea are including urea transport by DUR3 and NIP proteins, (Urea cycle) arginine degradation and ureides from purine catabolism [295] . Urea cycle derived urea is produced in mitochondria and then exported to the cytoplasm for hydrolysing into ammonium by urease [36] [219] . Assimilation of one urea molecule produces 0. 33H+ ion [296] .
9. Vacuolar Nitrogen Storage
In plants, vacuole occupies the major volume of mature cells and is used as a buffering system for maintaining homeostasis of cytoplasm. It is a major storage site for different forms of nitrogen. Vacuoles are acidic and contain hydrolytic enzymes. Vacuolar solute transport is driven by the proton-motive force generated by vacuolar ATPase/vacuolar pyrophosphatase [297] [298] . Vacuolar ion movement is mediated by aquaporins (water channel proteins) and other specific ion channels (Figure 4) (Table 7).
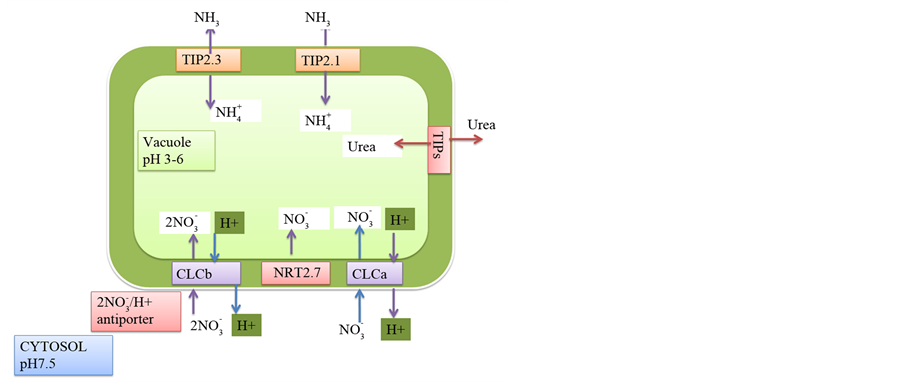
Vacuole (the nitrate storage pool) stores about 30 - 50 mol・m−3 nitrate when compared to just about 3 - 5 mol・m−3 nitrate present in the cytoplasm (the metabolic pool) and the stored vacuolar nitrate is transported into the cytoplasm for utilization or mobilization [299] [300] . Nitrate transporters like CLC family proteins and NRT2.7 are involved in vacuolar nitrate transport. Vacuolar nitrate storage is genotype and variety dependent regulated by photosynthates and energy [81] [176] [301] [302] .
Figure 4. Vacuolar nitrogen storage. Vacuolar nitrogen storage is carried out by nitrate transporters NRT2.7 and CLCa, b proteins which are tonoplast localised. Vacuolar ammonium loading is driven by TIP2.1 and TIP2.3 proteins and vacuolar urea loading is mediated by TIPs.
Table 7. Proteins associated with vacuolar storage of nitrogen.
In cereals, CLC proteins are the major vacuolar nitrate transporters [173] . Though absent in cereals, NRT2.7 is an important vacuolar nitrate transporter in Arabidopsis and is important for vascular nitrate loading during seed development [303] - [305] . The tonoplast localized CLCa and CLCb proteins possess high nitrate selectivity over chloride. They transport nitrate by the proton-nitrate exchange mechanism. About 50% of vacuolar nitrate storage is mediated by the mesophyll localized CLCa protein. Nitrate up regulates expression of CLCa and CLCb genes which are also diurnally regulated [180] [181] . NRT2.7 is expressed in the cotyledon tonoplasts during late embryonic developmental stage and in roots in the tonoplast of the main vacuole of mature roots [303] .
Vacuolar ammonium concentration ranges from 2 - 45 mol・m−3 [306] . Vacuolar ammonium transport is mediated by ammonium inducible aquaporins, TIP1; 2, TIP2; 1 and TIP2; 3 proteins [307] - [310] . The ammonium in root cytoplasm brought in by AMT transporters are deprotonated immediately leading to increase in vacuolar pH. The differential pH in cytoplasm and vacuole generates an electro chemical gradient that drives the transport of ammonia into the vacuole by the TIP proteins. The ammonia entering the vacuole is reprotonated and stored as ammonium (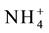
Vacuolar urea transport is mediated by the low affinity urea transporters, TIPa, TIP1; 1, TIP1; 2, TIP2; 1, TIP4; 1 and TIP4.4 [193] [242] [243] [313] . These transporters are constitutively expressed in tonoplast and involved in vacuolar loading and unloading of urea. Vacuolar urea transport is independent of pH gradient and is a passive transport [165] . Cytoplasm nitrogen depletion up regulates the expression of urea transporting TIP proteins leading to immediate remobilization of the vacuolar urea into the cytoplasm [241] . Over expression of TIP4; 1 gene increased not only vacuolar accumulation of urea, but also increased the urea uptake from soil [314] . Storage of urea in vacuoles avoids toxicity and prevents saturation of N-assimilation and stored urea is remobilized to cytoplasm when needed.
10. Nitrogen Remobilization
Remobilization is the process of moving nitrogen from old senescing plant parts to younger parts during vegetative phase or to the storage organs during reproductive phase [280] [315] - [319] . Remobilization facilitates efficient utilization of available nitrogen in the plant and also meets the inadequacy of the uptake system in providing enough nitrogen for storage in seeds. There is substantial quantity of nitrogen available in the senescing part of the plant, specially the leaves. Chloroplasts in leaves are one of the main sources of nitrogen for remobilization. Rubisco enzyme alone constitutes 50 - 56 percent of the total soluble proteins [320] [321] . The major contributor for seed nitrogen content is the leaf nitrogen remobilization and it varies from 50 - 90 percent in different crop plants [322] - [325] . So, the grain yield is influenced by the nitrate stored in the vegetative tissue and efficiency of remobilization of this stored nitrogen to the reproductive tissues. For efficient remobilization of leaf nitrogen there should be tight co-ordination between remobilization and senescence induced protein degradation. Fusion protein studies with rubisco and GFP/RFP showed that immediately after the onset of senescence the rubisco is taken inside the vacuole for processing. In autophagy deficient mutant this was not happening indicating the contribution of autophagy for degradation of substantial quantity of rubisco during senescence [326] .
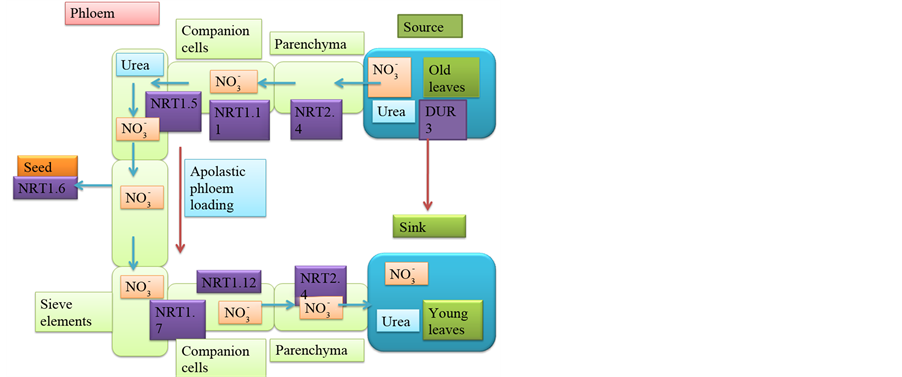
Nitrate transporters like NRT1.6, NRT1.7, NRT1.11, NRT1.12 and NRT2.4 are involved in N remobilization (Figure 5, Table 8) [227] [327] - [329] . Nitrogen starvation induces expression of NRT1.7 in shoots and the protein is targeted to the plasma membrane of mature leaf vascular tissue. It loads the excess nitrate available in mature leaves into phloem and remobilizes to younger leaves. Excess nitrate in older leaves is the vacuolar stored nitrate or the nitrate produced by senescence induced degradation of proteins. This vacuolar nitrate is likely to be transported by vacuolar nitrate transporters, which in turn is loaded into phloem by NRT1.7 [164] [328] .
NRT1.6 and NRT2.7 proteins transport nitrate for seed development, the former during early stages and the latter during late stages of seed development [303] [327] [328] . Nitrogen starvation induces expression of NRT1.6 in flowers, vascular bundle, and the epidermis of silique, seed funiculus and embryo sac wall [327] . NRT2.4 is involved in remobilization of nitrate from leaves by phloem loading [227] . NRT1.11 and NRT1.12 are involved in remobilization of nitrate from mature to young leaves by transferring nitrate from xylem to phloem. Expression of NRT1.11 gene is shoot specific, while NRT1.12 is expressed both in roots and shoots [329] . High affinity urea transporter DUR3 and low affinity urea transporters TIP1; 3 and TIP5; 1 are also asso-
Figure 5. Nitrogen Remobilization. Source to sink nitrogen remobilization is mediated by nitrate and urea transporter proteins. Nitrate is remobilized from old leaves to young leaves by NRT1.7, NRT1.11, NRT1.12, and NRT2.4. For seed development nitrate is remobilised from source tissues to developing seeds by NRT1.6 protein. Urea transporting DUR3 proteins also participated in nitrogen remobilization.
Table 8. Proteins associated with nitrogen remobilization.
ciated with nitrogen remobilization [239] [330] [331] . TIP1; 3 and TIP5; 1 are expressed in the mature pollen. TIP1; 3 is tonoplast localized while TIP5; 1 is mitochondria localized. These proteins remobilize nitrogen from source to sink tissues. Proteolysis of proteins during senescence results in production of free amino acids and the level of asparagine and glutamine increases in the phloem sap [332] - [334] . Amino acid transporting AAP-family proteins load free amino acids into phloem. In legumes ureides are used for translocation of nitrogen in the form of urea to seeds [335] - [337] .
11. Nitrogen Transport and Root Development
Plant root development is a complex process influenced by multiple genes/QTLs and the influence of nitrogen on root development is significant as it affects the nutrient and water absorption from the soil [338] [339] . The root surface area and the location of transporters in the roots determine nitrogen uptake. Nitrogen supply affects primary root growth, LR initiation and elongation. Nitrogen has a local stimulatory and systemic inhibitory effect on root development [340] - [344] .
11.1. Local Stimulatory Effect
Presence of local concentration of nitrate has a stimulatory effect on initiation of lateral root primordial and growth of lateral roots [48] [74] [345] [346] . This localized stimulatory effect is mediated by ANR1, NRT1.1 and auxin. NRT1.1 which can sense external nitrate has been suggested to stimulate localized lateral root growth via an ANR1-mediated pathway. ANR1 (Arabidopsis Nitrate Regulated1) is a nitrate inducible transcription factor which mediates induction of LR elongation in response to local supply of
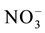
Ammonium, unlike nitrate, promotes lateral root initiation than elongation and local supply of ammonium inhibits elongation of existing roots and initiate new lateral roots leading to branching of existing roots. Nitrate and ammonium play complementary roles in promoting initiation and elongation of lateral roots, respectively. The ammonium sensing tranceptor AMT1; 3 mediates this local stimulatory effect [48] [58] [345] [347] [350] . The ammonium induced formation of new lateral roots is repressed by glutamine/glutamate. In the presence of ammonium as sole N source plants developed thinner and longer roots, with high root density [351] .
11.2. Systemic Inhibitory Effect
A uniform high soil nitrogen concentration inhibits root growth and development this is a reversible process. When the plants are transferred to normal nitrogen concentrations, this inhibitory effect can be relieved.
The high nitrate mediated systemic inhibitory effect on root development involves auxin, abscisic acid (ABA), ethylene and plant growth promoting bacteria. High nitrate condition leads to either suppression of auxin synthesis or prevention of shoot to root auxin movement. High shoot nitrate concentration suppresses lateral root development and involves root to shoot signaling [174] [345] . Auxin response factors and microRNAs are involved in this inhibition. Auxin induces expression of auxin receptor AFB3 gene and AFB3 has been found to regulate nitrate responsive primary and lateral root development [341] . The gene coding for miR393 is induced by nitrogen metabolites. The miR167/ARF8 regulatory module regulates the ratio between initiation and emergence of lateral roots [352] . Auxin response factor 8 (ARF8) is up regulated by high nitrate, while the microRNA that targets ARF8 was down regulated. The ARF8/miR167 module inhibits the repressing effect of high nitrate on lateral root elongation [352] . ABA plays an important role in high nitrate mediated lateral root growth inhibition [353] . Auxin and ethylene regulate lateral root development synergistically [354] . High nitrate stimulates ethylene production, which in turn regulates nitrate transporters (NRT1 and NRT2), resulting in alteration of root branching [355] . High nitrate mediated inhibition of lateral root deployment is also influenced by soil bacteria. Plant growth promoting bacteria protect plants from high nitrate mediated inhibition of lateral root growth by altering expression pattern of nitrate transporter genes. The exact mechanism behind this is not known [356] .
Under excess ammonium conditions, primary and lateral root growth is inhibited by decreasing cell elongation. Plant hormones auxin and ethylene have no role in this inhibition [250] [357] [358] . A direct contact of the root tip with external ammonium is needed for perceiving the inhibitory effect of excess ammonium and is mediated by GMPase (GDP-mannose pyrophosphorylase). This inhibition is due to altered ammonium metabolism and defect in N-Glycosylation of proteins in the root [58] [357] [359] [360] .
Along with the above dual effects, nitrogen also regulates the root growth by different ways. Under high carbon to nitrogen ratio, lateral root initiation is inhibited, while under external L-glutamate root branching is stimulated and primary root growth is inhibited. The suppression of LR initiation in response to high carbon to nitrogen ratio is due to prevention of shoot to root auxin translocation [83] . Due to high metabolic activities under high C/N conditions, there is a depletion of nitrate levels which is sensed by the nitrate transporter NRT2.1, leading to repression of LR initiation. NRT2.1 acts as the integrator of carbon and nitrogen metabolism and is regulated by both carbon and nitrogen signals [83] [154] [361] - [363] . Under N-limiting conditions, mutations in NRT2.1 reduced the number of lateral root primordia. Glutamine represses emergence of lateral root resulting in low level of third-order lateral root formation. In contrast, glutamate even at low concentrations inhibited growth of primary root but stimulated lateral root initiation and growth resulting in highly branched short roots [49] [83] [141] [246] [352] .
Nitrogen limitation increases the auxin content of roots by promoting shoot to root auxin translocation thereby inducing initiation and elongation of lateral roots [343] [364] . Low nitrogen induces lateral root growth by stimulating the auxin biosynthesis gene TAR2 (tryptophan aminotransferase related 2), which is needed for enhanced auxin accumulation in LR primordia [365] .
12. Regulation of Nitrogen Nutrition
Regulation of Nitrogen levels inside the plant is a necessity as limitation in nitrogen availability affects many developmental processes and excess nitrogen is detrimental to plants because of direct or indirect toxicity. Regulation occurs at uptake, assimilation, translocation, storage and remobilization steps (Figure 6). The N sources (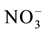

Imbalance in nitrate levels may lead to generation of superoxide, reactive nitrogen species (nitric oxide (NO); peroxynitrite (ONOO-)) resulting in oxidative stress [366] - [368] . So plants have multiple mechanisms to ensure nitrogen homeostasis. The nitrogen homeostasis is the result of balance among the following process: nitrogen uptake mediated by transport proteins, nitrogen assimilation, and storage in vacuole, translocation to other parts of the plant from roots, nitrogen efflux and downward transport of nitrogen into roots. The balance is achieved by regulating each of these processes individually and coordinated regulation of these processes. Fragmentary knowledge is available on individual regulation of these processes in which many transcription factor proteins (Table 9), small RNAs and kinases are involved. For understanding the co-ordinated regulation of these proce- sses more detailed research is necessary.
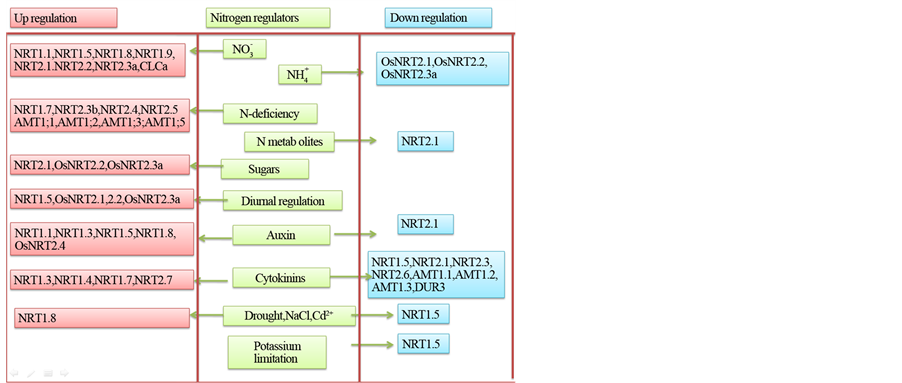
Figure 6. Regulation of nitrogen transporters. Nitrogen metabolism is regulated at many stages of plant development. The nitrogen transporting proteins are regulated by many factors like nitrate, ammonium, sugars, light, hormones, biotic and abiotic stresses, N-limitation and N metabolites.
Table 9. Transcription factors involved in regulation of nitrogen nutrition.
12.1. Regulation of Nitrogen Uptake
The first layer of regulation of nitrogen uptake is at the level of different N forms which induce or repress expression of nitrogen uptake genes. Nitrogen uptake is controlled by many factors, such as nitrogen level, energy status of the plant, assimilation status of imported nitrogen, nitrogen demand, and involves mobile signals between shoots and roots as well as between different parts of the root system [369] . Nitrate induces expression of genes coding for nitrate transport associated proteins, NRT1.1, NRT2.1, NRT2.2, NAR2.4 and H+-ATPases [178] [231] [232] [235] [288] [370] [371] . In rice, NAR2.1, NRT2.1, NRT2.2, and NRT2.3a genes are up regulated by nitrate and suppressed by ammonium. High temperature up regulates the expression of NRT2.2, NRT2.3b, and NRT2.4, but longer exposure to high temperature reduced the expression of NRT2.1, NRT2.2 and NRT2.3a [288] [372] . Both exogenous supply and internal increase of amino acids inhibit expression of nitrate transporters and nitrate uptake while decrease in level of root amino acids stimulated uptake of nitrogen [234] [373] [374] .
Though NRT2.1 gene is nitrate inducible, very high nitrate concentrations inhibited the expression of NRT2.1 and this inhibition was found to be mediated by NRT1.1 [76] (Munos, 2004). Promoter histone methylation mediated by HIN9/AtIWS1 has been found to be associated with this repression. Expression of NRT2.1 gene is suppressed by reduced nitrogen metabolites and ammonium [375] [376] . The N & C-terminals of NRT2.1 contain many protein kinase recognition motifs. Like NRT1.1, NRT2.1 is regulated by a phosphorylation/dephos- phorylation mechanism under high nitrogen conditions [377] [378] NRT2.1 gene expression is induced by light and sugars. Sugar and nitrate are coupled by glucose metabolism by oxidative pentose phosphate pathway (OPPP) [379] . Increased glucose levels and metabolites of OPPP pathway stimulate the expression of NRT2.1. The expression of different enzymes involved in OPPP pathway also directly regulates the expression of nitrogen transporters. Reduced expression of phosphogluconate dehydrogenase reduced glucose-mediated induction of NRT1.1, NRT1.5, NRT2.1 and AMT1; 3 [380] [381] . Antisense RNA mediated suppression of Hexokinase1 (HXK1) expression reduced NRT2.1 expression also. Sugar mediated nitrate uptake regulation involves glucose-6-phosphate and OPPP metabolites [382] .
The basic factor determining the regulation of ammonium uptake is the plant nitrogen status. Expression of genes coding for ammonium transporters is up regulated by low tissue nitrogen levels, while high tissue nitrogen down regulates the expression of these genes [150] [383] [384] . The negative membrane potential of roots drives ammonium uptake leading to high cytosolic ammonium concentration. As ammonium can be stored in vacuole, it indirectly influences the ammonium uptake by regulating the level of ammonium in tissues [192] [385] [386] . In wheat the ammonium uptake by AMT1; 1 was found to be dependent on extracellular pH [387] [388] .
Though ammonium represses most ammonium transporter genes, it has been found to up-regulate expression of AMT1; 1 of rice and AMT1; 2 of rice and tomato. In tomato AMT1; 2 gene expression has been found to be strongly up-regulated by nitrate [30] [218] . Genes coding for AMT1; 2 and AMT1; 3 transporters were found to be induced by sucrose and diurnal changes [382] . Nitrogen starvation derepressed nitrate, ammonium and urea uptake transporter genes in the roots [158] [168] [231] [361] .
Nitrogen starvation conditions in roots induce expression of genes coding for urea transporters, DUR3, TIP4; 4 and PIP1; 5b [166] [241] [242] . Presence of nitrate represses expression of DUR3 gene, but induces the low affinity urea transporter PIP1; 5b [158] [313] . Genes coding for low affinity NIP2; 1 and NIP2; 4 urea transporters are expressed constitutively and not affected by nitrogen starvation and availability [241] .
Post transcriptional regulation of proteins regulating expression of nitrate transporters is mediated by micro RNAs [389] . Micro RNA miR169 targets nuclear factor Y-A (NF-YA) subunits of the heterotrimeric NF-Ys transcription factors that directly regulate NRT1.1 and NRT2.1 expression [390] [391] - [393] . Nitrogen levels regulate expression of miR169 gene expression, nitrogen limitation down regulates expression of miR169 multigene family [394] [395] . Under nitrogen-sufficient conditions over expression of miR169a resulted in reduced expression of NRT1.1.
Nitrogen uptake is also regulated by hormones. Genes coding for uptake transporters, NRT1.1, NRT2.1, AMT1; 1, AMT1; 2 and AMT1; 3 and DUR3, were found to be repressed by cytokinin. The genes involved in cytokinin biosynthesis are induced by nitrate which is mediated by NRT1.1 [73] [75] [77] [396] . The cytokinin effect on nitrogen transporters is mediated by His?Asp phosphorelay system [397] [398] .
Phosphorylation/dephosphorylation of proteins have been found to regulate nitrogen uptake. The NRT1.1 transporter is phosphorylated at Thr101 position by CIPK23 kinase when the nitrate concentration is low and the phosphorylated NRT1.1performs high affinity transport of nitrate. High concentration of nitrate prohibits phosphorylation and the dephosphorylated NRT1.1 performs low affinity transport of nitrate [73] [78] [79] .
The ammonium channel made of trimeric form of AMT1; 1 protein is open and amenable for transport of
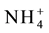
NLP7 protein is involved in
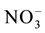
12.2. Regulation of Nitrogen Efflux
Only nitrate and ammonium efflux mechanisms are known in plants and there are no known efflux mechanisms for urea.
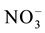
Like nitrate, excess ammonium is secreted out, to overcome ammonium toxicity and maintain the growth of the plants but the efflux mechanism involved in this is not known [247] . Ammonium can be effluxed in the form of NH3 and it is an energy dependent process. Intracellular ammonium concentrations range from 1 - 10 millimoles [250] . Ammonium efflux is linked to high ammonium mediated inhibition of primary root development [359] . Under high nitrogen to carbon conditions some of the uptake transporters are down regulated and nitrogen effluxed to maintain homeostasis [246] . In plants the transport of NH3 from leaves to outside contributes to overall ammonium efflux which is enhanced by high tissue ammonium concentrations [401] .
12.3. Regulation of Nitrogen Translocation and Assimilation
Nitrate induces genes coding for assimilatory enzymes, nitrate reductase and nitrite reductase, while glutamine repress these enzymes [276] . NLP7 transcription factor is involved in induction of these genes [85] . Transcription factor, 14-3-3 regulates nitrate reductase and glutamine synthetase in a N-dependent manner. The circadian clock regulating CCA1 transcription factor coordinates the response of nitrate assimilatory genes [402] .
There is a direct correlation between plant ammonium levels and expression of genes coding for assimilatory enzymes GS1.1 (shoots), GS1.2 (roots) GS2.1 (shoots and roots), Fd-GOGAT (shoots) NADH-GOGAT (roots) and GDH (shoots and roots). Ammonium assimilation genes are up regulated in the roots when there is an increase in ammonium levels due to over expression of AMT1; 1 [223] . Increased assimilation of nitrogen leads to increases in total amino acid content of the plant [403] [404] .
There are two glutamine synthases, GS1 (cytosolic) and GS2 (plastidic), induced by low and high nitrate conditions, respectively. These enzymes are regulated by phosphorylation and dephosphorylation. Phosphorylated enzymes are active and protected from degradation while dephosphorylated forms are inactive and degraded when bound with 14-3-3 proteins. GS1 isoforms 1 and 2 have high and low affinities for ammonium, respectively. GS1-2 isoform is involved in nitrogen remobilization [405] - [408] .
NRT1.5, the translocation and efflux protein, is involved in loading and unloading of nitrate in xylem. Gene coding for this protein is nitrate inducible but down regulated by abiotic stresses and mutation of this gene provided abiotic stress tolerance in Arabidopsis [245] [285] . The unloading of nitrate from xylem is mediated by the low-affinity transporter NRT1.8 and the expression of the gene coding for this protein is nitrate and cadmium stress inducible [286] . NRT1.9 is a phloem loader transporting nitrate downwards into roots. Arabidopsis plants with NRT1.9 mutation were found to transport more nitrate from root to shoot indicating a role for NRT1.9 in regulating nitrate levels in the shoot [292] . Cytokinin induces expression of nitrogen translocating NRT1.3, NRT1.4 genes and auxin upregulates the expression ofNRT1.3, NRT1.5, NRT1.8 genes [124] [409] . Ammonium translocation proteins are induced by nitrogen starvation. AMT3; 1 is induced by N starvation while AMT3; 2 and AMMT3; 3 are ammonium inducible. The pollen tube localized TIP1; 3 and TIP5; 1 urea transporters are associated with nitrogen translocation in pollen tubes and are regulated by pH and nitrogen limitation [331] .
12.4. Regulation of Vacuolar Nitrogen Transport
Different forms of nitrogen are stored in vacuoles with the twin aim of preventing toxicity and serving as a reserve so that it can be remobilized into cytoplasm when there is a limitation in nitrogen availability. Vacuolar nitrogen transporting CLCa and CLCb are diurnally regulated and up regulated by nitrate. Expression of vacuolar ammonium transporters TIP2; 1 and TIP2; 3 was found to be diurnally regulated in Arabidopsis roots. TIP1; 2 TIP2; 1 and TIP2; 3 proteins are ammonium inducible [309] . Ammonium supply increased the expression of these genes in a concentration dependent manner. Both nitrogen starvation and
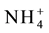
12.5. Regulation of Nitrogen Remobilization
Expression level of NRT1.7 gene, in the phloem tissues of older leaf, is correlated with the nitrate levels in these leaves. This expression was found to be induced by nitrogen starvation NRT1.6 gene is also induced under nitrogen starvation [327] [328] . NRT1.11 and NRT1.12 are companion cell localized low-affinity nitrate transporters which transport nitrate into phloem of mature leaves for redistribution to young tissues. Expression of these genes is not affected by levels of nitrate and ammonium [329] .
13. Genetic Engineering for Better Nitrogen Nutrition
Making the plant to utilize nitrogen efficiently is the primary goal of research on nitrogen metabolism which is aimed at reducing the fertilizer consumption and alleviating the environmental impact of excess nitrogen leached out. Nitrogen use efficiency is the “yield of grains per unit of available soil nitrogen” [410] . Though voluminous data has been generated in the past on nitrogen metabolism and nitrogen use efficiency, the genomic studies on nitrogen metabolism have shown that the nitrogen use efficiency is much more complex and more genes are involved than what was expected. Nitrogen use efficiency is influenced by the efficiency of the plant nitrogen uptake, efficiency of the plant in transporting nitrogen, efficiency of the plant in assimilating nitrogen, efficiency of the plant in storage (vacuolar) and efflux of nitrogen and efficiency of the plant in remobilization of nitrogen. Factors like soil nitrogen availability, photosynthetic carbon fixation, respiration and root architecture of the plant have a direct bearing of nitrogen use efficiency. Due to involvement of multiple processes in nitrogen use efficiency, it can be targeted at different levels depending on which process is limiting. Though nitrogen use efficiency is controlled by large number of genes, works carried out with individual pathways/genes have shown that targeting individual genes can alter nitrogen use efficiency of plants substantially (Table 10).
With respect to nitrogen uptake, it is not considered to be a limiting factor for improving nitrogen use efficiency because over expression studies with a number of nitrate uptake proteins did not alter the nitrogen use efficiency greatly. Over expression of Nicotiana NRT2.1 increased nitrate uptake but did not increase the overall NUE [411] . Over-expression of NRT1.1 improved growth of rice plant, but did not alter nitrogen uptake [288] [412] . Manipulation of nitrate assimilation genes also did not alter nitrogen use efficiency much. Though over expression of nitrate reductase and nitrite reductase enzymes increased the N uptake, no effect on growth and yield was seen [413] - [416] . Over expression of a hexose transporter STP13 resulted in increased expression of NRT2.2 leading to increased nitrate uptake and plant growth [417] .
Though manipulation of nitrate uptake and assimilation genes did not have serious effect on NUE, engineering ammonium uptake genes and ammonium assimilation genes have significant effect on nitrogen use efficiency. Over expression of lotus AMT1; 1 and AMT1; 3 genes in Nicotiana increased ammonium uptake but decreased the growth of the plant [57] . Over-expression of AMT1; 1 gene in rice under nitrogen-limiting condi-
Table 10. Genetic engineering for nitrogen nutrition in plants.
tions increased

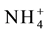
The nitrogen entering the plant through the uptake route has to be utilized immediately or translocated to other parts of the plant to meet the needs of the physiological and developmental processes. Assimilated nitrogen is also transported as amino acids [427] . Nitrogen is translocated mostly as nitrate and ammonium is translocated at very low concentrations [54] . Very few studies are available on manipulating the genes associated with translocation of nitrogen. In rice, over expression of NRT2.3b has been found to increase the nitrogen uptake and plant yield.
Vacuolar nitrogen storage is used by the plant as a buffer to balance the nitrogen level, prevent toxicity and as a source for remobilization for seed formation. Over expression of vascular H+-pyrophosphatase enhanced the enzyme activity and improved plant growth [428] . Over expression of Arabidopsis NRT2.7 resulted in enhanced seed nitrate content and improved seed germination.
The efficiency of nitrogen remobilization from vegetative tissues to reproductive tissues influences the seed development. Remobilization is a normal process happening at all stages of plant growth by which nitrogen in older leaves are mobilized to younger leaves. A huge quantity of nitrogen is stored in vegetative tissues in photosynthetic proteins (rubisco and PEFc) and GS. Rubisco alone accounts for close to 50% of store proteins in leaves [45] [95] [429] . Plants store substantial quantity of nitrate in their vacuoles and it is mobilized to seeds through phloem. Proteins associated with transport of nitrate through the tonoplast of plasma membranes to apoplast are yet to be identified.
Microarray and transcriptome studies on senescence have shown that there are many genes induced upon senescence some of which are associated with senescence induced remobilization. These genes include some isoforms of GS1, GDH, AS and proteases [427] . Over-expression of GS1 gene in lotus induced remobilization and early onset of senescence [430] . Nitrogen is remobilized as amino acids, glutamine, asparagine, glutamate and aspartame. Asparagine synthetase and aspartate aminotransferase are involved in conversion of glutamine to aspartame and glutamate to aspartate, respectively [228] [431] . Over expression ASN1 gene in Arabidopsis enhanced the soluble and total protein content of the plant, and facilitated better growth on N-limiting medium [136] [282] .
14. Genomics of Nitrogen Metabolism
14.1. Plant Nitrome (Nitro (Gen)ome)
The first important contribution of genomics to nitrogen nutrition is that it revealed the number of genes associated with different aspects of nitrogen metabolism (Table 11). Analysis of complete genomes revealed that Arabidopsis and rice contain 66 and 84 nitrate transporter genes, respectively [164] . Among the 66 Arabidopsis genes, 53 code for NRT1 transporters, 7 genes code for NRT2 transporters and 6 genes code for CLC transporters. In rice 80 genes code for NRT1 transporters and NRT2 transporters are coded by 4 genes. Though 5 genes coding for CLC transporters have been found in rice, they are found to be incapable of nitrate transport due to absence of conserved proline residues [154] [288] [432] .
Some of the NRT1 transporters are present in multiple copies. For example, three NRT1.1 genes are found in rice and sorghum, four in maize and Brachypodium. Single genes code for NRT1.2 transporter in Arabidopsis and grasses, three genes in cucumber and two genes in V. vinifera and Poplar. NRT1.3 is present in single copy in Arabidopsis, poplar and grass species but rice, maize and grape contains two copies of NRT1.3 in their genomes. NRT1.4 is present in two copies in rice maize, grape and cucumber genomes. NRT1.4 like gene is absent in poplar. Both NRT1.5 and NRT1.8 are coded by multiple genes in Brachypodium, cucumber, grape, maize, rice and sorghum NRT1.6 and NRT1.7 are coded by single genes in Brachypodium, poplar and rice. No NRT1.6 and NRT1.7 genes were found in Cucumber, grape, maize and sorghum genomes [305] [433] [434] .
There are six, twelve and fourteen genes coding for AMT proteins in Arabidopsis, rice and P. trichocarpa, respectively [167] [169] [435] . Presence of multiple genes coding for multiple forms of ammonium transporting proteins facilitate functional specialization through regulatory flexibility [436] . P. trichocarpa consists of 6 AMT1 and 8 AMT2 proteins [171] . Rice DUR3 gene codes for a 721 residue long membrane-protein and showed similar expression and regulation patterns as that of Arabidopsis DUR3 gene [239] [240] .
14.2. Genes Influenced by Nitrogen
The second contribution of genomics to nitrogen nutrition is that it revealed for the first time the number of genes
Table 11. Nitrome composition of plants.
influenced by nitrogen in crop plants. Plant transcriptome in response to nitrogen nutrition, level of nitrogen and tissue specific effects have been studied which identified large number of genes modulated by nitrogen (Table 12) [47] [283] [352] [437] - [439] . Microarray based gene expression profiling and RNA-seq analysis has been carried out in a). Nitrogen starvation conditions, b). Nitrogen limitation conditions c). In the presence of one or more nitrogen sources and d) different nitrogen and carbon combinations.
14.2.1. Nitrogen Starvation Conditions
Under nitrogen starvation conditions in Arabidopsis 509 genes were significantly differentially expressed. In roots up to 159 genes were significantly up regulated and up to 166 genes were significantly down-regulated. In shoots up to 104 genes were significantly up regulated whereas up to 80 genes were significantly down-regu- lated. These genes are associated with molecular transport, molecular metabolism, regulation and modification, organism development, stress stimuli and electron transport. In rice, 3518 genes are differentially expressed upon nitrogen starvation of which 462 genes showed differential expression in roots and shoots. In the same study a comparative study has been done between rice and Arabidopsis and shown that seventy three genes are expressed similarly in Arabidopsis and rice upon nitrogen starvation. Genes coding for miR399 and miR530 are differentially expressed upon nitrogen starvation [440] . RNA-Seq analysis of nitrogen-deficient cucumber plants showed that 196 genes associated with photosynthesis and cell wall metabolism are down regulated while 168 genes associated with antioxidant synthesis, flavonoid and phenylpropanoid biosynthesis are up regulated [441] .
14.2.2. Nitrogen Limitation Conditions
Both microarray and recent RNA-Seq experiments have shown that under nitrogen-limiting conditions increased expression of high affinity nitrogen transporter genes were seen while under nitrogen-sufficient conditions expression of low affinity nitrogen transporters was increased [442] .
Arabidopsis plants were grown in N-limiting (3 mM) and N-sufficient nitrate conditions (10 mM). In this study wild type plants and NLA mutant transcriptomes under different nitrate conditions were compared (nla mutants are plants with mutation in NLA gene which helps to cope with nitrogen limiting conditions). At early developmental stages wild type plants and NLA mutants under 3 mm and 10 mm nitrate conditions exhibited same transcriptome profiles but late developmental stages the expression profiles are different. Under nitrogen
Table 12. Transcriptome studies on nitrogen nutrition of in plants.
limitation conditions in wild type plants the expression 629 genes is altered. Among 629 genes, 340 genes involved in protein degradation, ubiquitin pathway gens and the biosynthesis of anthocyanin and phenylpropanoids are up-regulated and 289 genes involved in photosynthesis, signal transduction, hormone response, aminoacid and nucleotide biosynthesis genes and transcriptional regulation are down-regulated [443] (Under N-limiting conditions, NLA mutants altered the expression of 1744 genes, 1122 genes were up-regulated and 622 repressed. Nitrogen limitation conditions up regulates the expression of CLC-c protein while down regulates nitrate transporting CLC-a proteins in wild type plants [443] . Rice plants were transferred from high nitrogen (1.44 mM NH4NO3) to low nitrogen (0.24 mM NH4NO3) conditions to check the effect of nitrate on gene expression [444] . 471 genes showed differential expression upon nitrogen stress in the rice at seedling stage out of which 115 genes are up regulated and 358 genes are down regulated. Among down regulated genes are those associated with photosynthesis and energy metabolism while genes coding for some transcription factors, early stress response and signal transduction are up regulated. Expression of genes associated nitrogen uptake and assimilation is not altered [444] .
14.2.3. Nitrogen Addition Conditions
In Arabidopsis transcriptome study with 1 mM nitrate has induced 550 genes within 20 minutes. Nitrate addition induces genes associated with ribosomal proteins and oxidative pentose phosphate pathway genes and many other metabolic process associated genes. Ribosomal proteins and oxidative pentose phosphate pathway genes, regulate both nitrogen transport (NRT1.1, NRT1.5, NRT2.1, NRT2.4 and AMT1; 1) and assimilation. Among 550 genes, there are 146 genes which are constitutively induced upon nitrate addition, 120 genes are repressed by nitrate treatment and rest all genes are induced at different time courses. The early nitrate response on gene expression is mainly because of nitrate itself, but the delayed or prolonged nitrate response on the gene expression is due to the interaction between nitrate signalling and hormone signalling pathways [381] . In another study of Arabidopsis, plants were grown under both low (0.25 Mm) and high nitrate (5 Mm) condition and their transcriptome profiles were compared. At low nitrate concentration the expression of nitrogen transport, assimilation and metabolism genes were transiently increased compared to high nitrate conditions. In tomato resupply of nitrate to the nitrate starved plants increased the expression of 97 genes [445] [446] . In Arabidopsis nitrate addition to the nitrate starved plants significantly alters the expression of many genes involved in several metabolic processes within 30 min [282] .
RNA-Seq analysis of nitrogen deprived/treated plants have identified additional genes associated with nitrogen metabolism [441] [447] - [450] . Arabidopsis root transcriptome sequencing has revealed 40 novel nitrate responsive genes and a unique nitrate responsive microRNA5640/PPC3 module which regulates carbon and nitrogen balance and nitrate assimilation [449] . Hybrid vigour in terms of increased nitrogen transport and content under nitrogen limiting conditions in maize when compared to the parental strains is attributed to differential expression of 643 and 725 genes in leaves and roots, respectively [447] .
The number and type of genes influenced by nitrogen and their expression levels depend on the genotype of the plant. RNA-Seq analysis of two parental lines and the hybrid line in maize showed that the response of the plant to nitrogen starvation is genotype dependent. For example under nitrogen limiting conditions there was reduction in carbohydrate metabolic process due to reduction in expression of photosynthesis associated genes in SRG100 parental line but in the hybrid (SRG150) and the second parental line (SRG200), photosynthetic genes are not significantly down-regulated and they maintained significantly higher sugar content. Similarly, increased nitrogen transport under nitrogen limiting conditions was seen in both hybrid and SRG200 parental line which was not there in the other parental line [447] . Comparative RNA-Seq analysis of low nitrogen-tolerant and nitrogen-sensitive genotypes of Sorghum showed that they respond differently to N-limitation. Tolerant genotypes showed enhanced expression of genes coding for high affinity nitrate transporters and nitrate assimilation genes to enhance the plant nitrogen levels. While the sensitive genotypes showed enhanced expression of genes associated with flavonoid biosynthesis, cell wall metabolism, protein kinases and different transcription factors associated with auxin metabolism which is a sign of adaptation to low nitrogen levels.
14.3. Link between Nitrogen and Carbon Metabolism
The carbon and nitrogen metabolisms are complex and regulated by nitrate, ammonium, amino acids and signals from C metabolism. Nitrogen availability influences plant photosynthesis rate and yield [451] [452] . The integration of carbon metabolism with nitrogen mainly occurs at nitrogen assimilation and regulation of nitrogen metabolism. The nitrogen assimilation produces glutamine which is the primary source of nitrogen and carbon for amino acids [255] . C/N balance is regulated by senescence, which is dependent on nitrogen and carbon interactions [453] [454] . Maintaining an adequate photosynthetic rate under nitrogen starvation is necessary for keeping reduced C levels which favours efficient use of nitrogen [447] .
Proteins which can sense intracellular carbon and nitrogen levels are associated with maintenance and regulation of the integration of carbon and nitrogen metabolism [455] . PII protein and glutamate receptors are involved in nitrogen sensing. NRT2.1, TOR pathway, SNF1/AMP-dependent kinase, the ubiquitin ligases ATL31 and ATL6 and trehalose-6-phosphateare specific to internal carbon status sensors [456] - [459] . Transcription factors like Dof1, methyl transferase OSU1, GABA (γ-amino butyric acid), CCA1 and bZIP1 and plant hormones cytokinin and abscisic acid also play roles in signaling carbon and nitrogen status [402] [460] - [464] . GNC and CGA1 protein genes are inducible by nitrate and cytokinin and are also involved in chloroplast biogenesis [465] - [467] . Cytokinin mediates the enhanced photosynthesis by stimulation chloroplast biogenesis through GNC and CGA1 in a nitrate induced manner. The C/N balance influences plant growth in many ways, regulating lateral root growth, seed germination and cell wall biogenesis. NRT2.1 acts a sensor of high carbon to nitrogen response during formation of lateral roots. Methyltransferase (oversensitive to sugar (OSU1) negatively regulates both high and low carbon to nitrogen responses [468] .
Genome wide expression studies under different C/N condition have shown that, carbon sources such as sugars regulate the expression of nitrate, ammonium and urea transporters and also enzymes involved in nitrogen assimilation [361] . Even nitrogen sources (such as nitrate) have been shown to affect the expression of carbon metabolism genes [439] . Nitrogen modulates the expression of 8% of the glucose regulated genes. Some genes are up regulated by nitrogen and some other genes are down regulated by nitrogen. Carbon and nitrogen alone can repress or induce specific genes but when they are together they cannot do the same [455] [469] . Carbon/nitrogen balance is maintained by genes associated with hormone pathways, metabolism, signal transduction and protein synthesis and degradation.
14.4. Link between Senescence and Nitrogen Availability
Many earlier studies have shown that there is a direct connection between nitrogen depletion and early leaf senescence and the reversal of senescence upon resupply of nitrogen [470] [471] . But the actual mechanism behind this was not known until detailed transcriptome analysis was carried out, which revealed the participation of large number of genes [472] [473] . Senescence down regulates photosynthetic genes and up regulates genes associated with degradation and recycling of metabolic resources and regulatory genes [474] [475] .
14.5. Evolution of Nitrogen Metabolism Genes
Genome wide analysis for nitrogen transporters have revealed that the high affinity transport is mediated by fewer numbers of transporters and the genes coding for them are present in fewer copies when compared to low affinity nitrogen transporters. For example in rice and Arabidopsis there are only four and seven genes coding for NRT2 proteins (high affinity) while NRT1 proteins (low affinity) are coded by 53 and 80 genes in Arabidopsis and rice respectively. Compared to nitrate transporters, the ammonium transporters are coded by fewer genes. In Arabidopsis and rice there are only six and twelve ammonium transporters respectively when compared to 66 and 84 nitrate transporter genes in Arabidopsis and rice respectively. But there is no clear trend with respect to genes coding for low and high affinity ammonium transporters. In Arabidopsis there are five high affinity AMT1 transport genes and in rice three genes code for AMT1 protein. In rice the low affinity AMT2 is coded by nine genes while in Arabidopsis AMT2 is coded by a single gene. With respect to urea transporters, the high affinity DUR3 is coded by a single gene in rice, what, maize and Arabidopsis, while low affinity transporter coding genes are many.
Gene(s) coding for different nitrate transporters show different levels of functional specialization in different hosts. Some of them are coded by single genes in some plants and multiple genes in some other plants. Multiple genes coding for nitrate transporters allow a greater regulatory flexibility and tissue specificity. In some plants additional regulatory flexibility is enabled by the presence of multiple transporter proteins performing similar function. The multifunctional nitrate transporter NRT1.1 is coded by multiple genes in rice (3), Sorghum (3), maize (4) and Brachypodium (4) but the expression pattern of individual copies is not known [305] . Though rice NRT1.1A and NRT1.1B genes are expressed throughout the plant, OsNRT1.1A is predominantly expressed in the root. Though NRT1.2 is coded by single genes in Arabidopsis and grasses, it is coded by multiple genes in cucumber (3), grape (2) and poplar (2). In poplar the two copies, NRT1.2A and NRT1.2B show functional specialization and express specifically in leaves and roots, respectively. In Cucumber though all three copies are expressed only in the shoots (cotyledons and old leaves), NRT1.2B is additionally found to express specifically in young petioles [305] .
NRT1.3 is coded by single genes in Arabidopsis and many grass species and two genes in rice, maize, poplar and grape. But the expression pattern of multiple genes is not known. NRT1.4 gene is present in two copies in rice maize, grape and cucumber genomes. No NRT1.4 homolog could be found in poplar. In cucumber NRT1.4A is specifically expressed only in young and old petioles, while NRT1.4B is expressed in cotyledons and leaves [305] [434] . NRT1.5 is coded by multiple genes in poplar (3), rice (2), maize (2), sorghum (2), Brachypodium (2), grape (2) and cucumber (3) (Both in rice and cucumber NRT1.5A gene is expressed all over the plant while NRT1.5B in rice is expressed in panicle. In cucumber both NRT1.5B and NRT1.5C genes are expressed only in the reproductive tissues. The functional specialization of multiple copies of NRT1.8 genes in cucumber (3) and poplar (2) is not known [434] .
In addition to nitrate remobilization specific transporters, NRT1.6 and NRT1.7 were found to be involved innitrate remobilization and seed development in some plants (Arabidopsis, Brachypodium, poplar and rice) but absent in others (maize, sorghum, grape and cucumber) [305] [433] . Absence of these transporters in some plants indicates that they represent species specific functional specialization. Arabidopsis thaliana contains additional nitrate transporters NAXT1 (nitrate efflux), NRT1.9 (downward transport of nitrate through phloem), NRT1.10 (seed growth), NRT1.11 and NRT1.12 (nitrate remobilization from leaves). Cucumber also contains NRT1.9 and NRT1.10 genes. Poplar contains 4 genes coding for NRT1.9 transporter [433] . As there is no information regarding the presence of these transporters in other plants, probably these transporters represent Arabidopsis specific functional specialization.
Genome-wide analysis of NRT2 genes in monocots and dicots have revealed that NRT2 genes of dicots are intronic while the grass NRT genes are devoid of introns indicating that NRT2 genes must have developed after divergence of monocots and dicots [234] . NRT2.1 and NRT2.2 proteins are coded by single genes in many plants. No homologs to these genes could be found in poplar genome [432] . In rice NRT2.3 is coded by 2 genes, OsNRT2.3a and OsNRT2.3b are single exonic while in Arabidopsis a single copy of NRT2.3 is present and contains three exons. Though NRT2.3a and NRT2.3b genes of rice shared the same coding sequences, their untranslated regions (UTRs) are different. NRT2.3 coding genes have shown rice specific functional specialization, NRT2.3a gene showed root specific expression while NRT2.3b showed shoot specific expression [287] [288] . NRT2.3 showed nitrate inducible expression in shoots while in roots it is constitutive [232] .
NRT2.4 high affinity nitrate uptake and remobilization transporter is coded by single genes in Arabidopsis and rice. In Poplar NRT2.4 is coded by 2 genes. Poplar NRT2.4A is expressed in roots, leaves and wood, while the expression pattern of NRT2.4B is not known [433] . Grass NRT2 transporters are intron less but rice NRT2.4 has an intron and similar to Arabidopsis NRT2.7 gene [154] [432] . NRT2.4 and NRT2.3b proteins are functional without NAR2 [288] .
In Arabidopsis, Maize and Barley NRT2.5 is coded by a single gene while in poplar NRT2.5 is coded by 3 genes. Poplar NRT2.5A is expressed in the bark, NRT2.5B is expressed in leaves and the expression of NRT2.5C is not known [433] [476] . Nitrogen starvation induced the expression of NRT2.5 gene while supply of nitrogen (ammonium/nitrate) repressed the gene [232] . NRT2.5 is involved in plant growth promotion under the rhizospheric bacterium STM196 [477] . NRT2.6 is constitutive high affinity transporter involved in pathogen resistance in plants by producing reactive oxygen species and is coded by a single gene in Arabidopsis and barley [478] . NRT2.7 like transporters is specific to dicots and not found in grass species. NRT2.7 is coded by a single gene in Arabidopsis and poplar and is involved in vacuolar nitrate accumulation in the seeds and also oxidation of proanthocyanindin in seeds [303] [479] . In poplar NRT2.7 is expressed in the wood and leaves and maintains nitrate homeostasis in the leaves [433] .
15. Biotechnological Options for Improving Nitrogen Nutrition in Crop Plants
Extensive research data is available on the genes/proteins associated with nitrogen nutrition of crop plants and their regulation and these data can be used for generating inter plant species/crop variations which have little practical significance for biotechnological interventions as differences are known to exist between plant species which are totally different genetic entities. What is needed is generations of high throughput data using modern genomic approaches which address inter cultivar variations on nitrogen use efficiency. Research has just entered this imported arena and by next few years we should have enough data on inter cultivar variations at genomic/ proteomic levels. Availability of the genomes of cultivars and cultivar specific transcriptome and microRNA profiling data will enables us to understand the variations/lack of variations that facilitate better nitrogen utilization by specific high yileding crop cultivars. If such data are available we can employ biotechnological options like genetic engineering/gene silencing etc to improve the nitrogen utilization efficiency of crop plants. Another area of interest for biotechnological options would be to delink the root growth from nitrogen concentration in soil. Root growth has been found to be influenced by the nitrogen content of soil, inhibition/induction of root growth by high soil nitrogen content is a problem for crop plants which are grown in wide range of nitrogen concentrations. If higher nitrogen content inhibits lateral root growth it will have serious implications on mild drought conditions as the roots are shallow and cannot harvest waters from deep layers. More data is needed for understanding the link between soil nitrogen content and root growth which will facilitate delinking the two processes.
16. Conclusions
Having achieved substantial production increase in crop plants by applying nitrogenous fertilizers, it is essential for assessing the fertilizer use and the ability of the plant to utilize the applied fertilizer. Though more than 57.5 million tons of nitrogen fertilizer is applied each year for cereals alone, only 33 percent of it is utilized by the plant and the rest is lost. A deep understanding of the nitrogen nutrition is necessary for overcoming this loss.
Recent progress in genomics and genetic engineering studies of crop and other plants have revealed the composition of nitrome and the genes influenced by presence or absence of different forms of nitrogen.
Sensing the levels of nitrogen, external and internal, influences nitrogen nutrition greatly as expression/action of many genes involved in nitrogen nutrition is influenced by the level of nitrogen. Though the mechanism of sensing by the dual affinity transporter NRT1.1 is understood, the mechanism of sensing by NRT2.1 is not known. External ammonium sensing by AMT1; 1 and AMT1; 3 are understood but very little information is available with respect to sensing external urea levels. Despite the clear understanding of mechanisms of internal nitrogen sensing in bacteria and fungi and existence of homologues of these proteins in plants, their mechanism of sensing is not understood in plants.
Though plants face severe problems when nitrogen level is low, to overcome this plants have not developed more number of low affinity transporters when compared to high affinity nitrate transporters, indicating that the high affinity transporters are very efficient. The high number of low affinity transporters may not be due to selection of crop plants to suit high nitrogen conditions as non crop plants also have more number of low affinity transporters. This shows that nitrogen uptake may not be the limiting factor and the processes downstream to uptake need to be looked into for addressing efficiency in nitrogen nutrition.
Plants do possess mechanisms which favour efflux of nitrogen outside the plant for which evidences are there for nitrate and ammonium but so far no proof exists for efflux of urea. The mechanism of nitrate efflux by NAXT1 is understood, the role played by bidirectional transporters NRT1.1, NRT1.4 and NRT1.5 in efflux is not clear. Though ammonium efflux is known, the proteins involved and their mechanism of action are yet to be understood. Though the mechanisms of translocation and assimilation of different forms of nitrogen is understood, the regulation of these processes under differing nitrogen levels is yet to be understood. Over expression of nitrogen assimilation genes (GS, NADH-GOGAT, ASN and alanine amino transferase) increased the nitrogen use efficiency. This shows that probably the nitrogen assimilation is the limiting factor for understanding the nitrogen nutrition. Storage of different forms of nitrogen in vacuole might be playing an important role in maintaining nitrogen status in different tissues. The transporters associated with this have been studied in detail but the existence of variation in levels of accumulation of different forms nitrogen in different crop varieties and their implication on overall nitrogen use efficiency needs detailed examination.
Recent genomic studies have established the correlation between remobilization and senescence. Integrated regulation of these two processes merit detailed investigation. Correlation between soil nitrogen status and root growth is well understood. Further understanding of this process may facilitate delinking root growth from soil nitrogen level which is desirable for developing crop varieties to suit different soil nitrogen conditions. The influence of carbon level in relation to nitrogen level on nitrogen nutrition is revealed by recent transcriptome studies, understanding the regulation of which requires further investigation.
Overall, we have understood the machinery (nitrome) associated with nitrogen nutrition and the mechanism of functioning. But the major lacunae is the poor understanding of regulation of nitrogen nutrition in which many transcription factors and small RNAs are likely to be involved. Comparative genomics and transcriptomics studies in crop plants of differing genetic background are essential for this purpose.
Acknowledgements
Financial Assistance from Department of Science and Technology, Government of India through DST-PURSE programme to Osmania University is acknowledged. Madhavi Reddy is supported by Junior Research Fellowship from Department of Biotechnology, Government of India under DBT-OU-ISLARE programme.
Cite this paper
Mettu MadhaviReddy,KandasamyUlaganathan, (2015) Nitrogen Nutrition, Its Regulation and Biotechnological Approaches to Improve Crop Productivity. American Journal of Plant Sciences,06,2745-2798. doi: 10.4236/ajps.2015.618275
References
- 1. Marschner, H. (2012) Mineral Nutrition of Higher Plants. 5th Edition, Academic Press, London.
- 2. Mengel, K., Kirkby, E.A., Kosegarten, H.A. and Appel, T. (2001) Principles of Plant Nutrition. 5th Edition. Kluwer Academic, Dordrecht.
http://dx.doi.org/10.1007/978-94-010-1009-2 - 3. Sanhueza, E. (1982) The Role of the Atmosphere in Nitrogen Cycling. Plant and Soil, 67, 61-71.
http://dx.doi.org/10.1007/BF02182755 - 4. Bluementhal, J., Baltensperger, D., Cassman, K.G., Mason, S. and Pavlista, A. (2008) Importance and Effect of Nitrogen on Crop Quality and Ealth. In: Hatfield, J.L. and Follett, R.F., Eds., Nitrogen in the Environment: Sources, Problems, and Management, Elsevier, Amsterdam, 51-70.
- 5. Kraiser, T., Gras, D.E., Gutierrez, A.G., Gonzalez, B. and Gutierrez, R.A. (2011) A Holistic View of Nitrogen Acquisition in Plants. Journal of Experimental Botany, 62, 1455-1466.
http://dx.doi.org/10.1093/jxb/erq425 - 6. Cassman, K.G. (1999) Ecological Intensification of Cereal Production Systems: Yield Potential, Soil Quality, and Precision Agriculture. Proceedings of the National Academy of Sciences of the United States of America, 96, 5952-5959.
http://dx.doi.org/10.1073/pnas.96.11.5952 - 7. Delmer, D. (2005) Agriculture in the Developing World: Connecting Innovations in Plant Research to Downstream Applications. Proceedings of the National Academy of Sciences of the United States of America, 102, 15739-15746.
http://dx.doi.org/10.1073/pnas.0505895102 - 8. International Fertilizer Industry (2013) Assessment of Fertilizer Use by Crop at the Global Level 2010-2010/11.
Http://www.Fertilizer.Org/Ifa/Statistics.Asp - 9. Camarguo, J.A. and Alonso, A. (2006) Ecological and Toxicological Effects of Inorganic Nitrogen Pollution in Aquatic Ecosystems: A Global Assessment. Environment International, 32, 831-849.
http://dx.doi.org/10.1016/j.envint.2006.05.002 - 10. Choudhury, A.T.M.A. and Kennedy, I.R. (2005) Nitrogen Fertilizer Losses from Rice Soils and Control of Environmental Pollution Problems. Communications in Soil Science and Plant Analysis, 36, 1625-1639.
http://dx.doi.org/10.1081/CSS-200059104 - 11. Gruber, N. and Galloway, J.N. (2008) An Earth System Perspective of the Global Nitrogen Cycle. Nature, 451, 293-296.
http://dx.doi.org/10.1038/nature06592 - 12. Ladha, J.K., Pathak, H., Krupnik, T.J., Six, J. and Vankessel, C. (2005) Efficiency of Fertilizer Nitrogen in Cereal Production: Retrospects and Prospects. Advances in Agronomy, 87, 85-156.
http://dx.doi.org/10.1016/S0065-2113(05)87003-8 - 13. Schlesinger, W.H. (2009) On the Fate of Anthropogenic Nitrogen. Proceedings of the National Academy of Sciences of the United States of America, 106, 203-208.
http://dx.doi.org/10.1073/pnas.0810193105 - 14. Sylvester-Bradley, R. and Kindred, D.R. (2009) Analysin Nitrogen Responses of Cereals to Prioritize Routes to the Improvement of Nitrogen Use Efficiency. Journal of Experimental Botany, 60, 1939-1951.
http://dx.doi.org/10.1093/jxb/erp116 - 15. Tilman, D., Cassman, K.G., Matson, P.A., Naylor, R. and Polasky, S. (2002) Agricultural Sustainability and Intensive Production Practices. Nature, 418, 671-677.
http://dx.doi.org/10.1038/nature01014 - 16. Vitousek, P.M., Naylor, R., Crews, T., David, M.B., Drinkwater, L.E. and Holland, E. (2009) Agriculture: Nutrient Imbalances in Agricultural Development. Science, 324, 1519-1520.
http://dx.doi.org/10.1126/science.1170261 - 17. Ahlgren, S., Baky, A., Bernesson, S., Nodberg, A., Noren, O. and Hansson, P.A. (2008) Ammonium Nitrate FertilizerProduction Based on Biomass—Environmental Effects from a Life Cycle Perspective. Bioresource Technology, 99, 8034-8041.
http://dx.doi.org/10.1016/j.biortech.2008.03.041 - 18. Conley, D.J., Paerl, H.W. and Howarth, R.W. (2009) Controlling Eutrophication: Nitrogen and Phosphorus. Science, 323, 1014-1015.
http://dx.doi.org/10.1126/science.1167755 - 19. Engstrom, R., Wadeskog, A. and Finnveden, G. (2007) Environmental Assessment of Swedish Agriculture. Ecological Economics, 60, 550-563.
http://dx.doi.org/10.1016/j.ecolecon.2005.12.013 - 20. Giles, J. (2005) Nitrogen Study Fertilizes Fears of Pollution. Nature, 433, 791.
http://dx.doi.org/10.1038/433791a - 21. Smith, P., Martino, D. and Cai, Z. (2008) Greenhouse Gas Mitigation in Agriculture. Philosophical Transactions of the Royal Society, B363, 789-813.
http://dx.doi.org/10.1098/rstb.2007.2184 - 22. Zhang, W.F., Zheng, X.D., Pan, H., Ju, X.T., Davidpowlson, D. and Chadwick, D (2013) New Technologies Reduce Greenhouse Gas Emissions from Nitrogenous Fertilizer In China. Proceedings of the National Academy of Sciences of the United States of America, 110, 8375-8380.
http://dx.doi.org/10.1073/pnas.1210447110 - 23. Beatty, P.H., Anbessa, Y., Juskiw, P., Carroll, R.T., Wang, J. and Good, A.G. (2010) Nitrogen Use Efficiencies of Spring Barley Grown Under Varying Nitrogen Conditions in the Field and Growth Chamber. Annals of Botany, 105, 1171-1182.
http://dx.doi.org/10.1093/aob/mcq025 - 24. Mcallister, C.H., Beatty, P.H. and Good, A.G. (2012) Engineering Nitrogen Use Efficient Crop Plants: The Current Status. Plant Biotechnology Journal, 10, 1011-1025.
http://dx.doi.org/10.1111/j.1467-7652.2012.00700.x - 25. White, P.J. and Brown, P.H. (2010) Plant Nutrition for Sustainable Development and Global Health. Annals of Botany, 105, 1073-1080.
http://dx.doi.org/10.1093/aob/mcq085 - 26. De Macale, M.A.R. and Velk, P.L.G. (2004) The Role of Azolla Cover in Improving the Nitrogen Use Efficiency of Lowland Rice. Plant Soil, 263, 311-321.
http://dx.doi.org/10.1023/B:PLSO.0000047742.67467.50 - 27. Hirel, B., Le Gouis, J., Ney, B. and Gallais, A. (2007) The Challenge Of Improving Nitrogen Use Efficiency in Crop Plants: Towards a More Central Role for Genetic Variability and Quantitative Genetics within Integrated Approaches. Journal of Experimental Botany, 58, 2369-2387.
http://dx.doi.org/10.1093/jxb/erm097 - 28. Lea, P.J. and Azevedo, R.A. (2006) Nitrogen Use Efficiency. 1. Uptake of Nitrogen from the Soil. Annals of Applied Biology, 149, 243-247.
http://dx.doi.org/10.1111/j.1744-7348.2006.00101.x - 29. Lea, P.J. and Azevedo, R.A. (2007) Nitrogen Use Efficiency. 2. Amino Acid Metabolism. Annals of Applied Biology, 151, 269-275.
http://dx.doi.org/10.1111/j.1744-7348.2007.00200.x - 30. Von Wiren, N., Gazzarrini, S., Gojon, A. and Frommer, W.B. (2000) The Molecular Physiology Of Ammonium Uptake And Retrieval. Current Opinion in Plant Biology, 3, 254-261.
http://dx.doi.org/10.1016/S1369-5266(00)00073-X - 31. De Willigen, P. (1986) Supply of Soil Nitrogen to the Plant during the Growing Season. In: Lambers, H., Neeteson, J.J. and Stulen, I., Eds., Fundamental, Ecological and Agricultural Aspects of Nitrogen Metabolism in Higher Plants, Martinusnijhoff, Dordrecht, 417-432.
http://dx.doi.org/10.1007/978-94-009-4356-8_62 - 32. Miller, A.J. and Cramer, M.D. (2004) Root Nitrogen Acquisition and Assimilation. Plant Soil, 274, 1-36.
http://dx.doi.org/10.1007/s11104-004-0965-1 - 33. Barber, S.A. (1995) Soil Nutrient Bioavailability: A Mechanistic Approach. 2nd Ed. John Wiley, New York.
- 34. Crawford, N.M. and Glass, A.D.M. (1998) Molecular and Physiological Aspects of Nitrate Uptake in Plants. Trends in Plant Science, 3, 389-395.
http://dx.doi.org/10.1016/S1360-1385(98)01311-9 - 35. Owen, A.G. and Jones, D.L. (2001) Competition for Amino Acids between Wheat Roots and Rhizosphere Microorganisms and the Role of Amino Acids in Plant N Acquisition. Soil Biology & Biochemistry, 33, 651-657.
http://dx.doi.org/10.1016/S0038-0717(00)00209-1 - 36. Wang, W.H., Kohler, B., Cao, F.Q. and Liu, L.H. (2008a) Molecular and Physiological Aspects of Urea Transport in Higher Plants. Plant Science, 175, 467-477.
http://dx.doi.org/10.1016/j.plantsci.2008.05.018 - 37. Gaudin, R., Dupuy, J. and Bournat, P. (1987) Suivi Du Contenue En Azote De La Solution Du Sol D’unerizière Après Placement D’urée. Agron. Trop, 42, 13-19.
- 38. Polacco, J.C. and Holland, M.A. (1993) Roles of Urease in Plant Cells. International Review of Cytology, 145, 65-103.
http://dx.doi.org/10.1016/S0074-7696(08)60425-8 - 39. Krogmeier, M.J., Mccarty, G.W. and Bremner, J.M. (1989) Phytotoxicity of Foliar-Applied Urea. Proceedings of the National Academy of Sciences of the United States of America, 86, 8189-8191.
http://dx.doi.org/10.1073/pnas.86.21.8189 - 40. Colmer, T.D. and Bloom, A.J. (1998) A Comparison of and Net fluxes along Roots of Rice and Maize. Plant Cell Environ, 21, 240-246.
http://dx.doi.org/10.1046/j.1365-3040.1998.00261.x - 41. Lee, R. and Rudge, K. (1986) Effects of Nitrogen Deficiency on the Absorption of Nitrate and Ammonium by Barley Plants. Annals of Botany, 57, 471-486.
- 42. Drew, M.C. and Saker, L.R. (1975) Nutrient Supply and the Growth of the Seminal Root System in Barley. II. Localized, Compensatory Increases in Lateral Root Growth and Rates of Nitrate Uptake When Nitrate Supply Is Restricted to Only Part of the Root System. Journal of Experimental Botany, 26, 79-90.
http://dx.doi.org/10.1093/jxb/26.1.79 - 43. Glass, A.D.M. (2003) Nitrogen Use Efficiency of Crop Plants: Physiological Constraints upon Nitrogen Absorption. Critical Reviews in Plant Sciences, 22, 453-470.
http://dx.doi.org/10.1080/07352680390243512 - 44. Gruber, B.D., Giehl, R.F.H., Friedel, S. and Von Wirén, N. (2013) Plasticity of the Arabidopsis Root System under Nutrient Deficiencies. Plant Physiology, 163, 161-179.
http://dx.doi.org/10.1104/pp.113.218453 - 45. Kant, S., Bi, Y.M. and Rothstein, S.J. (2011) Understanding Plant Response to Nitrogen Limitation for the Improvement of Crop Nitrogen Use Efficiency. Journal of Experimental Botany, 62, 1499-1509.
http://dx.doi.org/10.1093/jxb/erq297 - 46. Krapp, A., Berthomé, R., Orsel, M., Mercey-Boutet, S., Yu, A. and Castaings, L. (2011) Arabidopsis Roots and Shoots Show Distinct Temporal Adaptation Patterns Toward Nitrogen Starvation. Plant Physiology, 157, 1255-1282.
http://dx.doi.org/10.1104/pp.111.179838 - 47. Krouk, G., Crawford, N.M., Coruzzi, G.M. and Tsay, Y.F. (2010a) Nitrate Signaling: Adaptation to Fluctuating Environments. Current Opinion in Plant Biology, 13, 266-273.
http://dx.doi.org/10.1016/j.pbi.2009.12.003 - 48. Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E. and Mounier, E. (2006) The Arabidopsis NRT1.1 Transporter Participates in the Signaling Pathway Triggering Root Colonization Of Nitrate-Rich Patches. Proceedings of the National Academy of Sciences of the United States of America, 103, 19206-19211.
http://dx.doi.org/10.1073/pnas.0605275103 - 49. Remans, T., Nacry, P., Pervent, M., Girin, T., Tillard, P., Lepetit, M. and Gojon, A. (2006) A Central Role for the Nitrate Transporter NRT2.1 in the Integrated Morphological and Physiological Responses of the Root System to Nitrogen Limitation in Arabidopsis. Plant Physiology, 140, 909-921.
http://dx.doi.org/10.1104/pp.105.075721 - 50. Zhang, H. and Forde, B.G. (2000) Regulation of Arabidopsis Root Development by Nitrate Availability. Journal of Experimental Botany, 51, 51-59.
http://dx.doi.org/10.1093/jexbot/51.342.51 - 51. Ho, C.H. and Tsay, Y.F. (2010) Nitrate, Ammonium, and Potassium Sensing and Signaling. Current Opinion in Plant Biology, 13, 604-610.
http://dx.doi.org/10.1016/j.pbi.2010.08.005 - 52. Glass, A.D.M., Brito, D.T., Kaiser, B.N., Kronzucker, H.J., Kumar, A. and Okamoto, M. (2001) Nitrogen Transport in Plants, with an Emphasis on the Regulation Of Fluxes to Match Plant Demand. Journal of Plant Nutrition and Soil Science, 164, 199-207.
http://dx.doi.org/10.1002/1522-2624(200104)164:2<199::AID-JPLN199>3.0.CO;2-K - 53. Hoffmann, A., Milde, S., Desel, C., Hümpel, A., Kaiser, H. and Hammes, E. (2007) N Form-Dependent Growth Retardation of Arabidopsis thaliana Seedlings as Revealed from Physiological and Microarray Studies. Journal of Plant Nutrition and Soil Science, 170, 87-97.
http://dx.doi.org/10.1002/jpln.200625032 - 54. Schjoerring, J.K., Husted, S., Mäck, G. and Mattsson, M. (2002) The Regulation of Ammonium Translocation in Plants. Journal of Experimental Botany, 53, 883-890.
http://dx.doi.org/10.1093/jexbot/53.370.883 - 55. Loque, D., Lalonde, S., Looger, L.L., Von Wiren, N. and Frommer, W.B. (2007) A Cytosolic Trans-Activation Domain Essential for Ammonium Uptake. Nature, 446, 195-198.
http://dx.doi.org/10.1038/nature05579 - 56. Neuhäuser, B., Dynowski, M., Mayer, M. and Ludewig, U. (2007) Regulation of NH4 + Transport by Essential Cross Talk between AMT Monomers through the Carboxyl Tails. Plant Physiology, 143, 1651-1659.
http://dx.doi.org/10.1104/pp.106.094243 - 57. Rogato, A., Apuzzo, E.D. and Chiurazzi, M. (2010) The Multiple Plant Response to High Ammonium Conditions: The Lotus japonicas AMT1; 3 Protein Acts as a Putative Transceptor. Plant Signaling & Behavior, 5, 1594-1596.
http://dx.doi.org/10.4161/psb.5.12.13856 - 58. Lima, J.E., Kojima, S., Takahashi, H. and Von Wiren, N. (2010) Ammonium Triggers Lateral Root Branching in Arabidopsis in an Ammonium Transporter1; 3-Dependent Manner. Plant Cell, 22, 3621-3633.
http://dx.doi.org/10.1105/tpc.110.076216 - 59. Loqué, D., Yuan, L., Kojima, S., Gojon, A., Wirth, J., Gazzarrini, S., Ishiyama, K., Takahashi, H. and Von Wirén, N. (2006) Additive Contribution Of AMT1; 1 and AMT1; 3 to High-Affinity Ammonium Uptake across the Plasma Membrane of Nitrogen-Deficient Arabidopsis Roots. The Plant Journal, 48, 522-534.
http://dx.doi.org/10.1111/j.1365-313X.2006.02887.x - 60. Khademi, S., O’Connell 3rd, J., Remis, J., Robles-Colmenares, Y., Miercke, L.J. and Stroud R.M. (2004) Mechanism of Ammonia Transport by Amt/MEP/Rh: Structure of Amtb at 1.35 &ARING. Science, 305, 1587-1594.
http://dx.doi.org/10.1126/science.1101952 - 61. Ludewig, U., Von Wirén, N. and Frommer, W.B. (2002) Uniport Of by the Root Hair Plasma Membrane Ammonium Transporter Leamt1-1. Journal of Biological Chemistry, 277, 13548-13555.
http://dx.doi.org/10.1074/jbc.M200739200 - 62. Ludewig, U., Wilken, S., Wu, B., Jost, W., Obrdlik, P. and El Bakkoury, M. (2003) Homo- and Hetero-Oligomerization of Ammonium Transporter-1 NH4 Uniporters. Journal of Biological Chemistry, 278, 45603-45610.
http://dx.doi.org/10.1074/jbc.M307424200 - 63. Benschop, J.J., Mohammed, S., O’Flaherty, M., Heck, A.J., Slijper, M. and Menke, F.L. (2007) Quantitative Phosphoproteomics Of Early Elicitor Signaling in Arabidopsis. Molecular & Cellular Proteomics, 6, 1198-1214.
http://dx.doi.org/10.1074/mcp.M600429-MCP200 - 64. Hem, S., Rofidal, V., Sommerer, N. and Rossignol, M. (2007) Novel Subsets of the Arabidopsis Plasmalemma Phosphoproteome Identify Phosphorylation Sites in Secondary Active Transporters. Biochemical and Biophysical Research Communications, 363, 375-380.
http://dx.doi.org/10.1016/j.bbrc.2007.08.177 - 65. Lanquar, V., Loqué, D., Hörmann, F., Yuan, L., Bohner, A., Engelsberger, W.R., Lalonde, S. and Schulze, W.X. (2009) Feedback Inhibition of Ammonium Uptake by a Phospho-Dependent Allosteric Mechanism in Arabidopsis. Plant Cell, 21, 3610-3622.
http://dx.doi.org/10.1105/tpc.109.068593 - 66. Severi, A., Javelle, M. and Merrick, M. (2007) The Conserved Carboxyterminal Region of the Ammonia Channel Amtb Plays a Critical Role in Channel Function. Molecular Membrane Biology, 24, 161-171.
http://dx.doi.org/10.1105/tpc.109.068593 - 67. Yuan, L., Loque, D., Ye, F., Frommer, W.B. and Von Wiren, N. (2007a) Nitrogen-Dependent Posttranscriptional Regulation of the Ammonium Transporter Atamt1; 1. Plant Physiology, 143, 732-744.
http://dx.doi.org/10.1104/pp.106.093237 - 68. Lanquar, V. And Frommer, W.B. (2010) Adjusting Ammonium Uptake via Phosphorylation. Plant Signaling & Behavior, 5, 736-738.
http://dx.doi.org/10.4161/psb.5.6.11696 - 69. Yuan, L., Gu, R., Xuan, Y., Smith-Valle, E., Loqué, D., Frommer, W.B. and Von Wirén, N. (2013) Allosteric Regulation of Transport Activity by Hetero-Trimerization of Ammonium Transporter Complexes in Vivo. Plant Cell, 25, 974-984.
http://dx.doi.org/10.1105/tpc.112.108027 - 70. Wang, Q., Zhao, Y., Luo, W., Li, R., He, Q., Fang, X., Michele, R.D., Ast, C., Von Wirén, N. and Lin, J. (2013a) Single-Particle Analysis Reveals Shutoff Control of the Arabidopsis Ammonium Transporter AMT1; 3 by Clustering and Internalization. Proceedings of the National Academy of Sciences of the United States of America, 110, 13204-13209.
http://dx.doi.org/10.1073/pnas.1301160110 - 71. Gojon, A., Krouk, G., Perrine-Walker, F. and Laugier, E. (2011) Nitrate Transceptor(S) in Plants. Journal of Experimental Botany, 62, 2299-2308.
http://dx.doi.org/10.1093/jxb/erq419 - 72. Guo, F.Q., Wang, R., Chen, M. and Crawford, N.M. (2001) The Arabidopsis Dual-Affinity Nitrate Transporter Gene Atnrt1.1 (CHL1) Is Activated And Functions in Nascent Organ Development During Vegetative and Reproductive Growth. Plant Cell, 13, 1761-1777.
http://dx.doi.org/10.1105/tpc.13.8.1761 - 73. Ho, C.H., Lin, S.H., Hu, H.C. and Tsay, Y.F. (2009) CHL1functions as a Nitrate Sensor in Plants. Cell, 138, 1184-1194.
http://dx.doi.org/10.1016/j.cell.2009.07.004 - 74. Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K. and Mounier, E. (2010b) Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Developmental Cell, 18, 927-937.
http://dx.doi.org/10.1016/j.devcel.2010.05.008 - 75. Liu, K.H., Huang, C.Y. and Tsay, Y.F. (1999) CHL1 Is a Dual-Affinity Nitrate Transporter of Arabidopsis Involved in Multiple Phases of Nitrate Uptake. Plant Cell. 11, 865-874.
http://dx.doi.org/10.1105/tpc.11.5.865 - 76. Munos, S., Cazettes, C., Fizames, C., Gaymard, F., Tillard, P., Lepetit, M., Lejay, L. and Gojon, A.(2004) Transcript Profiling in Thechl1-5 Mutant of Arabidopsis Reveals a Role of the Nitrate Transporter NRT1.1 in the Regulation of Another Nitrate Transporter, NRT2.1. Plant Cell, 16, 2433-2447.
http://dx.doi.org/10.1105/tpc.104.024380 - 77. Wang, R., Xing, X., Wang, Y., Tran, A. and Crawford, N.M. (2009) A Genetic Screen for Nitrate Regulatory Mutants Captures the Nitrate Transporter Gene NRT1.1. Plant Physiology, 151, 472-478.
http://dx.doi.org/10.1104/pp.109.140434 - 78. Hu, H.C., Wang, Y.Y. and Tsay, Y.F. (2009) Atcipk8, a CBL-Interacting Protein Kinase, Regulates the Low-Affinity Phase of the Primary Nitrate Response. The Plant Journal, 57, 264-278.
http://dx.doi.org/10.1111/j.1365-313X.2008.03685.x - 79. Liu, K.H. and Tsay, Y.F. (2003a) Switching Between the Two Actions Modes of the Dual-Affinity Nitrate Transporter CHL1 by Phosphorylation. The EMBO Journal, 22, 1005-1013.
http://dx.doi.org/10.1093/emboj/cdg118 - 80. Batistic, O. and Kulda, J. (2009) Plant Calcineurin B-Like Proteins and Their Interacting Protein Kinases. Biochimica et Biophysica Acta, 1793, 985-992.
http://dx.doi.org/10.1016/j.bbamcr.2008.10.006 - 81. Xu, J., Li, H.D., Chen, L.Q., Wang, Y., Liu, L.L., He, L. and Wu, W.H. (2006) A Protein Kinase, Interacting with Two Calcineurin B-Like Proteins, Regulates K+ Transporter AKT1 in Arabidopsis. Cell, 125, 1347-1360.
http://dx.doi.org/10.1016/j.cell.2006.06.011 - 82. Bouguyon, E., Gojon, A. and Narcy, P. (2012) Nitrate Sensing and Signaling in Plants. Seminars in Cell & Developmental Biology, 23, 648-654.
http://dx.doi.org/10.1016/j.semcdb.2012.01.004 - 83. Little, Y.D., Rao, H., Oliva, S., Daniel-Vedel, F., Krapp, A. and Malamy, J.E. (2005) The Putative High-Affinity Nitrate Transporter NRT2.1 Represses Lateral Root Initiation in Response to Nutritional Cues. Proceedings of the National Academy of Sciences of the United States of America, 102, 13693-13698.
http://dx.doi.org/10.1073/pnas.0504219102 - 84. Lam, H.M., Chiao, Y.A., Li, M.W., Yung, Y.K. and Sang, J.I. (2006) Putative Nitrogen Sensing Systems in Higher Plants. Journal of Integrative Plant Biology, 48, 873-888.
http://dx.doi.org/10.1111/j.1744-7909.2006.00313.x - 85. Castaings, L., Camargo, A., Pocholle, D., Gaudon, V., Texier, Y., Boutet-Mercey, S., Taconnat, L., Renou, J.P., Daniel-Vedele, F. and Fernandez, E. (2009) The Nodule Inception-Like Protein 7 Modulates Nitrate Sensing and Metabolism in Arabidopsis. The Plant Journal, 57, 426-435.
http://dx.doi.org/10.1111/j.1365-313X.2008.03695.x - 86. Rubin, G., Tohge, T., Matsuda, F., Saito, K. and Scheible, W.R. (2009) Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell, 21, 3567-3584.
http://dx.doi.org/10.1105/tpc.109.067041 - 87. Zhang, H. and Forde, B.G. (1998) An Arabidopsis MADS Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science, 279, 407-409.
http://dx.doi.org/10.1126/science.279.5349.407 - 88. Gan, Y., Bernreiter, A., Filleur, S., Abram, B. and Forde, B.G. (2012) Over Expressing the ANR1 MADS-Box Gene in Transgenic Plants Provides New Insights into Its Role in the Nitrate Regulation of Root Development. Plant & Cell Physiology, 53, 1003-1016.
http://dx.doi.org/10.1093/pcp/pcs050 - 89. Marchive, C., Roudier, F., Castaings, L., Bréhaut, V., Blondet, E., Colot, V., Meyer, C. and Krapp, A. (2013) Nuclear Retention of the Transcription Factor NLP7 Orchestrates the Early Response to Nitrate in Plants. Nature Communications, 4, 1713.
http://dx.doi.org/10.1038/ncomms2650 - 90. Rueda-Lopez, M., Crespillo, R., Canovas, F.M. and Avila, C. (2008) Differential Regulation of Two Glutamine Synthetase Genes by a Single Dof Transcription Factor. The Plant Journal, 56, 73-85.
http://dx.doi.org/10.1111/j.1365-313X.2008.03573.x - 91. Yanagisawa, S., Akiyama, A., Kisaka, H., Uchimiya, H. and Miwa, T. (2004) Metabolic Engineering with Dof1 Transcription Factor in Plants: Improved Nitrogen Assimilation and Growth under Low-Nitrogen Conditions. Proceedings of the National Academy of Sciences of the United States of America, 101, 7833-7838.
http://dx.doi.org/10.1073/pnas.0402267101 - 92. Archondeguy, T., Jack, R. and Merrick, M. (2001) PII Signal Transduction Proteins: Privotal Players in Microbial Nitrogen Control. Microbiology and Molecular Biology Reviews, 65, 80-105.
http://dx.doi.org/10.1128/MMBR.65.1.80-105.2001 - 93. Hsieh, M.H., Lam, H.M., Van De Loo, F.J. and Coruzzi, G.A. (1998) PII-Like Protein in Arabidopsis: Putative Role in Nitrogen Sensing. Proceedings of the National Academy of Sciences of the United States of America, 95, 13965-13970.
http://dx.doi.org/10.1073/pnas.95.23.13965 - 94. Ninfa, A.J. and Atkinson, M.R. (2000) PII Signal Transduction Protein. Trends in Microbiology, 8, 172-179.
http://dx.doi.org/10.1016/S0966-842X(00)01709-1 - 95. Nunes-Nesi, A., Fernie, A.R. and Stitt. M. (2010) Metabolic and Signaling Aspects Underpinning the Regulation of Plant Carbon Nitrogen Interactions. Molecular Plant, 3, 973-996.
http://dx.doi.org/10.1093/mp/ssq049 - 96. Schachtman, D. and Shin, R. (2007) Nutrient Sensing and Signaling: NPKS. Annual Review of Plant Biology, 58, 47-69.
http://dx.doi.org/10.1146/annurev.arplant.58.032806.103750 - 97. Sugiyama, K., Hayakawa, T., Kudo, T., Ito, T. and Yamaya, T. (2004) Interaction of N-Acetylglutamate Kinase with a PII-Like Protein in Rice. Plant & Cell Physiology, 45, 1768-1778.
- 98. Smith, C.S., Weljie, A.M. and Moorhead, G.B.G. (2003) Molecular Properties of the Putative Nitrogen Sensor PII from Arabidopsis thaliana. The Plant Journal, 33, 353-360.
http://dx.doi.org/10.1046/j.1365-313X.2003.01634.x - 99. Uhrig, R.G., Ng, K.K. and Greg, B.G.M. (2009) PII in Higher Plants: A Modern Role for an Ancient Protein. Trends in Plant Science, 14, 501-511.
http://dx.doi.org/10.1016/j.tplants.2009.07.003 - 100. De Marsac, N.T., Lee, H.M., Hisbergues, M., Castets, A.M. and Bedu, S. (2001) Control of Nitrogen and Carbon Metabolism in Cyanobacteria. Journal of Applied Phycology, 13, 287-292.
http://dx.doi.org/10.1023/A:1017518530599 - 101. Kobayashi, M., Takatani, N., Tanigawa, M. and Omata, T. (2005) Posttranslational Regulation of Nitrate Assimilation in the Cyanobacterium Synechocystis sp. Strain PCC 6803. Journal of Bacteriology, 187, 498-506.
http://dx.doi.org/10.1128/JB.187.2.498-506.2005 - 102. Stitt, M. and Krapp, A. (1999) The Interaction between Elevated Carbon Dioxide and Nitrogen Nutrition: The Physiological and Molecular Background. Plant Cell and Environment, 22, 583-621.
http://dx.doi.org/10.1046/j.1365-3040.1999.00386.x - 103. Li, M.W. and Lam, H.M. (2008) Searching for Nitrogen Sensing Systems in Higher Plants. Dynamic Soil, Dynamic Plant, 2, 13-22.
- 104. Smith, C.S., Morrice, N.A. and Moorhead, G.B.G. (2004) Lack of Evidence for Phosphorylation of Arabidopsis thaliana PII: Implications for Plastid Carbon and Nitrogen Signaling. Biochimica et Biophysica Acta, 1699, 145-154.
http://dx.doi.org/10.1016/s1570-9639(04)00053-6 - 105. Burillo, S., Luque, I., Fuentes, I. and Contreras, A. (2004) Interactions between the Nitrogen Signal Transduction Protein PII and N-Acetyl Glutamate Kinase in Organisms That Perform Oxygenic Photosynthesis. Journal of Bacteriology, 186, 3346-3354.
http://dx.doi.org/10.1128/JB.186.11.3346-3354.2004 - 106. Chen, Y.M., Ferrar, T.S., Lohmeir-Vogel, E.M., Morrice, N., Mizuno, Y., Berenger, B., Ng, K.K., Muench, D.G. and Moorhead, G.B. (2006) The PII Signal Transduction Protein of Arabidopsis thaliana Forms an Arginine-Regulated Complex with Plastid N-Acetyl Glutamate Kinase. Journal of Biological Chemistry, 281, 5726-5733.
http://dx.doi.org/10.1074/jbc.M510945200 - 107. Ferrario-Mery, S., Besin, E., Pichon, O., Meyer, C. and Hodges, M. (2006) The Regulatory PII Protein Controls Arginine Biosynthesis in Arabidopsis. FEBS Letters, 580, 2015-2020.
http://dx.doi.org/10.1016/j.febslet.2006.02.075 - 108. Ferrario-Mery, S., Bouvet, M., Leleu, O., Savino, G., Hodges, M. and Meyer, C. (2015) Physiological Characterization of Arabidopsis Mutants Affected in the Expression of the Putative Regulatory Protein PII. Planta, 1, 28-39.
- 109. Mizuno, Y., Moorhead, G.B.G. and Ng, K.K.S. (2007) Structural Basis for the Regulation of N-Acetylgutamate Kinase by PII in Arabidopsis thaliana. The Journal of Biological Chemistry, 282, 35733-35740.
http://dx.doi.org/10.1074/jbc.M707127200 - 110. Hinnebusch, A.G. (2005) Translational Regulation of GCN4 and the General Amino Acid Control of Yeast. Annual Review of Microbiology, 59, 407-450.
http://dx.doi.org/10.1146/annurev.micro.59.031805.133833 - 111. Marton, M.J., Crouch, D. and Hinnebusch, A.G. (1993) GCN1, a Translational Activator of GCN4 in Saccharomyces cerevisiae, Is Required for Phosphorylation of Eukaryotic Translation Initiation Factor 2 by Protein Kinase GCN2. Molecular and Cellular Biology, 13, 3541-3556.
- 112. Chang, Y., Yang, W.Y., Browning, K. and Roth, D. (1999) Specific in Vitro Phosphorylation of Plant Eif2 Alpha by Eukaryotic Eif2 Alpha Kinases. Plant Molecular Biology, 41, 363-370.
http://dx.doi.org/10.1023/A:1006379623534 - 113. Chang, L.Y., Yang, W.Y. and Roth, D. (2000) Functional Complementation by Wheat Eif2 Alpha in the Yeast GCN2-Mediated Pathway. Biochemical and Biophysical Research Communications, 279, 468-474.
http://dx.doi.org/10.1006/bbrc.2000.3964 - 114. Guyer, D., Patton, D. and Ward, E. (1995) Evidence for Cross Pathway Regulation of Metabolic Gene Expression in Plants. Proceedings of the National Academy of Sciences of the United States of America, 92, 4997-5000.
http://dx.doi.org/10.1073/pnas.92.11.4997 - 115. Kato, T., Sato, S. and Tabata, S. (2004) A Soluble ABC Protein, GCN, Regulate Root Development in Arabidopsis. Plant and Cell Physiology, 45, S2.
- 116. Bhat, R.A., Riehl, M., Santandrea, G., Velasco, R., Slocombe, S., Donn, G., Steinbiss, H.H., Thompson, R.D. and Becker, H.A. (2003) Alteration of GCN5 Levels in Maize Reveals Dynamic Responses to Manipulating Histone Acetylation. The Plant Journal, 33, 455-469.
http://dx.doi.org/10.1046/j.1365-313X.2003.01642.x - 117. Kawaguchi, R. and Bailey-Serres, J. (2005) Mrna Sequence Features That Contribute to Translational Regulation in Arabidopsis. Nucleic Acids Research, 33, 955-965.
http://dx.doi.org/10.1093/nar/gki240 - 118. Rogozin, I.B., Kochetov, A.V., Kondrashov, F.A., Koonin, E.V. and Milanesi, L. (2001) Presence of ATG Triplets in 5’Untranslated Regions of Eukaryotic Cdnas Correlates with “Weak” Context of the Start Codon. Bioinformatics, 17, 890-900.
http://dx.doi.org/10.1093/bioinformatics/17.10.890 - 119. Stockinger, E.J., Mao, Y., Regier, M.K., Triezenberg, S.J. and Thomashow, M.F. (2001) Transcriptional Adaptor and Histone Acetyltransferase Proteins in Arabidopsis and Their Interactions with CBF1, a Transcriptional Activator Involved in Cold-Regulated Gene Expression. Nucleic Acids Research, 29, 1524-1533.
http://dx.doi.org/10.1093/nar/29.7.1524 - 120. Tsuda, K., Tsuji, T., Hirose, S. and Yamazaki, K. (2005) Three Arabidopsis MBF1 Homologs with Distinct Expression Profiles Plays Roles as Transcriptional Co-Activators. Plant and Cell Physiology, 45, 225-231.
http://dx.doi.org/10.1093/pcp/pch017 - 121. Langland, J.O., Langland, L.A., Browning, K.S. and Roth, D.A. (1996) Phosphorylation of Plant Eukaryotic Initiation Factor-2 by the Plant-Encoded Double-Stranded RNA-Dependent Protein Kinase, Ppkr, and Inhibition of Protein Synthesis in Vitro. Journal of Biological Chemistry, 271, 4539-4544.
http://dx.doi.org/10.1074/jbc.271.8.4539 - 122. Kakimoto, T. (2003) Perception and Signal Transduction of Cytokinins. Annual Review of Plant Physiology, 54, 605-627.
http://dx.doi.org/10.1146/annurev.arplant.54.031902.134802 - 123. Sakakibara, H., Suzuki, M., Takei, K., Deji, A., Taniguchi, M. and Sugiyama, T. (1998) A Response-Regulator Homologue Possibly Involved in Nitrogen Signal Transduction Mediated by Cytokinin in Maize. The Plant Journal, 14, 337-344.
http://dx.doi.org/10.1046/j.1365-313X.1998.00134.x - 124. Sakakibara, H., Takei, K. and Hirose, N. (2006) Interactions between Nitrogen and Cytokinin in the Regulation of Metabolism and Development. Trends in Plant Science, 11, 440-448.
http://dx.doi.org/10.1016/j.tplants.2006.07.004 - 125. Takei, K., Sakakibara, H., Taniguchi, M. and Sugiyama, T. (2001) Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species That Induces Gene Expression of Maize Response Regulator. Plant and Cell Physiology, 42, 85-93.
http://dx.doi.org/10.1093/pcp/pce009 - 126. Takei, K., Ueda, N., Aoki, K., Kuromori, T., Hirayama, T., Shinozaki, K., Yamaya, T. and Sakakibara, H. (2005) Atipt3 Is a Key Determinant of Nitrate-Dependent Cytokinin Biosynthesis in Arabidopsis. Plant and Cell Physiology, 45, 1053-1062.
http://dx.doi.org/10.1093/pcp/pch119 - 127. Collier, M.D., Fotelli, M.N., Nahm, M., Kopriva, S., Rennenberg, H., Hanke, D.E. and Geßler, A. (2003) Regulation of Nitrogen Uptake by Fagussyl Vatica on a Whole Plant Level-Interaction between Cytokinins and Soluble N Compounds. Plant, Cell & Environment, 26, 1549-1560.
http://dx.doi.org/10.1046/j.1365-3040.2003.01079.x - 128. Takei, K., Takahashi, T., Sugiyama, T., Yamaya, T. and Sakakibara, H. (2002) Multiple Routes Communicating Nitrogen Availability from Roots to Shoots: A Signal Transduction Pathway Mediated by Cytokinin. Journal of Experimental Botany, 53, 971-977.
http://dx.doi.org/10.1093/jexbot/53.370.971 - 129. Miyawaki, K., Matsumoto-Kitano, M. and Kakimoto, T. (2004) Expression of Cytokinin Biosynthetic Isopentenyltransferase Genes in Arabidopsis: Tissue Specificity and Regulation by Auxin, Cytokinin, and Nitrate. The Plant Journal, 37, 128-138.
http://dx.doi.org/10.1046/j.1365-313X.2003.01945.x - 130. Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., Ueguchi, C., Sugiyama, T. and Mizuno, T. (1999) Compilation and Characterization of Arabidopsis thaliana Response Regulators Implicated in His-Asp Phosphorelay Signal Transduction. Plant and Cell Physiology, 40, 733-742.
http://dx.doi.org/10.1093/oxfordjournals.pcp.a029600 - 131. Kiba, T., Naitou, T., Koizumi, N., Yamashino, T., Sakakibara, H. and Mizuno, T. (2005) Combinatorial Microarray Analysis Revealing Arabidopsis Genes Implicated in Cytokinin Responses through the His->Asp Phosphorelay Circuitry. Plant and Cell Physiology, 466, 339-355.
http://dx.doi.org/10.1093/pcp/pci033 - 132. Appleby, J.L., Parkinson, J.S. and Bourret, R.B. (1996) Signal Transduction via the Multi-Step Phosphorelay: Not Necessarily a Road Less Travelled. Cell, 86, 845-848.
http://dx.doi.org/10.1016/S0092-8674(00)80158-0 - 133. Mizuno, T. (1998) His-Asp Phosphotransfer Signal Transduction. Journal of Biological Chemistry, 123, 555-563.
http://dx.doi.org/10.1093/oxfordjournals.jbchem.a021972 - 134. Sakakibara, H., Hayakawa, A., Deji, A., Gawronska, S.W. and Sugiyama, T. (1999) His-Asp Phosphotransfer Possibly Involved in a Nitrogen Signal Transduction Mediated by Cytokinin in Maize: Molecular Cloning of Cdnas for Two-Component Regulatory Factors and Demonstration of Phosphotransfer Activity in Vitro. Plant Molecular Biology, 41, 563-573.
http://dx.doi.org/10.1023/A:1006391304881 - 135. Anantharaman, V. and Aravnd, L. (2001) The CHASE Domain: A Predicted Ligand-Binding Module in Plant Cytokinin Receptors and Other Eukaryotic and Bacterial Receptors. Trends in Biochemical Sciences, 26, 579-582.
http://dx.doi.org/10.1016/S0968-0004(01)01968-5 - 136. Wang, R., Tischner, R., Gutierrez, R.A., Hoffman, M., Xing, X. and Chen, M. (2004) Genomic Analysis of the Nitrate Response Using a Nitrate Reductase-Null Mutant of Arabidopsis. Plant Physiology, 136, 2512-2522.
http://dx.doi.org/10.1104/pp.104.044610 - 137. Chiu, J., Desalle, R., Lam, H.M., Meisel, L. and Coruzzi, G. (1996) Molecular Evolution of Glutamate Receptors: A Primitive Signaling Mechanism That Existed before Plants and Animals Diverged. Molecular Biology and Evolution, 16, 826-838.
http://dx.doi.org/10.1093/oxfordjournals.molbev.a026167 - 138. Dingledine, R., Borges, K., Bowie, D. and Traynelis, S.F. (1999) The Glutamate Receptor Ion Channels. Pharmacological Reviews, 51, 7-61.
- 139. Lacombe, B., Becker, D., Hedrich, R., Desalle, R., Hollmann, M., Kwak, J.M., Schroeder, J.I., Le Novère, N., Nam, H.G., Spalding, E.P., Tester, M., Turano, F.J., Chiu, J. and Coruzzi, G. (2002) The Identity of Plant Glutamate Receptors. Science, 292, 1486-1487.
http://dx.doi.org/10.1126/science.292.5521.1486b - 140. Kang, J.M. and Turano, F.J. (2003) The Putative Glutamate Receptor 1.1 (Atglr1.1) Functions as a Regulator of Carbon and Nitrogen Metabolism in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 100, 6872-6877.
http://dx.doi.org/10.1073/pnas.1030961100 - 141. Filleur, S., Walch-Liu, P., Gan, Y. and Forde, B.G. (2005) Nitrate and Glutamate Sensing by Plant Roots. Biochemical Society Transactions, 33, 283-286.
http://dx.doi.org/10.1042/BST0330283 - 142. Qi, Z., Stephens, N.R. and Spalding, E.P. (2006) Calcium Entry Mediated by GLR3.3, an Arabidopsis Glutamate Receptor with a Broad Agonist Profile. Plant Physiology, 142, 963-971.
http://dx.doi.org/10.1104/pp.106.088989 - 143. Chiang, T.Y. and Marzluf, G.A. (1992) DNA Recognition by the NIT2 Nitrogen Regulatory Protein—Importance of the Number, Spacing, and Orientation of GATA Core Elements and Their Flanking Sequences upon NIT2 Binding. Biochemistry, 33, 576-582.
http://dx.doi.org/10.1021/bi00168a024 - 144. Danielvedele, F. and Caboche, M.A. (1993) Tobacco Cdna Clone Encoding a GATA-1 Zinc-Finger Protein Homologous to Regulators of Nitrogen-Metabolism in Fungi. Molecular and General Genetics, 240, 365-373.
- 145. Fu, Y.H. and Marzluf, G.A. (1990) Site-Directed Mutagenesis of the Zinc Finger DNA Binding Domain of the Nitrogen-Regulatory Protein NIT2 of Neurospora. Molecular Microbiology, 4, 1847-1852.
http://dx.doi.org/10.1111/j.1365-2958.1990.tb02033.x - 146. Rastogi, R., Bate, N.J., Sivasankar, S. and Rothstein, S.J. (1997) Foot Printing of the Spinach Nitrite Reductase Gene Promoter Reveals the Preservation of Nitrate Regulatory Elements between Fungi and Higher Plants. Plant Molecular Biology, 34, 465-476.
http://dx.doi.org/10.1023/A:1005842812321 - 147. Bi, Y.M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T. and Rothstein, S. (2005) Genetic Analysis of Arabidopsis GATA Transcription Factor Gene Family Reveals a Nitrate-Inducible Member Important for Chlorophyll Synthesis and Glucose Sensitivity. The Plant Journal, 44, 680-692.
http://dx.doi.org/10.1111/j.1365-313X.2005.02568.x - 148. Filleur, S., Dorbe, M.F., Cerezo, M., Orsel, M., Granier, F., Gojon, A. and Daniel-Vedele, F. (2001) An Arabidopsis T-DNA Mutant Affected in Nrt2 Genes Is Impaired in Nitrate Uptake. FEBS Letters, 489, 220-224.
http://dx.doi.org/10.1016/S0014-5793(01)02096-8 - 149. Forde, G. and Clarkson, D.T. (1990) Nitrate and Ammonium Nutrition of Plants: Physiological and Molecular Perspectives. Advances in Botanical Research, 30, 1-90.
http://dx.doi.org/10.1016/S0065-2296(08)60226-8 - 150. Ludewig, U., Neuhauser, B. and Dynowski, M. (2007) Molecular Mechanisms of Ammonium Transport and Accumulation in Plants. FEBS Letters, 581, 2301-2308.
http://dx.doi.org/10.1016/j.febslet.2007.03.034 - 151. Mack, G. and Tischner, R. (1994) Constitutive and Inducible Net NH4+ Uptake of Barley (Hordeum vulgare L.) Seedlings. Journal of Plant Physiology, 144, 351-357.
http://dx.doi.org/10.1016/S0176-1617(11)81198-3 - 152. Miller, A.J., Fan, X., Orsel, M., Smith, S.J. and Wells, D.M. (2007) Nitrate Transport and Signaling. Journal of Experimental Botany, 58, 2297-2306.
http://dx.doi.org/10.1093/jxb/erm066 - 153. Orsel, M., Filleur, S., Fraisier, V. and Daniel-Vedele, F. (2002) Nitrate Transport in Plants: Which Gene and Which Control? Journal of Experimental Botany, 53, 825-833.
http://dx.doi.org/10.1093/jexbot/53.370.825 - 154. Orsel, M., Krapp, A. and Daniel-Vedele, F. (2002) Analysis of the NRT2 Nitrate Transporter Family in Arabidopsis. Structure and Gene Expression. Plant Physiology, 129, 886-896.
http://dx.doi.org/10.1104/pp.005280 - 155. Wang, M.Y., Siddiqi, M.Y., Ruth, T.J. and Glass, A.D.M. (1993) Ammonium Uptake by Rice Roots. I. Fluxes and Subcellular Distribution of 13 . Plant Physiology, 103, 1249-1258.
- 156. Witte, C.P. (2011) Urea Metabolism in Plants. Plant Science, 180,431-438.
http://dx.doi.org/10.1016/j.plantsci.2010.11.010 - 157. Huang, N.C., Liu, K.H., Lo, H.J. and Tsay, Y.F. (1999) Cloning and Functional Characterization of an Arabidopsis Nitrate Transporter Gene That Encodes a Constitutive Component of Low-Affinity Uptake. Plant Cell, 11, 381-1392.
http://dx.doi.org/10.1105/tpc.11.8.1381 - 158. Kojima, S. Bohner, A., Gassert, B., Yuan, L. and Von Wiren, N. (2007) AtDUR3 Represents the Major Transporter for High-Affinity Urea Transport across the Plasma Membrane of Nitrogen-Deficient Arabidopsis Roots. The Plant Journal, 52, 30-40.
http://dx.doi.org/10.1111/j.1365-313X.2007.03223.x - 159. Li, W., Wang, Y., Okamoto, M., Crawford, N.M., Siddiqi, M.Y. and Glass, A.D. (2007) Dissection of the AtNRT2.1: AtNRT2.2 Inducible High-Affinity Nitrate Transporter Gene Cluster. Plant Physiology, 143, 425-433.
http://dx.doi.org/10.1104/pp.106.091223 - 160. Ninnemann, O., Jauniaux, J.C. and Frommer, W.B. (1994) Identification of a High Affinity Transporter from Plants. EMBO Journal, 1, 3464-3471.
- 161. Tsay, Y.F., Schroeder, J.I., Feldmann, K.A. and Crawford, N.M. (1993) The Herbicide Sensitivity Gene CHL1 of Arabidopsis Encodes a Nitrate-Inducible Nitrate Transporter. Cell, 72, 705-713.
http://dx.doi.org/10.1016/0092-8674(93)90399-B - 162. Yuan, L., Loque, E.D., Kojima, S., Rauch, S., Ishiyama, K. and Finoue, E. (2007) The Organization of High-Affinity Ammonium Uptake in Arabidopsis Roots Depends on the Spatial Arrangement and Biochemical Properties of AMT1- Type Transporters. Plant Cell, 19, 2636-2652.
http://dx.doi.org/10.1105/tpc.107.052134 - 163. Mayer, M. and Ludewig, U. (2006) Role of AMT1; 1 in Acquisition in Arabidopsis thaliana. Plant Biology, 8, 522-528.
http://dx.doi.org/10.1055/s-2006-923877 - 164. Tsay, Y.F. (2007) Nitrate Transporters and Peptide Transporters. FEBS Letters, 581, 2290-2300.
http://dx.doi.org/10.1016/j.febslet.2007.04.047 - 165. Kojima, S., Bohner, A. and Von Wiren, N. (2006) Molecular Mechanisms of Urea Transport in Plants. Journal of Membrane Biology, 212, 83-91.
http://dx.doi.org/10.1007/s00232-006-0868-6 - 166. Liu, L.H., Ludewig, U., Frommer, W.B. and Von Wiren, N. (2003) AtDUR3 Encodes a New Type of High Affinity Urea H+Symporter in Arabidopsis. Plant Cell, 15, 790-800.
http://dx.doi.org/10.1105/tpc.007120 - 167. Deng, R.L., Gu, J.T., Lu, W.J., Xu, H.R., Cao, Y.F. and Xiao, K. (2007) Characterization, Function and Expression Analysis of Ammonium Transporter Gene OsAMT1.4 and OsAMT5 in Rice (Oryza sativa). Scientia Agricultura Sinica, 40, 2395-2402.
- 168. Gazzarini, S., Lejay, L., Gojon, A., Ninnemann, O. and Frommer, W.B. (1999) Three Functional Transporters for Constitutive, Diurnally Regulated and Starvation-Induced Uptake of Ammonium into Arabidopsis Roots. Plant Cell, 11, 937-948.
http://dx.doi.org/10.1105/tpc.11.5.937 - 169. Li, B.Z., Merrick, M., Li, S.M., Li, H.Y., Zhu, S.U., Shi, W.M. and Su, Y.H. (2009) Molecular Basis and Regulation of Ammonium Transporter in Rice. Rice Science, 16, 314-322.
http://dx.doi.org/10.1016/S1672-6308(08)60096-7 - 170. Sohlenkamp, C., Wood, C.C., Roeb, G.W. and Udvardi, M.K. (2002) Characterization of Arabidopsis Atamt2, a High-Affinity Ammonium Transporter of the Plasma Membrane. Plant Physiology, 130, 1788-1796.
http://dx.doi.org/10.1104/pp.008599 - 171. Suenaga, A., Moriya, K., Sonoda, Y., Ikeda, A., Von Wirén, N., Hayakawa, T., Yamaguchi, J. and Yamaya, T. (2003) Constitutive Expression of a Novel-Type Ammonium Transporter OsAMT2 in Rice Plants. Plant and Cell Physiology, 44, 206-211.
http://dx.doi.org/10.1093/pcp/pcg017 - 172. Barbier-Brygoo, H., De Angeli, A., Filleur, S., Frachisse, J.M., Gambale, F., Thomine, S. and Wege, S. (2011) Anion Channels/Transporters in Plants: From Molecular Bases to Regulatory Networks. Annual Review of Plant Biology, 62, 25-51.
http://dx.doi.org/10.1146/annurev-arplant-042110-103741 - 173. Dechorgnat, J., Nguyen, C.T., Armengaud, P., Jossier, M., Diatloff, E., Filleur, S. and Daniel-Vedele, F. (2011) From the Soil to the Seeds: The Long Journey of Nitrate in Plants. Journal of Experimental Botany, 62, 1349-1359.
http://dx.doi.org/10.1093/jxb/erq409 - 174. Negi, J. (2008) CO2 Regulator SLAC1 and Its Homologues Are Essential for Anion Homeostasis in Plant Cells. Nature, 452, 483-486.
http://dx.doi.org/10.1038/nature06720 - 175. Sasaki, T., Mori, I.C., Furuichi, T., Munemasa, S., Toyooka, K., Matsuoka, K., Murata, Y. and Yamamoto, Y. (2010) Closing Plant Stomata Requires a Homologue of an Aluminium-Activated Malate Transporter. Plant and Cell Physiology, 51, 354-365.
http://dx.doi.org/10.1093/pcp/pcq016 - 176. Wang, Y.Y., Hsu, P.K. and Tsay, Y.F. (2012) Uptake, Allocation and Signaling of Nitrate. Trends in Plant Sciences, 17, 458-467.
http://dx.doi.org/10.1016/j.tplants.2012.04.006 - 177. Morère-Le Paven, M.C., Vlau, L., Hamon, A., Vandecasteele, C., Pellizzaro, C.B., Laffont, C. and Lapied, B. (2011) Characterization of a Dual-Affinity Nitrate Transporter MtNRT1.3 in the Model Legume Medicago truncatula. Journal of Experimental Botany, 62, 5595-5605.
http://dx.doi.org/10.1093/jxb/err243 - 178. Okamoto, M., Kumar, A., Li, W., Wang, Y., Siddiqi, M.Y., Crawford, N.M. and Glass, A.D. (2006) High-Affinity Nitrate Transport in Roots of Arabidopsis Depends on Expression of the NAR2-Like Gene AtNRT3.1. Plant Physiology, 140, 1036-1046.
http://dx.doi.org/10.1104/pp.105.074385 - 179. Yong, Z., Kotur, Z. and Glass, A.D. (2010) Characterization of an Intact Two-Component High-Affinity Nitrate Transporter from Arabidopsis Roots. The Plant Journal, 63, 739-748.
http://dx.doi.org/10.1111/j.1365-313X.2010.04278.x - 180. De Angeli, A., Monachello, D., Ephritikhine, G., Frachisse, J.M., Thomine, S., Gambale, F. and Barbier-Brygoo, H. (2006) The Nitrate/Proton Antiporter Atclca Mediates Nitrate Accumulation in Plant Vacuoles. Nature, 442, 939-942.
http://dx.doi.org/10.1038/nature05013 - 181. Von Der Fecht-Bartenbach, J., Bogner, M., Dynowski, M. and Ludewig, U. (2010) CLC-B-Mediated /H+ Exchange across the Tonoplast of Arabidopsis Vacuoles. Plant and Cell Physiology, 51, 960-968.
http://dx.doi.org/10.1093/pcp/pcq062 - 182. Geiger, D., Maierhofer, T., Al-Rasheid, K.A., Scherzer, S., Mumm, P., Liese, A. and Ache, P. (2011) Stomatal Closure by Fast Abscisic Acid Signaling Is Mediated by the Guard Cell Anion Channel SLAH3 and the Receptor RCAR1. Science Signaling, 4, Ra32.
http://dx.doi.org/10.1126/scisignal.2001346 - 183. Lee, S.C., Lan, W., Buchanan, B.B. and Luan, S.A. (2009) Protein Kinase-Phosphatase Pair Interacts with an Ion Channel to Regulate ABA Signaling in Plant Guard Cells. Proceedings of the National Academy of Sciences of the United States of America, 106, 21419-21424.
http://dx.doi.org/10.1073/pnas.0910601106 - 184. Andrade, S.L., Dickmanns, A., Ficner, R. and Einsle, O. (2005) Crystal Structure of the Archaeal Ammonium Transporter Amt-1 from Archaeoglobus fulgidus. Proceedings of the National Academy of Sciences of the United States of America, 102, 14994-14999.
http://dx.doi.org/10.1073/pnas.0506254102 - 185. Loque, D. and Von Wiren, N. (2004) Regulatory Levels for the Transport of Ammonium in Plant Roots. Journal of Experimental Botany, 55, 1293-1305.
http://dx.doi.org/10.1093/jxb/erh147 - 186. Forrest, L.R. (2013) Structural Biology. (Pseudo-) Symmetrical Transport. Science, 339, 399-401.
http://dx.doi.org/10.1126/science.1228465 - 187. Shi, Y. (2013) Common Folds and Transport Mechanisms of Secondary Active Transporters. Annual Review of Biophysics, 42, 51-72.
http://dx.doi.org/10.1146/annurev-biophys-083012-130429 - 188. De Michele, R., Ast, C., Loqué, D., Ho, C.H., Andrade, S.L., Lanquar, V., Grossmann, G. and Gehne, S. (2013) Fluorescent Sensors Reporting the Activity of Ammonium Transceptors in Live Cells. Elife, 2, Article ID: E00800.
http://dx.doi.org/10.7554/elife.00800 - 189. Pantoja, O. (2013) High Affinity Ammonium Transporters and Mechanism of Action. Frontiers in Plant Science, 3, 34.
- 190. Dynowski, M., Schaaf, G., Loque, D., Moran, O. and Ludewig, U. (2008) Plant Plasma Membrane Water Channels Conduct the Signaling Molecule H2O2. Biochemical Journal, 414, 53-61.
http://dx.doi.org/10.1042/BJ20080287 - 191. Jahn, T.P., Moller, A.L., Zeuthen, T., Holm, L.M., Klærke, D.A., Mohsin, B., Kühlbrandt, W. and Schjoerring, J.K. (2004) Aquaporin Homologues in Plants and Mammals Transport Ammonia. FEBS Letters, 574, 31-36.
http://dx.doi.org/10.1016/j.febslet.2004.08.004 - 192. Loqué, D., Ludewig, U., Yuan, L. and Von Wirén, N. (2005) Tonoplast Intrinsic Proteins AtTIP2; 1 and AtTIP2; 3 Facilitate NH3 Transport into the Vacuole. Plant Physiology, 137, 671-680.
http://dx.doi.org/10.1104/pp.104.051268 - 193. Léran, S., Varala, K., Boyer, J.C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., David, L. and Dickstein, R. (2014) A Unified Nomenclature of Nitrate Transporter1/Peptide Transporter Family Members in Plants. Trends in Plant Science, 19, 5-9.
http://dx.doi.org/10.1016/j.tplants.2013.08.008 - 194. Boursiac, Y. (2013) ABA Transport and Transporters. Trends in Plant Science, 18, 325-333.
http://dx.doi.org/10.1016/j.tplants.2013.01.007 - 195. Criscuolo, G., Valkov, V.T., Parlati, A., Alves, L.M. and Chiurazzi, M. (2012) Molecular Characterization of the Lotus japonicus NRT1 (PTR) and NRT2 Families. Plant, Cell & Environment, 35, 1567-1581.
http://dx.doi.org/10.1111/j.1365-3040.2012.02510.x - 196. Nour-Eldin, H.H. (2012) NRT/PTR Transporters Are Essential for Translocation of Glucosinolate Defence Compounds to Seeds. Nature, 488, 531-534.
http://dx.doi.org/10.1038/nature11285 - 197. Tegeder, M. and Rentsch, D. (2010) Uptake and Partitioning of Aminoacids and Peptides. Molecular Plant, 3, 997-1011.
http://dx.doi.org/10.1093/mp/ssq047 - 198. Sun, J., Bankston, J.R., Payandeh, J., Hinds, T.R., Zagotta, W.N. and Zheng, N. (2014) Crystal Structure of the Plant Dual-Affinity Nitrate Transporter NRT1.1. Nature, 507, 73-77.
http://dx.doi.org/10.1038/nature13074 - 199. Pao, S.S., Paulsen, I.T. and Saier, M.H. (1998) Major Facilitator Super Family. Microbiology and Molecular Biology Reviews, 62, 1-34.
- 200. Bergsdorf, E.Y., Zdebik, A.A. and Jentsch, T.J. (2009) Residues Important for Nitrate/Proton Coupling in Plant and Mammalian CLC Transporters. Journal of Biological Chemistry, 284, 11184-11193.
http://dx.doi.org/10.1074/jbc.M901170200 - 201. Nguitragool, W. and Miller, C. (2006) Uncoupling of a CLC Cl-/H+ Exchange Transporter by Polyatomic Anions. Journal of Molecular Biology, 362, 682-690.
http://dx.doi.org/10.1016/j.jmb.2006.07.006 - 202. Zifarelli, G. and Pusch, M. (2009) Conversion of the 2 Cl-/1 H+ Antiporter Clc-5in A /H+ Antiporter by a Single Point Mutation. EMBO Journal, 28, 175-182.
http://dx.doi.org/10.1038/emboj.2008.284 - 203. Dutzler, R., Campbell, E.B., Cadene, M., Chait, B.T. and Mackinnon, R. (2002) X-Ray Structure of a Clc Chloride Channel at 3.0 a Reveals the Molecular Basis of Anion Selectivity. Nature, 415, 287-294.
http://dx.doi.org/10.1038/415287a - 204. Picollo, A., Malvezzi, M., Houtman, J.C. and Accardi, A. (2009) Basis of Substrate Binding and Conservation of Selectivity in the CLC Family of Channels and Transporters. Nature Structural & Molecular Biology, 16, 1294-1301.
http://dx.doi.org/10.1038/nsmb.1704 - 205. De Angeli, A., Moran, O., Wege, S., Filleur, S., Ephritikhine, G. and Thomine, S. (2009) ATP Binding to the C Terminus of the Arabidopsis thaliana Nitrate/Proton Antiporter, Atclca, Regulates Nitrate Transport into Plant Vacuoles. Journal of Biological Chemistry, 284, 26526-26532.
http://dx.doi.org/10.1074/jbc.M109.005132 - 206. Geelen, D., Lurin, C., Bouchez, D., Frachisse, J.M., Lelièvre, F. and Courtial, B. (2000) Disruption of Putative Anion Channel Gene Atclc-A in Arabidopsis Suggests a Role in the Regulation of Nitrate Content. The Plant Journal, 21, 259-267.
http://dx.doi.org/10.1046/j.1365-313x.2000.00680.x - 207. Harada, H., Kuromori, T., Hirayama, T., Shinozaki, K. and Leigh, R.A. (2004) Quantitative Trait Loci Analysis of Nitrate Storage in Arabidopsis Leading to an Investigation of the Contribution of the Anion Channel Gene, Atclc-C, to Variation in Nitrate Levels. Journal of Experimental Botany, 405, 2005-2014.
http://dx.doi.org/10.1093/jxb/erh224 - 208. Zifarell, G. and Pusch, M. (2010) CLC Transport Proteins in Plants. FEBS Letters, 584, 2122-2127.
http://dx.doi.org/10.1016/j.febslet.2009.12.042 - 209. Kollist, H., Jossier, M., Laanemets, K. and Thomine, S. (2011) Anion Channels in Plant Cells. FEBS Journal, 278, 4277-4292.
http://dx.doi.org/10.1111/j.1742-4658.2011.08370.x - 210. Schwacke, R., Schneider, A., Van Der Graaff, E., Fischer, K., Catoni E. and Desimone, M. (2003) ARAMEMNON, a Novel Database for Arabidopsis Integral Membrane Proteins. Plant Physiology, 131, 16-26.
http://dx.doi.org/10.1104/pp.011577 - 211. Saraste, M., Shibbald, P.R. and Wittinghofer, A. (1999) The P-Loop—A Common Motif in ATP- and GTP-Binding Proteins. Trends in Biochemical Sciences, 15, 430-434.
http://dx.doi.org/10.1016/0968-0004(90)90281-F - 212. Forrest, K.L. and Bhave, M. (2007) Major Intrinsic Proteins (Mips) in Plants: A Complex Gene Family with Major Impacts on Plant Phenotype. Functional & Integrative Genomics, 7, 263-289.
http://dx.doi.org/10.1007/s10142-007-0049-4 - 213. Maurel, C., Verdoucq, L., Luu, D.T. and Santoni, V. (2008) Plant Aquaporins: Membrane Channels with Multiple Integrated Functions. Annual Review of Plant Physiology, 59, 595-624.
http://dx.doi.org/10.1146/annurev.arplant.59.032607.092734 - 214. Fujiyoshi, Y., Mitsuoka, K., De Groot, B.L., Philippsen, A., Grubmüller, H. and Agre, P. (2002) Structure and Function of Water Channels. Current Opinion in Structural Biology, 12, 509-515.
http://dx.doi.org/10.1016/S0959-440X(02)00355-X - 215. Wudick, M., Luu, D.T. and Maurel, C. (2009) A Look Inside: Localization Patterns and Functions of Intracellular Plant Aquaporins. New Phytologist, 184, 289-302.
http://dx.doi.org/10.1111/j.1469-8137.2009.02985.x - 216. Hoque, M.S., Masle, J., Udvardi, M.K., Ryan, P.R. and Upadhyaya, N.M. (2006) Over-Expression of the Rice Osamt1-1gene Increases Ammonium Uptake and Content, but Impairs Growth and Development of Plants under High Ammonium Nutrition. Functional Plant Biology, 33, 153-163.
http://dx.doi.org/10.1071/FP05165 - 217. Sonoda, Y., Ikeda, A., Saiki, S., Von Wirén, N., Yamaya, T. and Yamaguchi, J. (2003) Distinct Expression and Function of Three Ammonium Transporter Genes (Osamt1; 1-1; 3) in Rice. Plant and Cell Physiology, 44, 726-734.
http://dx.doi.org/10.1093/pcp/pcg083 - 218. Gu, R., Duan, F., An, X., Zhang, F., Von Wirén, N. and Yuan, L. (2013) Characterization of AMT-Mediated High-Affinity Ammonium Uptake in Roots of Maize (Zea mays L.). Plant and Cell Physiology, 54, 515-524.
http://dx.doi.org/10.1093/pcp/pct099 - 219. Li, S.M., Li, B.Z. and Shi, W.M. (2012) Expression Patterns of Nine Ammonium Transporters in Rice in Response to N Status. Pedosphere, 22, 860-869.
http://dx.doi.org/10.1016/S1002-0160(12)60072-1 - 220. Yao, S.G. and Sonada, Y. (2009) Promoter Analysis of OsAMT1; 2 and OsAMT1; 3 Implies Their Distinct Roles in Nitrogen Utilization. Breeding Science, 58, 201-207.
http://dx.doi.org/10.1270/jsbbs.58.201 - 221. Lauter, F.R., Ninnemann, O., Bucher, M., Riesmeier, J. and Frommer, W.B. (1996) Preferential Expression of an Ammonium Transporter and Two Putative Nitrate Transporters in Root Hairs of Tomato. Proceedings of the National Academy of Sciences of the United States of America, 93, 8139-8144.
http://dx.doi.org/10.1073/pnas.93.15.8139 - 222. Tabuchi, M., Abiko, T. and Yamaya, T. (2007) Assimilation of Ammonium Ions and Reutilization of Nitrogen in Rice (Oryza sativa L.). Journal of Experimental Botany, 8, 2319-2327.
http://dx.doi.org/10.1093/jxb/erm016 - 223. Lin, C.M., Koh, S., Stacey, G., Yu, S.M., Lin, T.Y. and Tsay, Y.F. (2000) Cloning and Functional Characterization of a Constitutively Expressed Nitrate Transporter Gene Osnrt1 from Rice. Plant Physiology, 122, 379-388.
http://dx.doi.org/10.1104/pp.122.2.379 - 224. Cerezo, M., Tillard, P., Filleur, S., Munos, S., Daniel-Vedele, F. and Gojon, A. (2001) Major Alterations of the Regulation of Root Uptake Are Associated with the Mutation of Nrt2.1 and Nrt2.2 Genes in Arabidopsis. Plant Physiology, 127, 262-271.
http://dx.doi.org/10.1104/pp.127.1.262 - 225. Orsel, M., Eulenburg, K., Krapp, A. and Daniel-Vedele, F. (2004) Disruption of the Nitrate Transporter Genes Atnrt2.1 and Atnrt2.2 Restrict Growth at Low External Nitrate Concentration. Planta, 219, 714-721.
http://dx.doi.org/10.1007/s00425-004-1266-x - 226. Kiba, T., Feria-Bourrellier, A.B., Lafouge, F., Lezhneva, L., Boutet-Mercey, S., Orsel, M. and Bréhaut, V. (2012) The Arabidopsis Nitrate Transporter NRT2.4 Plays a Double Role in Roots and Shoots of Nitrogen-Starved Plants. Plant Cell, 24, 245-258.
http://dx.doi.org/10.1105/tpc.111.092221 - 227. Masclaux-Daubresse, C., Daniel-Vedele, F., Dechorgnat, J., Chardon, F, Gaufichon, L. and Suzuki, A. (2010) Nitrogen Uptake, Assimilation and Remobilization in Plants: Challenges for Sustainable and Productive Agriculture. Annals of Botany, 105, 1141-1157.
http://dx.doi.org/10.1093/aob/mcq028 - 228. Araki, R. and Hasegawa, H. (2006) Expression of Rice (Oryza Sativa L.) Genes Involved in High-Affinity Nitrate Transport during the Period of Nitrate Induction. Breeding Science, 56, 295-302.
http://dx.doi.org/10.1270/jsbbs.56.295 - 229. Krapp, A., Fraisier, V., Scheible, W.-R., Quesada, A., Gojon, A., Stitt, M. and Caboche, M. (1998) Expression Studies of Nrt2: 1Np, a Putative High-Affinity Nitrate Transporter: Evidence for Its Role in Nitrate Uptake. The Plant Journal, 14, 723-731.
http://dx.doi.org/10.1046/j.1365-313x.1998.00181.x - 230. Zhuo, D., Okamoto, M., Vidmar, J.J. and Glass, A.D. (1999) Regulation of a Putative High-Affinity Nitrate Transporter (Nrt2; 1At) in Roots of Arabidopsis thaliana. The Plant Journal, 17, 563-568.
http://dx.doi.org/10.1046/j.1365-313X.1999.00396.x - 231. Nazoa, P., Videmar, J., Tranbarger, T.J., Mouline, K., Damiani, I., Tillard, P. and Glass, A.D.M. (2003) Regulation of the Nitrate Transporter Gene Atnrt2.1 in Arabidopsis thaliana, Responses to Nitrate, Amino Acids, and Developmental Stage. Plant Molecular Biology, 52, 689-703.
http://dx.doi.org/10.1023/A:1024899808018 - 232. Okamoto, M., Vidmar, J.J. and Glass, A.D. (2003) Regulation of NRT1 and NRT2 Gene Families of Arabidopsis thaliana: Responses to Nitrate Provision. Plant and Cell Physiology, 44, 4-17.
http://dx.doi.org/10.1093/pcp/pcg036 - 233. Vidmar, J.J., Zhuo, D., Siddiqi, M.Y., Schoerring, J.K., Touraine, B. and Glass, A.D.M. (2000) Regulation of High Affinity Nitrate Transporter Genes and High Affinity Nitrate Influx by Nitrogen Pools in Plant Roots. Plant Physiology, 123, 307-318.
http://dx.doi.org/10.1104/pp.123.1.307 - 234. Santi, G., Locci, R., Monte, R., Pinton, Z. and Varanini, Z. (2003) Induction of Nitrate Uptake in Maize Roots: Expression of a Putative High-Affinity Nitrate Transporter and Plasma Membrane H+-Atpase Isoforms. Journal of Experimental Botany, 54, 1851-1864.
http://dx.doi.org/10.1093/jxb/erg208 - 235. Ullrich, W.R. (1992) Transport of Nitrate and Ammonium through Plant Membranes. In: Mengel, K. and Pilbeam, D.J., Eds., Nitrogen Metabolism of Plants, Clarendon Press, Oxford, 121-137.
- 236. Parker, J.L. and Newstead, S. (2014) Molecular Basis of Nitrate Uptake by the Plant Nitrate Transporter NRT1.1. Nature, 507, 68-72.
http://dx.doi.org/10.1038/nature13116 - 237. Elberry, H.M., Majumdar, M.L., Cunningham, T.S., Sumrada, R.A. and Cooper, T.G. (1993) Regulation of the Urea Active Transporter Gene (DUR3) in Saccharomyces cerevisiae. Journal of Bacteriology, 175, 4688-4698.
- 238. Wang, W.H., Köhler, B., Cao, F.Q., Liu, G.W., Gong, Y.Y. and Sheng, S. (2012b) Rice DUR3 Mediates High-Affinity Urea Transport and Plays an Effective Role in Improvement af Urea Acquisition and Utilization When Expressed in Arabidopsis. New Phytologist, 193, 432-444.
http://dx.doi.org/10.1111/j.1469-8137.2011.03929.x - 239. Wu, Q., Chen, F., Chen, Y., Yuan, L., Zhang, F. and Mi, G. (2011) Root Growth in Response to Nitrogen Supply in Chinese Maize Hybrids Released between 1973 and 2009. Science China Life Sciences, 54, 642-650.
http://dx.doi.org/10.1007/s11427-011-4186-6 - 240. Gu, R., Chen, X., Zhou, Y. and Yuan, L. (2012) Isolation and Characterization of Three Maize Aquaporin Genes, Zmnip2; 1, Zmnip2; 4 and Zmtip4; 4 Involved in Urea Transport. Biochemistry and Molecular Biology Reports, 4, 96-101.
http://dx.doi.org/10.5483/BMBRep.2012.45.2.96 - 241. Liu, L.H., Ludewig, U., Gassert, B., Frommer, W.B. and Von Wiren, N. (2003) Urea Transport by Nirogen-Regulated Tonoplast Intrinsic Proteins in Arabidopsis. Plant Physiology, 133, 1220-1228.
http://dx.doi.org/10.1104/pp.103.027409 - 242. Wallace, I.S. and Roberts, D.M. (2005) Distinct Transport Selectivity of Two Structural Subclasses of the Nodulin-Like Intrinsic Protein Family of Plant Aquaglyceroporin Channels. Biochemistry, 44, 16826-16834.
http://dx.doi.org/10.1021/bi0511888 - 243. Wallace, I.S., Choi, W.G. and Roberts, D.M. (2006) The Structure, Function and Regulation of the Nodulin 26-Like Intrinsic Protein Family of Plant Aquaglyceroporins. Biochimica et Biophysica Acta, 1758, 1165-1175.
http://dx.doi.org/10.1016/j.bbamem.2006.03.024 - 244. Lin, S.H., Kuo, H.F. and Canivenc, G. (2008) Mutation of the Arabidopsis NRT1.5 Nitrate Transporter Causes Defective Root-to-Shoot Nitrate Transport. The Plant Cell, 20, 2514-2528.
http://dx.doi.org/10.1105/tpc.108.060244 - 245. Segonzac, C., Boyer, J.C., Ipotesi, E., Szponarski, W., Tillard, P. and Touraine, B. (2007) Nitrate Efflux at the Root Plasma Membrane: Identification of an Arabidopsis Excretion Transporter. Plant Cell, 19, 3760-3777.
http://dx.doi.org/10.1105/tpc.106.048173 - 246. Britto, D.T., Glass, A.D.M., Kronzucker, H.J. and Siddiqi, M.Y. (2001) Cytosolic Concentrations and Transmembrane Fluxes of NH4+/NH3. An Evaluation of Recent Proposals. Plant Physiology, 125, 523-526.
http://dx.doi.org/10.1104/pp.125.2.523 - 247. Chen, G., Guo, S., Kronzucker, H.J. and Shi, W. (2013) Nitrogen Use Efficiency (NUE) in Rice Links to Toxicity and Futile Cycling in Roots. Plant Soil, 369, 351-363.
http://dx.doi.org/10.1007/s11104-012-1575-y - 248. Li, B., Li, G., Kronzucker, H.J., Baluska, F. and Shi, W. (2014) Ammonium Stress in Arabidopsis: Signaling, Genetic Loci, and Physiological Targets. Trends in PlantScience, 19, 107-114.
http://dx.doi.org/10.1016/j.tplants.2013.09.004 - 249. Britto, D.T. and Kronzucker, H.J. (2002) Toxicity in Plants: A Critical Review. Journal of Plant Physiology, 159, 567-584.
http://dx.doi.org/10.1078/0176-1617-0774 - 250. Crawford, N.M. and Forde, B.G. (2002) Molecular and Developmental Biology of Inorganic Nitrogen Nutrition. The Arabidopsis Book: American Society of Plant Biologists, 1, Article ID: E0011.
http://dx.doi.org/10.1199/tab.0011 - 251. Hirel, B. and Lea, P. (2001) Ammonia Assimilation. In: Lea, P.J. and Morot-Gaudry, J.F., Eds., Plant Nitrogen, Springer-Verlag, Berlin, 79-100.
http://dx.doi.org/10.1007/978-3-662-04064-5_4 - 252. Terce-Laforgue, T., Mäck, G. and Hirel, B. (2004) New Insights towards the Function of Glutamate Dehydrogenase Revealed during Source-Sink Transition of Tobacco (Nicotiana tabacum) Plants Grown under Different Nitrogen Regimes. Physiologia Plantarum, 120, 220-228.
http://dx.doi.org/10.1111/j.0031-9317.2004.0241.x - 253. Linka, M. and Weber, A.P. (2005) Shuffling Ammonia between Mitochondria and Plastids during Photorespiration. Trends in Plant Sciences, 10, 461-465.
http://dx.doi.org/10.1016/j.tplants.2005.08.002 - 254. Forde, B.G. and Lea, P.J. (2007) Glutamate in Plants: Metabolism, Regulation, and Signaling. Journal of Experimental Botany, 58, 2339-2358.
http://dx.doi.org/10.1093/jxb/erm121 - 255. Oliveira, I.C., Brenner, E., Chiu, J., Hsieh, M.H., Kouranov, A., Lam, H.M., Shin, M.J. and Coruzzi, G.J. (2001) Metabolite and Light Regulation of Metabolism in Plants Lessons from the Study of a Single Biochemical Pathway. Brazilian Journal of Medical and Biological Research, 34, 567-575.
http://dx.doi.org/10.1590/s0100-879x2001000500003 - 256. Dluzniewka, P., Gessler, A., Kopriva, S., Strnad, M., Novák, O., Rennenberg, H. and Dietrich, H. (2006) Exogenous Supply of Glutamine and Active Cytokinin to the Roots Reduces Uptake Rates in Poplar. Plant, Cell & Environment, 29, 1284-1297.
http://dx.doi.org/10.1111/j.1365-3040.2006.01507.x - 257. Wingler, A., Lea, P.J., Quick, W.P. and Leegood, R.C. (2000) Photorespiration: Metabolic Pathways and Their Role in Stress Protection. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 355, 1517- 1529.
http://dx.doi.org/10.1098/rstb.2000.0712 - 258. Kichey, T., Heumez, E., Pocholle, D., Pageau, K., Vanacker, H., Dubois, F.D.R., Le Gouis, J. and Hirel, B. (2006) Combined Agronomic and Physiological Aspects of Nitrogen Management in Wheat Highlight a Central Role for Glutamine Synthetase. The New Phytologist, 169, 265-278.
http://dx.doi.org/10.1111/j.1469-8137.2005.01606.x - 259. Suzuki, A. and Knaff, D.B. (2005) Glutamate Synthase: Structural, Mechanistic and Regulatory Properties, and Role in the Amino Acid Metabolism. Photosynthesis Research, 85, 191-217.
http://dx.doi.org/10.1007/s11120-004-3478-0 - 260. Andrews, M., Lea, P.J., Raven, J.A. and Lindsey, K. (2004) Can Genetic Manipulation of Plant Nitrogen Assimilation Enzymes Result in Increased Crop Yield and Greater N-Use Efficiency? An Assessment. Annals of Applied Biology, 145, 25-40.
http://dx.doi.org/10.1111/j.1744-7348.2004.tb00356.x - 261. Raven, J.A. (1985) Regulation of Ph and Generation of Osmolarity in Vascular Plants: A Cost and Benefit Analysis in Relation to Efficiency of Use of Energy, Nitrogen and Water. New Phytologist, 101, 25-77.
http://dx.doi.org/10.1111/j.1469-8137.1985.tb02816.x - 262. Hecht, U. and Mohr, H. (1990) Factors Controlling Nitrate and Ammonium Accumulation in Mustard (Sinapis alba) Seedlings. Physiologia Plantarum, 78, 379-387.
http://dx.doi.org/10.1111/j.1399-3054.1990.tb09052.x - 263. Yuan, L., Graff, L., Loque, D., Kojima, S., Tsuchiya, Y.N., Takahashi, H. and Von Wiren, N. (2009) Atamt1.4, a Pollen-Specific High-Affinity Ammonium Transporter of the Plasma Membrane in Arabidopsis. Plant and Cell Physiology, 50, 13-25.
http://dx.doi.org/10.1093/pcp/pcn186 - 264. Bu, Y., Takano, T., Nemoto, K. and Liu, S. (2011) Research Progress of Ammonium Transporter in Rice Plants. Genomics and Applied Biology, 2, 19-23.
- 265. Li, S.M. and Shi, W.M. (2006) Quantitative Characterization of Nitrogen Regulation of Osamt1;1, Osamt1;2, and Osamt2;2 Expression in Rice Seedlings. Russian Journal of Plant Physiology, 6, 837-843.
http://dx.doi.org/10.1134/S102144370606015X - 266. Gaur, V.S., Singh, U.S., Gupta, A.K. and Kumar, A. (2012) Understanding the Differential Nitrogen Sensing Mechanism in Rice Genotypes through Expression Analysis of High and Low Affinity Ammonium Transporter Genes. Molecular Biology Reports, 39, 2233-2241.
http://dx.doi.org/10.1007/s11033-011-0972-2 - 267. Morgan, M.A. and Jackson, W.A. (1998) Inward and Outward Movement of Ammonium in Root Systems: Transient Responses during Recovery from Nitrogen Deprivation in Presence of Ammonium. Journal of Experimental Botany, 39, 179-191.
http://dx.doi.org/10.1093/jxb/39.2.179 - 268. Shelden, M.C., Dong, B., De Bruxelles, G.L., Trevaskis, B., Whelan, J., Ryan, P.R. and Howitt, S.M. (2001) Arabidopsis Ammonium Transporter, Atamt1;1 and Atamt1;2, Have Different Biochemical Properties and Functional Roles. Plant Soil, 231, 151-160.
http://dx.doi.org/10.1023/A:1010303813181 - 269. Pearson, J.N., Finnemann, J. and Schjoerring, J.K. (2005) Regulation of the High-Affinity Ammonium Transporter (Bnamt1; 2) in the Leaves of Brassica napus by Nitrogen Status. Plant Molecular Biology, 4, 483-490.
- 270. Bussell, J.D., Keech, O., Fenske, R. and Smith, S.M. (2013) Requirement for the Plastidial Oxidative Pentose Phosphate Pathway for Nitrate Assimilation in Arabidopsis. The Plant Journal, 75, 578-591.
http://dx.doi.org/10.1111/tpj.12222 - 271. Murphy, A.T. and Lewis, O.A.M. (1987) Effect of Nitrogen Feeding Source on the Supply of Nitrogen from Root to Shoot and the Site of Nitrogen Assimilaton in Maize (Zea maize). New Phytologist, 107, 327-333.
http://dx.doi.org/10.1111/j.1469-8137.1987.tb00184.x - 272. Scheurwater, I., Koren, M., Lambers, H. and Atkin, O.K. (2002) The Contribution of Roots and Shoots to Whole Plant Nitrate Reduction in Fast and Slow Growing Grass. Journal of Experimental Botany, 53, 1635-1642.
- 273. Cookson, S.J., Williams, L.E. and Miller, A.J. (2005) Light-Dark Changes in Cytosolic Nitrate Pools Depend on Nitrate Reductase Activity in Arabidopsis Leaf Cells. Plant Physiology, 138, 1097-1105.
http://dx.doi.org/10.1104/pp.105.062349 - 274. Fedorova, E., Greenwood, J.S. and Oaks, A. (2005) In-Situ Lotalization of Nitrate Reductase in Maize Roots. Planta, 194, 279-2815.
http://dx.doi.org/10.1007/BF01101689 - 275. Meyer, C. and Stitt, M. (2001) Nitrate Reduction and Signaling. In: Lea, P.J. and Morot-Gaudry, J.F., Eds., Plant Nitrogen, Springer-Verlag, Berlin, 61-78.
- 276. Kaiser, W.M. and Huber, S.C. (2001) Post-Translational Regulation of Nitrate Reductase: Mechanism, Physiological Relevance and Environmental Triggers. Journal of Experimental Botany, 52, 1981-1989.
http://dx.doi.org/10.1093/jexbot/52.363.1981 - 277. Fan, X., Gordon-Weeks, R., Shen, Q. and Miller, A.J. (2006) Glutamine Transport and Feedback Regulation of Nitrate Reductase Activity in Barley Roots Leads to Changes in Cytosolic Nitrate Pools. Journal of Experimental Botany, 57, 1333-1340.
http://dx.doi.org/10.1093/jxb/erj110 - 278. Fritz, C., Mueller, C., Matt, P., Feil, R. and Stitt, M. (2006) Impact of the C-N Status on the Amino Acid Profile in Tobacco Source Leaves. Plant, Cell & Environment, 29, 2055-2076.
http://dx.doi.org/10.1111/j.1365-3040.2006.01580.x - 279. Lemaitre, T., Gaufichon, L., Boutet-Mercey, S., Christ, A. and Masclaux Daubresse, C. (2008) Enzymatic and Metabolic Diagnostic of Nitrogen Deficiency in Arabidopsis thaliana Wassileskija Accession. Plant and Cell Physiology, 49, 1056-1065.
http://dx.doi.org/10.1093/pcp/pcn081 - 280. Redinbaugh, M.G. and Campbell, W.H. (1998) Nitrate Regulation of the Oxidative Pentose Phosphate Pathway in Maize (Zea mays L. Root Plastids Induction of 6-Phosphogluconate Dehydrogenase Activity Protein and Transcript Levels. Plant Science, 134, 129-140.
http://dx.doi.org/10.1016/S0168-9452(98)00048-X - 281. Wang, R., Tischner, R., Gutiérrez, R.A., Hoffman, M., Xing, X., Chen, M. and Coruzzi, G. (2004) Genomic Analysis of the Nitrate Response Using a Nitrate Reductase-Null Mutant of Arabidopsis. Plant Physiology, 136, 2512-2522.
http://dx.doi.org/10.1104/pp.104.044610 - 282. Wang, R., Xing, X. and Crawford, N. (2007) Nitrite Acts as a Transcriptome Signal at Micromolar Concentrations in Arabidopsis Roots. Plant Physiology, 145, 1735-1745.
http://dx.doi.org/10.1104/pp.107.108944 - 283. Peuke, A.D. (2010) Correlations in Concentrations, Xylem and Phloem Flows, and Partitioning of Elements and Ions in Intact Plants: A Summary and Statistical Re-Evaluation of Modelling Experiments in Ricinus communis. Journal of Experimental Botany, 61, 635-655.
http://dx.doi.org/10.1093/jxb/erp352 - 284. Chen, C.Z., Lv, X.F., Li, J.Y., Yi, H.Y. and Gong, J.M. (2012) Arabidopsis NRT1.5 Is Another Essential Component in the Regulation of Nitrate Reallocation and Stress Tolerance. Plant Physiology, 159, 1582-1590.
http://dx.doi.org/10.1104/pp.112.199257 - 285. Li, J.Y., Fu, Y.L., Pike, S.M., Bao, J., Tian, W. and Zhang, Y. (2010) The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. The Plant Cell, 22, 1633-1646.
http://dx.doi.org/10.1105/tpc.110.075242 - 286. Tang, Z., Fan, X., Li, Q., Feng, H., Miller, A.J. and Shen, Q. (2012) Knockdown of a Rice Stelar Nitrate Transporter Alters Long-Distance Translocation but Not Root Influx. Plant Physiology, 160, 2052-2063.
http://dx.doi.org/10.1104/pp.112.204461 - 287. Feng, H., Yan, M., Fan, X., Li, B., Shen, Q., Miller, A.J. and Xu, G. (2011) Spatial Expression and Regulation of Rice High-Affinity Nitrate Transporters by Nitrogen and Carbon Status. Journal of Experimental Botany, 62, 2319-2332.
http://dx.doi.org/10.1093/jxb/erq403 - 288. Chiu, C.C., Lin, C.S., Hsia, A.P., Su, R.C., Lin, H.L. and Tsay, Y.F. (2004) Mutation of a Nitrate Transporter, Atnrt1:4, Results in a Reduced Petiole Nitrate Content and Altered Leaf Development. Plant & Cell Physiology, 45, 1139-1148.
http://dx.doi.org/10.1093/pcp/pch143 - 289. Hayashi, H. and Chino, M. (1985) Nitrate and Other Anions in the Rice Phloem Sap. Plant and Cell Physiology, 26, 325-330.
- 290. Hayashi, H. and Chino, M. (1986) Collection of Pure Phloem Sap from Wheat and Its Chemical Composition. Plant & Cell Physiology, 27, 1387-1393.
- 291. Wang, Y.Y. and Tsay, Y.F. (2011) Arabidopsis Nitrate Transporter NRT1. 9 Is Important in Phloem Nitrate Transport. The Plant Cell, 23, 1945-1957.
http://dx.doi.org/10.1105/tpc.111.083618 - 292. Hine, J.C. and Sprent, J.I. (1988) Growth of Phaseolus Vulgaris on Various Nitrogen Sources: The Importance of Urease. Journal of Experimental Botany, 39, 1505-1512.
http://dx.doi.org/10.1093/jxb/39.11.1505v - 293. Witte, C.P., Tiller, S., Isidore, E., Davies, H.V. and Taylor, M.A. (2005) Analysis of Two Alleles of the Urease Gene from Potato: Polymorphisms, Expression, and Extensive Alternative Splicing of the Corresponding Mrna. Journal of Experimental Botany, 56, 91-99.
http://dx.doi.org/10.1038/nchembio.265 - 294. Werner, A.K., Romeis, T. and Witte, C.P. (2010) Ureide Catabolism in Arabidopsis thaliana and Escherichia Coli. Nature Chemical Biology, 6, 19-21.
http://dx.doi.org/10.1038/nchembio.265 - 295. Merigout, M., Lelandais, F., Bitton, X., Briand, C. and Meyer, C. (2008) Physiological and Transcriptomic Aspects of Urea Uptake and Assimilation in Arabidopsis Plants. Plant Physiology, 147, 1225-1238.
http://dx.doi.org/10.1104/pp.108.119339 - 296. Kluge, C., Lahr, J., Hanitzsch, M., Bolte, S., Golldack, D. and Dietz, K.J. (2003) New Insight into the Structure and Regulation of the Plant Vacuolar H+-Atpase. Journal of Bioenergetics and Biomembranes, 35, 377-388.
http://dx.doi.org/10.1023/A:1025737117382 - 297. Maeshima, M. (2000) Vacuolar H+-Pyrophosphatase. Biochimica et Biophysica Acta, 1465, 37-51.
http://dx.doi.org/10.1016/S0005-2736(00)00130-9 - 298. Chen, J.K., Luo, X.M., Yin, J.L., Zhan, P.W. and Shen, Q.R. (2004) Distribution and Remobilization of Nitrate in Two Cultivars of Pakchoi Plant. Scientia Agricultura Sinica, 38, 2277-2282.
- 299. Martinoia, U., Heck, A. and Wiemken, A. (1981) Vacuoles as Storage Compartments of Nitrate in Barley Leaves. Nature, 289, 292-294.
http://dx.doi.org/10.1038/289292a0 - 300. Wang, T., Jia, J.L. and Shen, Q.R. (2008b) Relationship between Nitrate Remobilization in Root Vacuoles and Plant Growth of Two Genotypes of Lettuce. Acta Pedologica Sinica, 45, 555-560.
- 301. Zhao, S.P., Ye, X.Z., Zhang, Y.Z. and Zheng, J.C. (2010) The Contribution of Bnnrt1 and Bnnrt2 to Nitrate Accumulation Varied According to Genotypes in Chinese Cabbage. African Journal of Biotechnology, 9, 4910-4917.
- 302. Chopin, F., Orsel, M., Dorbe, M.F., Chardon, F., Truong, H.N., Miller, A.J. and Krapp, A. (2007) The Arabidopsis ATNRT2.7 Nitrate Transporter Controls Nitrate Content in Seeds. The Plant Cell, 19, 1590-1602.
http://dx.doi.org/10.1105/tpc.107.050542 - 303. Kotur, Z., Mackenzie, N., Ramesh, S., Tyerman, S.D., Kaiser, B.N. and Glass, A.A.M. (2012) Nitrate Transport Capacity of the Arabidopsis thaliana NRT2 Family Members and Their Interactions with Atnar2.1. New Phytologist, 194, 724-731.
http://dx.doi.org/10.1111/j.1469-8137.2012.04094.x - 304. Plett, D., Toubia, J., Garnett, T., Tester, M., Kaiser, B.N. and Baumann, U. (2010) Dichotomy in the NRT Gene Families of Dicots and Grass Species. PLoS ONE, 5, e15289.
http://dx.doi.org/10.1371/journal.pone.0015289 - 305. Miller, A.J., Cookson, S.J., Smith, S.J. and Wells, D.M. (2001) The Use of Microelectrodes to Investigate Compartmentation and the Transport of Metabolized Inorganic Ions in Plants. Journal of Experimental Botany, 52, 541-549.
http://dx.doi.org/10.1093/jexbot/52.356.541 - 306. Bertl, R. and Kaldenhoff, R. (2007) Function of a Separate NH3-Pore in Aquaporin TIP2; 2 from Wheat. FEBS Letters, 581, 5413-5417.
http://dx.doi.org/10.1016/j.febslet.2007.10.034 - 307. Holm, L.M., Jahn, T.P., Moller, A.L., Schjoerring, J.K., Ferri, D. and Klaerke, D.A. (2005) NH3 and Permeability in Aquaporin-Expressing Xenopus Oocytes. Pflügers Archiv, 450, 415-428.
http://dx.doi.org/10.1007/s00424-005-1399-1 - 308. Klebl, F., Wolf, M. and Sauer, N. (2003) A Defect in the Yeast Plasma Membrane Urea Transporter Dur3p Is Complemented by Cpnip1, a Nod26-Like Protein from Zucchini (Cucurbita pepo L.), and by Arabidopsis thaliana Delta- TIP or Gamma-TIP. FEBS Letters, 547, 69-74.
http://dx.doi.org/10.1016/S0014-5793(03)00671-9 - 309. Uehlein, N., Lovisolo, C., Siefritz, F. and Kaldenhoff, R. (2003) The Tobacco Aquaporin Ntaqp1 Is a Membrane CO2 Pore with Physiological Functions. Nature, 425, 734-737.
http://dx.doi.org/10.1038/nature02027 - 310. Howitt, S.M. and Udvardi, M.K. (2005) Structure, Function and Regulation of Ammonium Transporters in Plants. Biochimica et Biophysica Acta, 1465, 152-170.
http://dx.doi.org/10.1016/S0005-2736(00)00136-X - 311. Wood, C.C., Poree, F., Dreyer, I., Koehler, G.J. and Udvardi, M.K. (2006) Mechanisms of Ammonium Transport, Accumulation, and Retention in Ooyctes and Yeast Cells Expressing Arabidopsis Atamt1; 1. FEBS Letters, 580, 3931- 3936.
http://dx.doi.org/10.1016/j.febslet.2006.06.026 - 312. Gaspar, M., Sissoëff, I., Bousser, A., Roche, O., Hoarau, J. and Mahé, A. (2003) Cloning and Characterization of Zmpip1-5, an Aquaporin Transporting Water and Urea. Plant Science, 165, 21-31.
http://dx.doi.org/10.1016/S0168-9452(03)00117-1 - 313. Kim, S.H., Kim, K., Ju, H.W., Lee, H. and Hong, S.W. (2008) Over Expression of Gene Encoding Tonoplast Intrinsic Aquaporin Promotes Urea Transport in Arabidopsis. Journal of Applied Biological Chemistry, 51, 102-110.
http://dx.doi.org/10.3839/jabc.2008.017 - 314. Malagoli, P., Laine, P., Rossato, L. and Ourry, A. (2005) Dynamics of Nitrogen Uptake and Mobilization in Field-Grown Winter Oilseed Rape (Brassica napus) from Stem Extension to Harvest. Annals of Botany, 95, 853-861.
http://dx.doi.org/10.1093/aob/mci091 - 315. Rossato, L., Laine, P. and Ourry, A. (2001) Nitrogen Storage and Remobilization in Brassica napus L. during the Growth Cycle: Nitrogen Fluxes within the Plant and Changes in Soluble Protein Patterns. Journal of Experimental Botany, 52, 1655-1663.
http://dx.doi.org/10.1093/jexbot/52.361.1655 - 316. Schiltz, S., Munier-Jolain, N., Jeudy, C., Burstin, J. and Salon, C. (2005) Dynamics of Exogenous Nitrogen Partitioning and Nitrogen Remobilization from Vegetative Organs in Pea Revealed by 15N in Vivo Labelling throughout Seed Filling. Plant Physiology, 137, 1463-1473.
http://dx.doi.org/10.1104/pp.104.056713 - 317. Simpson, R.J., Lambers, H. and Dalling, M.J. (1993) Nitrogen Redistribution during Grain Growth in Wheat (Triticum aestivum L.): IV. Development of a Quantitative Model of the Translocation of Nitrogen to the Grain. Plant Physiology, 71, 7-14.
http://dx.doi.org/10.1104/pp.71.1.7 - 318. Wendler, R., Carvalho, P., Pereira, J. and Millard, P. (1995) Role of Nitrogen Remobilization from Old Leaves for New Leaf Growth of Eucalyptus Globulus Seedlings. Tree Physiology, 15, 679-683.
http://dx.doi.org/10.1093/treephys/15.10.679 - 319. Izumi, M., Wada, S., Makino, A. and Ishida, H. (2010) The Autophagic Degradation of Chloroplasts via Rubisco-Containing Bodies Is Specifically Linked to Leaf Carbon Status but Not Nitrogen Status in Arabidopsis. Plant Physiology, 154, 1196-1209.
http://dx.doi.org/10.1104/pp.110.158519 - 320. Mae, T., Makino, A. and Ohira, K. (1984) Changes in the Amounts of Ribulose Biphosphate Carboxylase Synthesized and Degraded during the Life Span of Rice Leaf (Oryza sativa L.). Plant and Cell Physiology, 24, 1079-1086.
- 321. Feller, U. and Keist, M. (1986) Senescence and Nitrogen Metabolism in Annual Plants. In: Lambers, H., Neetson, J.J. and Stulen, I., Eds., Fundamental, Ecological and Agricultural Aspects of Nitrogen Metabolism, Martinus Nijhoff Publishers, Dordrecht, 219-234.
http://dx.doi.org/10.1007/978-94-009-4356-8_34 - 322. Gallais, A., Coque, M., Quilléré, I., Le Gouis, J., Prioul, J.L. and Hirel, B. (2007) Estimating the Proportion of Nitrogen Remobilization and of Postsilking Nitrogen Uptake Allocated to Maize Kernels by Nitrogen-15 Labeling. Crop Science, 47, 685-691.
http://dx.doi.org/10.2135/cropsci2006.08.0523 - 323. Kichey, T., Hirel, B., Heumez, E., Dubois, F. and Le Gouis, J. (2007) In Winter Wheat (Triticum aestivum L.), Post-Anthesis Nitrogen Uptake and Remobilisation to the Grain Correlates with Agronomic Traits and Nitrogen Physiological Markers. Field Crops Research, 102, 22-32.
http://dx.doi.org/10.1016/j.fcr.2007.01.002 - 324. Masclaux-Daubresse, C., Quilleré, I., Gallais, A. and Hirel, B. (2001) The Challenge of Remobilisation in Plant Nitrogen Economy. A Survey of Physio-Agronomic and Molecular Approaches. Annals of Applied Biology, 138, 68-81.
- 325. Ono, Y., Wada, S., Izumi, M., Makino, A. and Ishida, H. (2013) Evidence for Contribution of Autophagy to Rubisco Degradation during Leaf Senescence in Arabidopsis thaliana. Plant Cell Environment, 36, 1147-1159.
http://dx.doi.org/10.1111/pce.12049 - 326. Almagro, A., Lin, S.H. and Tsay, Y.F. (2008) Characterization of the Arabidopsis Nitrate Transporter NRT1.6 Reveals a Role of Nitrate in Early Embryo Development. The Plant Cell, 20, 3289-3299.
http://dx.doi.org/10.1105/tpc.107.056788 - 327. Fan, S.C., Lin, C.S., Hsu, P.K., Lin, S.H. and Tsay, Y.F. (2009) The Arabidopsis Nitrate Transporter NRT1.7, Expressed in Phloem, Is Responsible for Source-to-Sink Remobilization of Nitrate. The Plant Cell, 21, 2750-2761.
http://dx.doi.org/10.1105/tpc.109.067603 - 328. Hsu, P.K. and Tsay, Y.F. (2013) Two Phloem Nitrate Transporters, NRT1.11 and NRT1.12, Are Important for Redistributing Xylem-Borne Nitrate to Enhance Plant Growth. Plant Physiology, 163, 844-856.
http://dx.doi.org/10.1104/pp.113.226563 - 329. Soto, G., Alleva, K., Mazzella, M.A., Amodeo, G. and Muschietti, J.P. (2008) Attip1;3 and Attip5;1, the Only Highly Expressed Arabidopsis Pollen-Specific Aquaporins, Transport Water and Urea. FEBS Letters, 582, 4077-4082.
http://dx.doi.org/10.1016/j.febslet.2008.11.002 - 330. Soto, G., Fox, R., Ayub, N., Alleva, K., Guaimas, F., Erijman, E.J., Mazzella, A. and Amodeo, G. (2010) TIP5; 1 Is an Aquaporin Specifically Targeted to Pollen Mitochondria and Is Probably Involved in Nitrogen Remobilization in Arabidopsis thaliana. The Plant Journal, 64, 1038-1047.
http://dx.doi.org/10.1111/j.1365-313X.2010.04395.x - 331. Masclaux-Daubresse, C., Reisdorf-Cren, M. and Pageau, K. (2006) Glutamine Synthetase-Glutamate Synthase Pathway and Glutamate Dehydrogenase Play Distinct Roles in the Sink-Source Nitrogen Cycle in Tobacco. Plant Physiology, 140, 444-456.
http://dx.doi.org/10.1104/pp.105.071910 - 332. Patrick, J.W. and Offler, C.E. (2001) Compartmentation of Transport and Transfer Events in Developing Seeds. Journal of Experimental Botany, 52, 551-564.
http://dx.doi.org/10.1093/jexbot/52.356.551 - 333. Rochat, C. and Boutin, J.P. (1991) Metabolism of Phloem-Borne Amino Acids in Maternal Tissues of Fruit of Nodulated or Nitrate-Fed Pea Plants (Pisum sativum L.). Journal of Experimental Botany, 42, 207-214.
http://dx.doi.org/10.1093/jxb/42.2.207 - 334. Bailey, C.J. and Boulter, D. (1971) Urease, a Typical Seed Protein of the Leguminosae. In: Harborne, J., Boulter, D. and Turner, B., Eds., Chemotaxonomy of the Leguminosae, Academic Press, New York, 485-502.
- 335. Stebbins, N.E. and Polacco, J.C. (1995) Urease Is Not Essential for Ureide Degradation in Soybean. Plant Physiology, 109, 169-175.
- 336. Zonia, L.E., Stebbins, N.E. and Polacco, J.C. (1995) Essential Role of Urease in Germination of Nitrogen-Limited Arabidopsis thaliana Seeds. Plant Physiology, 107, 1097-1103.
http://dx.doi.org/10.1104/pp.107.4.1097 - 337. Suryapriya, P., Snehalatha, A., Kayalvili, U., Krishna, R., Singh, S. and Ulaganathan, K. (2009) Genome-Wide Analyses of Rice Root Development Qtls and Development of an Online Resource, Rootbrowse. Bioinformation, 3, 279-281.
http://dx.doi.org/10.6026/97320630003279 - 338. Rani, S. (2010) Rnai Mediated Functional Validation of Root Development QTL Associated Candidate Genes in Rice. PhD Thesis, Osmania University, Hyderabad.
- 339. Forde, B.G. and Lorenzo, H. (2011) The Nutritional Control of Root Development. Plant Soil, 232, 51-68.
http://dx.doi.org/10.1023/A:1010329902165 - 340. Roycewicz, P. and Malamy, J.E. (2012) Dissecting the Effects of Nitrate, Sucrose and Osmotic Potential on Arabidopsis Root and Shoot System Growth in Laboratory Assays. Philosophical Transactions of the Royal Society B, 367, 1489-1500.
http://dx.doi.org/10.1098/rstb.2011.0230 - 341. Vidal, E.A., Araus, V., Lu, C., Parry, G. and Green, P.J. (2010) Nitrate-Responsive Mir393/AFB3 Regulatory Module Controls Root System Architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 107, 4477-4482.
http://dx.doi.org/10.1073/pnas.0909571107 - 342. Walch-Liu, P., Ivanov, I.I., Filleur, S., Gan, Y., Remans, T. and Forde, B. (2006) Nitrogen Regulation of Root Branching. Annals of Botany, 97, 875-881.
http://dx.doi.org/10.1093/aob/mcj601 - 343. Walch-Liu, P. and Forde, B.G. (2008) Nitrate Signaling Mediated by the NRT1.1 Nitrate Transporter Antagonises L-Glutamate-Induced Changes in Root Architecture. The Plant Journal, 54, 820-828.
http://dx.doi.org/10.1111/j.1365-313X.2008.03443.x - 344. Zhang, H., Jennings, A., Barlow, P.W. and Forde, B.G. (1999) Dual Pathways for Regulation of Root Branching by Nitrate. Proceedings of the National Academy of Sciences of the United States of America, 96, 6529-6534.
http://dx.doi.org/10.1073/pnas.96.11.6529 - 345. Zhang, H., Rong, H. and Pilbeam, D. (2007) Signaling Mechanisms Underlying the Morphological Responses of the Root System to Nitrogen in Arabidopsis thaliana. Journal of Experimental Biology, 58, 2329-2338.
- 346. De Smet, I., Tetsumura, T., De Rybel, B., Frey, N.F., Laplaze, L., Casimiro, I. and Swarup, R. (2007) Auxin-Dependent Regulation of Lateral Root Positioning in the Basal Meristem of Arabidopsis. Development, 134, 681-690.
http://dx.doi.org/10.1242/dev.02753 - 347. Laskowski, M., Grieneisen, V.A., Hofhuis, H., Hove, C.A., Hogeweg, P., Maree, A.F. and Scheres, B. (2008) Root System Architecture from Coupling Cell Shape to Auxin Transport. PLoS Biology, 6, E307.
http://dx.doi.org/10.1371/journal.pbio.0060307 - 348. Mounier, E., Pervent, M., Ljung, K., Gojon, A. and Nacry, P. (2014) Auxin-Mediated Nitrate Signaling by NRT1.1 Participates in the Adaptive Response of Arabidopsis Root Architecture to the Spatial Heterogeneity of Nitrate Availability. Plant Cell And Environment, 37, 162-174.
http://dx.doi.org/10.1111/pce.12143 - 349. Péret, B., Larrieu, A. and Bennet, M.J. (2009) Lateral Root Emergence: A Difficult Birth. Journal of Experimental Biology, 60, 3637-3643.
http://dx.doi.org/10.1093/jxb/erp232 - 350. Bloom, A.J., Paul, A., Meyerhoff, P.A., Taylor, A.R. and Rost, T.L. (2002) Root Development and Absorption of Ammonium and Nitrate from the Rhizosphere. Journal of Plant Growth Regulation, 21, 416-431.
http://dx.doi.org/10.1007/s00344-003-0009-8 - 351. Gifford, M.L., Dean, A., Gutierrez, R.A., Coruzzi, G.M. and Birnbaum, K.D. (2008) Cell-Specific Nitrogen Responses Mediate Developmental Plasticity. Proceedings of the National Academy of Sciences of the United States of America, 105, 803-808.
http://dx.doi.org/10.1073/pnas.0709559105 - 352. De Smet, I., Signora, L., Beeckman, T., Inze, D., Foyer, C.H. and Zhang, H. (2003) An Abscisic Acid-Sensitive Checkpoint in Lateral Root Development of Arabidopsis. The Plant Journal, 33, 543-555.
http://dx.doi.org/10.1046/j.1365-313X.2003.01652.x - 353. Ivanchenko, M.G., Muday, G.K. and Dubrovsky, J.G. (2008) Ethylene-Auxin Interactions Regulate Lateral Root Initiation and Emergence in Arabidopsis thaliana. The Plant Journal, 55, 335-347.
http://dx.doi.org/10.1111/j.1365-313X.2008.03528.x - 354. Tian, Q.Y., Sun, P. and Zhang, W.H. (2009) Ethylene Is Involved in Nitrate-Dependent Root Growth and Branching in Arabidopsis thaliana. New Phytologist, 184, 918-931.
http://dx.doi.org/10.1111/j.1469-8137.2009.03004.x - 355. Mantelin, S., Desbrosses, G., Larcher, M., Tranbarger, T.J., Cleyet-Marel, J.C. and Touraine, B. (2006) Nitrate-Dependent Control of Root Architecture and N Nutrition Are Altered by a Plant Growth-Promoting Phyllobacterium sp. Planta, 223, 591-603.
http://dx.doi.org/10.1007/s00425-005-0106-y - 356. Barth, C., Gouzd, Z.A., Steelt, H.P. and Imperio, R.M. (2010) A Mutation in GDP-Mannose Pyrophosphorylase Causes Conditional Hypersensitivity to Ammonium, Resulting in Arabidopsis Root Growth Inhibition, Altered Ammonium Metabolism, and Hormone Homeostasis. Journal of Experimental Botany, 61, 379-394.
http://dx.doi.org/10.1093/jxb/erp310 - 357. Cao, Y.W., Glass, A.D.M. and Crawford, N.M. (1993) Ammonium Inhibition of Arabidopsis Root Growth Can Be Reversed by Potassium and by Auxin Resistance Mutations Aux1, Axr1, and Axr2. Plant Physiology, 102, 983-989.
http://dx.doi.org/10.1104/pp.102.3.983 - 358. Li, Q., Li, B., Kronzucker, H.J. and Shi, W. (2010) Root Growth Inhibition by in Arabidopsisis Mediated by the Root Tip and Is Linked to Efflux and Gmpase Activity. Plant Cell and Environment, 33, 1529-1542.
- 359. Qin, C., Qian, W., Wang, Y., Wu, C., Yu, X., Jiang, D., Wang, P. and Wu, P. (2008) GDP-Mannose Pyrophosphorylase Is a Genetic Determinant of Ammonium Sensitivity in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 105, 18308-18313.
http://dx.doi.org/10.1073/pnas.0806168105 - 360. Lejay, L., Tillard, P., Lepetit, M., Olife, F., Filleur, S., Daniel-Vedele, F. and Gojon, A. (1999) Molecular and Functional Regulation of Two Uptake Systems by N- and C-Status of Arabidopsis Plants. The Plant Journal, 18, 509-519.
http://dx.doi.org/10.1046/j.1365-313x.1999.00480.x - 361. Malamy, J. and Ryan, K. (2001) Environmental Regulation of Lateral Root Initiation in Arabidopsis. Plant Physiology, 127, 899-909.
http://dx.doi.org/10.1104/pp.010406 - 362. Walch-Liu, P., Liu, L.H., Remans, T., Tester, M. and Forde, B.G. (2006) Evidence That L-Glutamate Can Act as an Exogenous Signal to Modulate Root Growth and Branching in Arabidopsis thaliana. Plant and Cell Physiology, 47, 1045-1057.
http://dx.doi.org/10.1093/pcp/pcj075 - 363. Tian, Q., Chen, F., Liu, J., Zhang, F. and Mi, G. (2008) Inhibition of Maize Root Growth by High Nitrate Supply Is Correlated with Reduced IAA Levels in Roots. Journal of Plant Physiology, 165, 942-951.
http://dx.doi.org/10.1016/j.jplph.2007.02.011 - 364. Ma, W., Li, J., Qu, B., He, X., Zhao, X., Li, B., Fu, X. and Tong, Y. (2014) Auxin Biosynthetic Gene TAR2 Is Involved in Low Nitrogen Mediated Reprogramming of Root Architecture in Arabidopsis. The Plant Journal, 78, 70-79.
http://dx.doi.org/10.1111/tpj.12448 - 365. Barber, M.J. and Kay, C.J. (1996) Superoxide Production during Reduction of Molecular Oxygen by Assimilatory Nitrate Reductase. Archives of Biochemistry and Biophysics, 326, 227-232.
http://dx.doi.org/10.1006/abbi.1996.0069 - 366. Meyer, C., Lea, U.S., Provan, F., Kaiser, W.M. and Lillo, C. (2005) Is Nitrate Reductase a Major Player in the Plant NO (Nitric Oxide) Game? Photosynthesis Research, 83, 181-189.
http://dx.doi.org/10.1007/s11120-004-3548-3 - 367. Yamasaki, H. and Sakihama, Y. (2000) Simultaneous Production of Nitric Oxide and Peroxynitrite by Plant Nitrate Reductase: In Vitro Evidence for the NR-Dependent Formation of Active Nitrogen Species. FEBS Letters, 468, 89-92.
http://dx.doi.org/10.1016/S0014-5793(00)01203-5 - 368. Von Wirén, N., Gazzarrini, S. and Frommer, W.B. (1997) Regulation of Mineral Nitrogen Uptake in Plants. Plant and Soil, 196, 191-199.
http://dx.doi.org/10.1023/A:1004241722172 - 369. Filleur, S. and Daniel-Vedele, F. (1997) Expression Analysis of a High-Affinity Nitrate Transporter Isolated from Arabidopsis thaliana by Differential Display. Planta, 207, 461-469.
http://dx.doi.org/10.1007/s004250050505 - 370. Santi, S., Locci, G., Pinton, R. and Varanini, Z. (2003)Induction of Nitrate Uptake in Maize Roots: Expression of a Putative High-Affinity Nitrate Transporter and Plasma Membrane H+-Atpase Isoforms. Journal of Experimental Botany, 54, 1851-1864.
http://dx.doi.org/10.1093/jxb/erg208 - 371. Yan, M., Fan, X.R., Feng, H.M., Miller, A.J., Shen, Q. and Xu, G.H. (2011) Rice OsNAR2.1 Interacts with OsNRT2.1, OsNRTt2.2 and OsNRT2.3a Nitrate Transporters to Provide Uptake over High and Low Concentration Ranges. Plant, Cell & Environment, 34, 1360-1372.
http://dx.doi.org/10.1111/j.1365-3040.2011.02335.x - 372. Dluzniewska, P., Gessler, A., Kopriva, S., Strand, M., Novak, O., Dietrich, H. and Rennenberg, H. (2006) Supply of Glutamine and Active Cytokinin to the Roots Reduces NO3 Uptake Rates in Poplar. Plant Cell and Environment, 29, 1284-1297.
http://dx.doi.org/10.1111/j.1365-3040.2006.01507.x - 373. Muller, B., Tilliard, P. and Touraine, V. (1995) Nitrate Fluxes in Soybean Seedling Roots and Their Response to Amino Acids: An Approach Using 15N. Plant Cell and Environment, 18, 1267-1279.
http://dx.doi.org/10.1111/j.1365-3040.1995.tb00186.x - 374. Gansel, X., Muños, S., Tillard, P. and Gojon, A. (2001) Differential Regulation of The NO3- and Transporter Genes Atnrt2.1 and Atamt1;1 in Arabidopsis: Relation with Long-Distance and Local Controls by N Status of the Plant. The Plant Journal, 26, 143-155.
http://dx.doi.org/10.1046/j.1365-313x.2001.01016.x - 375. Widiez, T., Kafafiel, E.L., Girin, T., Berr, A., Ruffel, S., Krouk, G. and Vayssières, A. (2011) High Nitrogen Insensitive 9 (HNI9)-Mediated Systemic Repression of Root Uptake Is Associated with Changes in Histone Methylation. Proceedings of the National Academy of Sciences of the United States of America, 108, 13329-13334.
http://dx.doi.org/10.1073/pnas.1017863108 - 376. Engelsberger, W.R. and Schulze, W.X. (2012) Nitrate and Ammonium Lead to Distinct Global Dynamic Phosphorylation Patterns When Resupplied to Nitrogen-Starved Arabidopsis Seedlings. The Plant Journal, 69, 978-995.
http://dx.doi.org/10.1111/j.1365-313x.2011.04848.x - 377. Vialaret, J., Pietro, M.D., Hem, S., Maurel, C., Rossignol, M. and Santoni, V. (2014) Phosphorylation Dynamics of Membrane Proteins from Arabidopsis Roots Submitted to Salt Stress. Proteomics, 14, 1058-1070.
http://dx.doi.org/10.1002/pmic.201300443 - 378. De Jong, F., Thodey, K., Lejay, L.V. and Bevan, M.V. (2013) Glucose Elevates NRT2.1 Protein Levels and Nitrate Transport Activity Independently of Its HXK1-Mediated Stimulation of NRT2.1 Expression. Plant Physiology, 164, 308-320.
- 379. Kruger, N.J. and Von Schaewen, A. (2003) The Oxidative Pentose Phosphate Pathway: Structure and Organisation. Current Opinion in Plant Biology, 16, 236-246.
http://dx.doi.org/10.1016/S1369-5266(03)00039-6 - 380. Lejay, L., Wirth, V., Pervent, M., Cross, J.M.F., Tillard, P. and Gojon, A. (2008) Oxidative Pentose Phosphate Pathway-Dependent Sugar Sensing as a Mechanism for Regulation of Root Ion Transporters by Photosynthesis. Plant Physiology, 146, 2036-2053.
http://dx.doi.org/10.1104/pp.107.114710 - 381. Lejay, L., Gansel, X., Cerezo, M., Tillard, P., Muller, C., Krapp, A. And Von Wiren, N. (2003) Regulation of Root Ion Transporters by Photosynthesis: Functional Importance and Relation with Hexokinase. The Plant Cell, 5, 2218-2232.
http://dx.doi.org/10.1105/tpc.013516 - 382. Kronzucker, H.J., Siddiqi, M.Y. and Glass, A.D.M. (1995) Compartmentation and Flux Characteristics of Ammonium in Spruce. Planta, 196, 691-698.
http://dx.doi.org/10.1007/BF01106762 - 383. Rawat, S.R., Silim, S.N., Kronzucker, H.J., Siddiqi, M.Y. and Glass, A.D.M. (1999) Atamt1 Gene Expression and Uptake in Roots of Arabidopsis thaliana: Evidence for Regulation by Root Glutamine Levels. The Plant Journal, 19, 143-152.
http://dx.doi.org/10.1046/j.1365-313X.1999.00505.x - 384. Loque, D. and Wiren, N. (2004) Regulatory Levels for the Transport of Ammonium in Plant Roots. Journal of Experimental Botany, 55, 1293-1305.
http://dx.doi.org/10.1093/jxb/erh147 - 385. Loque, D., Ludewig, U., Yuan, L. and Von Wiren, N. (2005) Tonoplast Intrinsic Proteins AtTIP2; 1 and AtTIP2; 3 Faciliate NH3 Transport into the Vacuole. Plant Physiology, 137, 671-680.
http://dx.doi.org/10.1104/pp.104.051268 - 386. Ortiz-Ramirez, C., Mora, S.I., Trejo, J. and Pantoja, O. (2011) PvAMT1; 1, a Highly Selective Ammonium Transporter That Functions as H+/ Symporter. Journal of Biological Chemistry, 286, 31113-31122.
http://dx.doi.org/10.1074/jbc.M111.261693 - 387. Sogaard, R., Alsterfjord, M., Macaulay, N. and Thomas, Z. (2009) Ammonium Ion Transport by the AMT/Rh Homolog TaAMT1; 1 Is Stimulated by Acidic pH. Pflügers Archiv—European Journal of Physiology, 458, 733-743.
http://dx.doi.org/10.1007/s00424-009-0665-z - 388. Zhao, M., Ding, H., Zhu, J.K., Zhang, F. and Li, W.X. (2011) Involvement of miR169 in the Nitrogen-Starvation Responses in Arabidopsis. New Phytologist, 190, 906-915.
http://dx.doi.org/10.1111/j.1469-8137.2011.03647.x - 389. Li, W.X., Oono, Y., Zhu, J., He, X.J., Wu, J.M., Iida, K., Lu, X.Y., Cui, X., Jin, H. and Zhu, J.K. (2008) The Arabidopsis NFYA5 Transcription Factor Is Regulated Transcriptionally and Post transcriptionally to Promote Drought Resistance. Plant Cell, 20, 2238-2251.
http://dx.doi.org/10.1105/tpc.108.059444 - 390. Mantovani, R. (1999) The Molecular Biology of the CCAAT-Binding Factor NF-Y. Gene, 239, 15-27.
http://dx.doi.org/10.1016/s0378-1119(99)00368-6 - 391. Nelson, D.E. (2007) Plant Nuclear Factor Y (NF-Y) B Subunits Confer Drought Tolerance and Lead to Improved Corn Yields on Water-Limited Acres. Proceedings of the National Academy of Sciences of the United States of America, 104, 16450-16455.
http://dx.doi.org/10.1073/pnas.0707193104 - 392. Zanetti, M.E., Blanco, F.A., Beker, M.P., Battaglia, M. and Aguilar, O.M. (2010) A C Subunit of the Plant Nuclear Factor NF-Y Required for Rhizobial Infection and Nodule Development Affects Partner Selection in the Common Bean-Rhizobium Etli Symbiosis. Plant Cell, 22, 4142-4157.
http://dx.doi.org/10.1105/tpc.110.079137 - 393. .Hsieh, L.C., Lin, S.I., Shih, A.C., Chen, J.W., Lin, W.Y., Tseng, C.Y., Li, W.H. and Chiou, T.J. (2009) Uncovering Small RNA-Mediated Responses to Phosphate Deficiency in Arabidopsis by Deep Sequencing. Plant Physiology, 151, 2120-2132.
http://dx.doi.org/10.1104/pp.109.147280 - 394. Pant, B.D., Musialak-Lange, M., Nuc, P., May, P., Buhtz, A., Kehr, J. and Walther, D. (2009) Identification of Nutrient-Responsive Arabidopsis and Rapeseed Micrornas by Comprehensive Real-Time Polymerase Chain Reaction Profiling and Small RNA Sequencing. Plant Physiology, 150, 1541-1555.
http://dx.doi.org/10.1104/pp.109.139139 - 395. Kiba, T., Kudo, T., Kojima, M. and Sakakibara, H. (2011) Hormonal Control of Nitrogen Acquisition: Roles of Auxin, Abscisic Acid, and Cytokinin. Journal of Experimental Botany, 62, 1399-1409.
http://dx.doi.org/10.1093/jxb/erq410 - 396. Argyros, R.D., Mathews, D.E., Chiang, Y.H., Palmer, C.M., Thibault, D.M., Etheridge, N. and Argyros, D.A. (2008) Type B Response Regulators of Arabidopsis Play Key Roles in Cytokinin Signaling and Plant Development. Plant Cell, 20, 2102-2116.
http://dx.doi.org/10.1105/tpc.108.059584 - 397. Séguéla, M., Briat, J.F., Vert, G. and Curie, C. (2008) Cytokinins Negatively Regulate the Root Iron Uptake Machinery in Arabidopsis through a Growth Dependent Pathway. The Plant Journal, 55, 289-300.
http://dx.doi.org/10.1111/j.1365-313X.2008.03502.x - 398. Aslam, M., Travis, R.L. and Rains, D.W. (1996) Evidence for Substrate Lnduction of a Nitrate Efflux System in Barley Roots. Plant Physiology, 112, 1167-1175.
- 399. Leran, S., Muños, S., Brachet, C., Tillard, P., Gojon, A. and Lacombe, B. (2013) The Arabidopsis NRT1.1 Is a Bidirectional Transporter Involved in Root-to-Shoot Nitrate Translocation. Molecular Plant, 6, 1984-1987.
- 400. Mattsson, M. and Schjoerring, J.K. (2002) Dynamic and Steady-State Responses of Inorganic Nitrogen Pools and NH3 Exchange in Leaves of Lolium perenne and Bromus erectus to Changes in Root Nitrogen Supply. Plant Physiology, 128, 742-750.
http://dx.doi.org/10.1104/pp.010602 - 401. Gutierrez, R.A., Stokes, T.L., Thum, K., Xu, X., Obertello, M. and Katari, M.S. (2008) Systems Approach Identifies an Organic Nitrogen-Responsive Gene Network That Is Regulated by the Master Clock Control Gene CCA1. Proceedings of the National Academy of Sciences of the United States of America, 105, 4939-4944.
http://dx.doi.org/10.1073/pnas.0800211105 - 402. Lam, H.M., Coschigano, K.T., Oliveira, I.C., Melo-Oliveira, R. and Coruzzi, G.M. (1996) The Molecular-Genetics of Nitrogen Assimilation into Amino Acids in Higher Plants. Annual Review of Plant Physiology and Plant Molecular Biology, 47, 569-593.
http://dx.doi.org/10.1146/annurev.arplant.47.1.569 - 403. Ranathunge, K., El-Kereamy, A., Gidda, S., Bi, Y.M. and Rothstein, S.J. (2014) Osamt1;1 Transgenic Rice Plants with Enhanced Permeability Show Superior Growth and Higher Yield under Optimal and Suboptimal Conditions. Journal of Experimental Botany, 65, 965-979.
- 404. Ishiyama, K., Inoue, E., Watanabe-Takahashi, A., Obara, M., Yamaya, T. and Takahashi, H. (2004) Kinetic Properties and Ammonium-Dependent Regulation of Cytosolic Isoenzymes of Glutamine Synthetase in Arabidopsis. Journal of Biological Chemistry, 27916, 598-605.
http://dx.doi.org/10.1074/jbc.m313710200 - 405. Lima, L., Seabra, A., Melo, P., Cullimore, J. and Carvalho, H. (2006) Post-Translational Regulation of Cytosolic Glutamine Synthetase of Medicago truncatula. Journal of Experimental Botany, 57, 2751-2761.
http://dx.doi.org/10.1093/jxb/erl036 - 406. Lothier, J., Gaufichon, L., Sormani, R., Lemaitre, T., Azzopardi, M. and Morin, H. (2011) The Cytosolic Glutamine Synthetase GLN1;2 Plays a Role in the Control of Plant Growth and Ammonium Homeostasis in Arabidopsis Rosettes When Nitrate Supply Is Not Limiting. Journal of Experimental Botany, 62, 1375-1390.
http://dx.doi.org/10.1093/jxb/erq299 - 407. Li, J.Y., Fu, Y.L., Pike, S.M., Bao, J., Tian, W. and Zhang, Y. (2010) The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell, 22, 1633-1646.
http://dx.doi.org/10.1105/tpc.110.075242 - 408. Krapp, A. (2015) Plant Nitrogen Assimilation and Its Regulation: A Complex Puzzle with Missing Pieces. Current Opinion in Plant Biology, 25, 115-122.
- 409. Brenner, W.G., Romanov, G.A., Kollmer, I., Burkle, L. and Schmülling, T. (2005) Immediate-Early and Delayed Cytokinin Response Genes of Arabidopsis thaliana Identified by Genome-Wide Expression Profiling Reveal Novel Cytokinin-Sensitive Processes and Suggest Cytokinin Action through Transcriptional Cascades. The Plant Journal, 44, 314- 333.
http://dx.doi.org/10.1111/j.1365-313X.2005.02530.x - 410. Good, A.G., Shrawat, A.K. and Muench, D.G. (2004) Can Less Yield More? Is Reducing Nutrient Input into the Environment Compatible with Maintaining Crop Production? Trends in Plant Science, 9, 597-605.
http://dx.doi.org/10.1016/j.tplants.2004.10.008 - 411. Fraisier, V., Dorbe, M.F. and Daniel-Vedele, F. (2001) Identification and Expression Analyses of Two Genes Encoding Putative Low-Affinity Nitrate Transporters from Nicotiana plumbaginifolia. Plant Molecular Biology, 45, 181-190.
http://dx.doi.org/10.1023/A:1006426616760 - 412. Katayama, H., Mori, M., Kawamura, Y., Tanaka, T., Mori, M. and Hasegawa, H. (2009) Production and Characterization of Transgenic Rice Plants Carrying a High-Affinity Nitrate Transporter Gene (Osnrt2.1). Breeding Science, 59, 237-243.
http://dx.doi.org/10.1270/jsbbs.59.237 - 413. Djennane, S., Chauvin, J.E. and Meyer, C. (2002) Glasshouse Behaviour of Eight Transgenic Potato Clones with a Modified Nitrate Reductase Expression under Two Fertilization Regimes. Journal of Experimental Botany, 53, 1037-1045.
http://dx.doi.org/10.1093/jexbot/53.371.1037 - 414. Djennane, S., Chauvin, J.E., Quillere, I., Meyer, C. and Chupeau, Y. (2002) Introduction and Expression of a Deregulated Tobacco Nitrate Reductase Gene in Potato Lead to Highly Reduced Nitrate Levels in Transgenic Tubers. Transgenic Research, 11, 175-184.
http://dx.doi.org/10.1023/A:1015299711171 - 415. Lea, U.S., Hoopen, F., Provan, F., Kaiser, W.M. and Meyer, C. (2009) Mutation of the Regulatory Phosphorylation Site of Tobacco Nitrate Reductase Results in High Nitrite Excretion and No Emission from Leaf and Root Tissue. Planta, 219, 59-65.
http://dx.doi.org/10.1007/s00425-004-1209-6 - 416. Lillo, C. (2008) Signaling Cascades Integrating Light-Enhanced Nitrate Metabolism. Biochemical Journal, 415, 11-19.
http://dx.doi.org/10.1042/BJ20081115 - 417. Schofield, R.A., Bi, Y.M., Kant, S. and Rothstein, S.J. (2009) Over-Expression Ofstp13, a Hexose Transporter, Improves Plant Growth and Nitrogen Use in Arabidopsis thaliana Seedlings. Plant, Cell & Environment, 32, 271-285.
http://dx.doi.org/10.1111/j.1365-3040.2008.01919.x - 418. Brauer, E.K., Rochon, A., Bi, Y.M., Bozzo, G.G., Rothstein, S.J. and Shelp, B.J. (2011) Reappraisal of Nitrogen Use Efficiency in Rice over Expressing Glutamine Synthetase1. Physiologia Plantarum, 141, 361-372.
http://dx.doi.org/10.1111/j.1399-3054.2011.01443.x - 419. Gallardo, F., Fu, J., Cantón, F., Gutierrez, A., Cánovas, F.M. and Kirby, E.G. (1999) Expression of a Conifer Glutamine Synthetase Gene in Transgenic Poplar. Planta, 210, 19-26.
http://dx.doi.org/10.1007/s004250050649 - 420. Habash, D.Z., Massiah, A.J., Rong, H.L., Wallsgrove, R.M. and Leigh, R.A. (2001) The Role of Cytosolic Glutamine Synthetase in Wheat. Annals of Applied Biology, 138, 83-89.
- 421. Migge, A., Bork, C., Hell, R. and Becker, T.W. (2000) Negative Regulation of Nitrate Reductase Gene Expression by Gluta Mate or Asparagine Accumulation in Leaves of Sulfur Deprived Tobacco. Planta, 211, 587-595.
http://dx.doi.org/10.1007/s004250000322 - 422. Suerez, R.V., Marquez, J., Shishkova, S. and Hernandez, G. (2003) Over Expression of Alfalfa Cytosolic Glutamine Synthetase in Nodules and Flowers of Transgenic Lotus japonicus Plants. Physiologia Plantarum, 117, 326-336.
http://dx.doi.org/10.1034/j.1399-3054.2003.00053.x - 423. Good, A.G., Johnson, S.J., De Pauw, M., Carroll, R.T. and Savidov, N. (2007) Engineering Nitrogen Use Efficiency with Alanine Aminotransferase. Canadian Journal of Botany, 85, 252-262. http://dx.doi.org/10.1139/B07-019
- 424. Good, A. and Beatty, P. (2011) Biotechnological Approaches to Improving Nitrogen Use Efficiency in Plants: Alanine Aminotransferase as a Case Study. In: Hawkesford, M.J. and Barraclough, P., Eds., The Molecular and Physiological Basis of Nutrient Use Efficiency in Crops, Wiley-Blackwell, Oxford, 165-192.
http://dx.doi.org/10.1002/9780470960707.ch9 - 425. Strange, S., Park, J., Bennett, R. and Phipps, R. (2008) The Use of Life-Cycle Assessment to Evaluate the Environmental Impacts Ogrowing Genetically Modified Nitrogen Use-Efficient Canola. Plant Biotechnology Journal, 6, 337- 345.
http://dx.doi.org/10.1111/j.1467-7652.2008.00323.x - 426. Shrawat, A.K., Carroll, R.T., Depauw, M., Taylor, G.J. and Good, A.G. (2008) Genetic Engineering of Improved Nitrogen Use Efficiency in Rice by the Tissue-Specific Expression of Alanine Amino Transferase. Plant Biotechnology Journal, 6, 722-732.
http://dx.doi.org/10.1111/j.1467-7652.2008.00351.x - 427. Okumoto, S. and Pilot, G. (2011) Amino Acid Export in Plants: A Missing Link in Nitrogen Cycling. Molecular Plant, 4, 453-463.
http://dx.doi.org/10.1093/mp/ssr003 - 428. Li, X., Guo, C., Gu, J., Duan, W., Zhao, M., Ma, C., Du, X., Lu, W. and Xiao, K. (2014) Overexpression of VP, a Vacuolar H+-Pyrophosphatase Gene in Wheat (Triticum aestivum L.), Improves Tobacco Plant Growth under Pi and N Deprivation, High Salinity, and Drought. Journal of Experimental Botany, 65, 683-696.
http://dx.doi.org/10.1093/jxb/ert442 - 429. Bally, J., Nadai, M., Vitel, M., Rolland, A., Dumain, R. and Dubald, M. (2009) Plant Physiological Adaptations to the Massive Foreign Protein Synthesis Occurring in Recombinant Chloroplasts. Plant Physiology, 150, 1474-1481.
http://dx.doi.org/10.1104/pp.109.139816 - 430. Vincent, R., Fraisier, V., Chaillou, S., Limami, M.A. and Deleens, E. (1997) Overexpression of a Soybean Gene Encoding Cytosolic Glutamine Synthetase in Shoots of Transgenic Lotus corniculatus L. Plants Triggers Changes in Ammonium and Plant Development. Planta, 201, 424-433.
http://dx.doi.org/10.1007/s004250050085 - 431. Hodges, M. (2002) Enzyme Redundancy and the Importance of 2-Oxoglutarate in Plant Ammonium Assimilation. Journal of Experimental Botany, 53, 905-916.
http://dx.doi.org/10.1093/jexbot/53.370.905 - 432. Cai, C., Wang, J.Y., Zhu, Y.G., Shen, Q.R., Li, B. and Tong, Y.P. (2008) Gene Structure and Expression of the High-Affinity Nitrate Transport System in Rice Roots. Journal of Integrative Plant Biology, 50, 443-451.
http://dx.doi.org/10.1111/j.1744-7909.2008.00642.x - 433. Bai, H., Euring, D., Volmerz, K., Janz, D. and Polle, A. (2013) The Nitrate Transporter (NRT) Gene Family in Poplar. PLoS ONE, 8, E72126.
http://dx.doi.org/10.1371/journal.pone.0072126 - 434. Migocka, M., Warzybok, A. and Klobus, G. (2013) The Genomic Organization and Transcriptional Pattern of Genes Encoding Nitrate Transporters 1 (NRT1) in Cucumber. Plant Soil, 364, 245-260.
http://dx.doi.org/10.1007/s11104-012-1345-x - 435. Couturier, J., Montanini, B., Martin, F., Brun, A., Blaudez, D. and Chalot, M. (2007) The Expanded Family of Ammonium Transporters in the Perennial Poplar Plant. New Phytologist, 174, 137-150.
http://dx.doi.org/10.1111/j.1469-8137.2007.01992.x - 436. D’Apuzzo, E., Rogato, A., Simon-Rosin, U., El Alaoui, H., Barbulova, A. and Betti, M. (2004) Characterisation of Three Functional High Affinity Ammonium Transporters in Lotus japonicus with Differential Transcriptional Regulation and Spatial Expression. Plant Physiology, 134, 1763-1774.
http://dx.doi.org/10.1104/pp.103.034322 - 437. Alvarezj, M., Vidal, E.A. and Gutiérrez, R.A. (2012) Integration of Local and Systemic Signaling Pathways for Plant N Responses. Current Opinion in Plant Biology, 15, 185-191.
http://dx.doi.org/10.1016/j.pbi.2012.03.009 - 438. Vidal, E.A. and Gutiérrez, R.A. (2008) A Systems View of Nitrogen Nutrient and Metabolite Responses in Arabidopsis. Current Opinion in Plant Biology, 11, 521-529.
http://dx.doi.org/10.1016/j.pbi.2008.07.003 - 439. Wang, R., Okamoto, M., Xing, X. and Crawford, N.M. (2003) Microarray Analysis of the Nitrate Response in Arabidopsis Roots and Shoots Reveals over 1,000 Rapidly Responding Genes and New Linkages to Glucose, Trehalose-6-Phosphate, Iron, and Sulfate Metabolism. Plant Physiology, 132, 556-567.
http://dx.doi.org/10.1104/pp.103.021253 - 440. Cai, H., Lui, Y., Xie, W., Zhu, T. and Lian, X. (2012) Transcriptome Response to Nitrogen Starvation in Rice. Journal of Biosciences, 37, 731-747.
http://dx.doi.org/10.1007/s12038-012-9242-2 - 441. Zhao, W., Yang, X., Yu, H., Jiang, W., Sun, N. and Liu, X.X. (2014) RNA-Seq-Based Transcriptome Profiling of Early Nitrogen Deficiency Response in Cucumber Seedlings Provides New Insight into the Putative Nitrogen Regulatory Network. Plant and Cell Physiology, 56, 455-467.
http://dx.doi.org/10.1093/pcp/pcu172 - 442. O’Rourke, J.A., Iniguez, L.P., Fu, F., Bucciarelli, B., Miller, S.S. and Jackson S.A. (2014) An RNA-Seq Based Gene Expression Atlas of the Common Bean. BMC Genomics, 15, 866.
http://dx.doi.org/10.1186/1471-2164-15-866 - 443. Peng, M., Bi, Y.M., Zhu, T. and Rothstein, S.J. (2007) Genome-Wide Analysis of Arabidopsis Responsive Transcriptome Response to Nitrogen Limitation and Its Regulation by the Ubiquitin Ligas Gene NLA. Plant Molecular Biology, 65, 775-797.
http://dx.doi.org/10.1007/s11103-007-9241-0 - 444. Lian, X., Wang, S., Zhang, J., Feng, Q., Zhang, L. and Fan, D. (2006) Expression Profiles of 10,422 Genes at Early Stage of Low Nitrogen Stress in Rice Assayed Using a cDNA Microarray. Plant Molecular Biology, 60, 617-631.
http://dx.doi.org/10.1007/s11103-005-5441-7 - 445. Wang, R., Guegler, K., Labrie, S.T. and Crawford, N.M. (2000) Genomic Analysis of a Nutrient Response in Arabidopsis Reveals Diverse Expression Patterns and Novel Metabolic and Potential Regulatory Genes That Are Induced by Nitrate. Plant Cell, 12, 1491-510.
http://dx.doi.org/10.1105/tpc.12.8.1491 - 446. Wang, Y.H., Garvin, D.F. and Kochian, L.V. (2001) Nitrate-Induced Genes in Tomato Roots. Array Analysis Reveals Novel Genes That May Play a Role in Nitrogen Nutrition. Plant Physiology, 127, 345-359.
http://dx.doi.org/10.1104/pp.127.1.345 - 447. Bi, Y.M., Meyer, A., Downs, G.S., Shi, X., El-Kereamy, A. and Lukens, L. (2014) High Throughput RNA Sequencing of Hybrid Maize and Its Parents Shows Different Mechanisms Responsive to Nitrogen Limitation. BMC Genomics, 15, 77.
http://dx.doi.org/10.1186/1471-2164-15-77 - 448. Cabeza, R., Koester, B., Liese, R., Lingner, A., Baumgarten, V. and Dirks, J. (2014) An RNA Sequencing Transcriptome Analysis Reveals Novel Insights into Molecular Aspects of the Nitrate Impact on the Nodule Activity of Medicago truncatula. Plant Physiology, 164, 400-411.
http://dx.doi.org/10.1104/pp.113.228312 - 449. Vidal, E.A., Moyano, T.C., Krouk, G., Katari, M.S., Tanurdzic, M. and Mccombie, W.R. (2013) Integrated RNA-Seq and Srna-Seq Analysis Identifies Novel Nitrate-Responsive Genes in Arabidopsis thaliana Roots. BMC Genomics, 14, 701.
http://dx.doi.org/10.1186/1471-2164-14-701 - 450. Lei, B., Lu, K., Ding, F., Zhang, K., Chen, Y. and Zhao, H. (2014) RNA Sequencing Analysis Reveals Transcriptomic Variations in Tobacco (Nicotiana tabacum) Leaves Affected by Climate, Soil, and Tillage Factors. International Journal of Molecular Sciences, 15, 6137-6160.
http://dx.doi.org/10.3390/ijms15046137 - 451. Boussadia, O., Steppeb, K., Zgallaid, H., Hadjc, S.B.E., Brahama, M. and Lemeurb, R. (2010) Effects of Nitrogen Deficiency on Leaf Photosynthesis, Carbohydrate Status and Biomass Production in Two Olive Cultivars “Meski” and “Koroneiki”. Scientia Horticulturae, 123, 336-342.
http://dx.doi.org/10.1016/j.scienta.2009.09.023 - 452. Foyer, C.H., Noctor, G. and Verrier, P. (2006) Photosynthetic Carbon-Nitrogen Interactions: Modelling Inter-Pathway Control and Signaling. In: Plaxton, W. and Mcmanus, M.T., Eds., Annual Plant Reviews: Control of Primary Metabolism in Plants, Blackwell, Oxford, 325-347.
http://dx.doi.org/10.1002/9780470988640.ch12 - 453. Taylor, L., Nunesnesi, A., Parsley, K., Leiss, A., Leach, G. and Coates, S. (2010) Cytosolic Pyruvate, Orthophosphate Dikinase Functions in Nitrogen Remobilization during Leaf Senescence and Limits Individual Seed Growth and Nitrogen Content. The Plant Journal, 62, 641-652.
http://dx.doi.org/10.1111/j.1365-313x.2010.04179.x - 454. Wingler, A., Purdy, S., Maclean, A. and Pourtau, N. (2006) The Role of Sugars in Integrating Environmental Signals during the Regulation of Leaf Senescence. Journal of Experimental Botany, 57, 391-399.
http://dx.doi.org/10.1093/jxb/eri279 - 455. Gutierrez, R.A., Lejay, L.V., Dean, A., Chiaromonte, F., Shasha, D.E. and Coruzzi, G.M. (2007) Qualitative Network Models and Genome-Wide Expression Data Define Carbon/Nitrogen-Responsive Molecular Machines in Arabidopsis. Genome Biology, 8, R7.
http://dx.doi.org/10.1186/gb-2007-8-1-r7 - 456. Diaz-Troya, S., Perez-Perez, M.E., Florencio, F.J. and Crespo, J.L. (2008) TOR in Autophagy Regulation from Yeast to Plants and Mammals. Autophagy, 4, 851-865.
http://dx.doi.org/10.4161/auto.6555 - 457. Kapahi, P., Chen, D., Rogers, A.N., Katewa, S.D., Li, P.W.L. and Thomas, E.L. (2010) With TOR, Less Is More: A Key Role for the Conserved Nutrient-Sensing TOR Pathway in Aging. Cell Metabolism, 11, 453-465.
http://dx.doi.org/10.1016/j.cmet.2010.05.001 - 458. Maekawa, S., Sato, T., Asada, Y., Yasuda, S., Yoshida, M., Chiba, Y. and Yamaguchi, J. (2012) The Arabidopsis Ubiquitin Ligases ATL31 and ATL6 Control the Defense Response as Well as the Carbon/Nitrogen Response. Plant Molecular Biology, 79, 217-227.
http://dx.doi.org/10.1007/s11103-012-9907-0 - 459. Malamy, J.E. (2005) Intrinsic and Environmental Response Pathways That Regulate Root System Architecture. Plant, Cell & Environment, 28, 67-77.
http://dx.doi.org/10.1111/j.1365-3040.2005.01306.x - 460. Fait, A., Nesi, A.N., Angelovici, R., Lehmann, M., Pham, P.A., Song, L. and Haslam, R.P. (2011) Targeted Enhancement of Glutamate-to-Γ-Aminobutyrate Conversion in Arabidopsis Seeds Affects Carbon-Nitrogen Balance and Storage Reserves in a Development-Dependent Manner. Plant Physiology, 157, 1026-1042.
http://dx.doi.org/10.1104/pp.111.179986 - 461. Kurai, T., Wakayama, M., Abiko, T., Yanagisawa, S., Aoki, N. and Ohsugi, R. (2011) Introduction of the Zmdof1 Gene into Rice Enhances Carbon and Nitrogen Assimilation under Low-Nitrogen Conditions. Plant Biotechnology Journal, 9, 826-837.
http://dx.doi.org/10.1111/j.1467-7652.2011.00592.x - 462. Nero, D., Krouk, G., Tranchina, D. and Coruzzi, G.M. (2009) A System Biology Approach Highlights a Hormonal Enhancer Effect on Regulation of Genes in a Nitrate Responsive “Biomodule”. BMC Systems Biology, 3, 59.
http://dx.doi.org/10.1186/1752-0509-3-59 - 463. Gao, P., Xin, Z. and Zheng, Z.L. (2008) The OSU1/QUA2/TSD2-Encoded Putative Methyltransferase Is a Critical Modulator of Carbon and Nitrogen Nutrient Balance Response in Arabidopsis. PLoS ONE, 3, E1387.
http://dx.doi.org/10.1371/journal.pone.0001387 - 464. Price, J., Laxmi, A., St Martin, S.K. and Jang, J.C. (2004) Global Transcription Profiling Reveals Multiple Sugar Signal Transduction Mechanisms in Arabidopsis. Plant Cell, 16, 2128-2150.
http://dx.doi.org/10.1105/tpc.104.022616 - 465. Bi, Y.M., Zhang, Y., Signorelli, T., Zhao, R., Zhu, T. and Rothstein, S. (2005) Genetic Analysis of Arabidopsis GATA Transcription Factor Gene Family Reveals a Nitrate-Inducible Member Important for Chlorophyll Synthesis and Glucose Sensitivity. The Plant Journal, 44, 680-692.
http://dx.doi.org/10.1111/j.1365-313X.2005.02568.x - 466. Chiang, Y.H., Zubo, Y.O., Tapken, W., Kim, H.J., Lavanway, A.M. and Howard, L. (2012) Functional Characterization of the GATA Transcription Factors GNC and CGA1 Reveals Their Key Role in Chloroplast Development, Growth, and Division in Arabidopsis. Plant Physiology, 160, 332-348.
http://dx.doi.org/10.1104/pp.112.198705 - 467. Naito, T., Kiba, T., Koizumi, N., Yamashino, T. and Mizuno, T. (2007) Characterization of a Unique GATA Family Gene That Responds to Both Light and Cytokinin in Arabidopsis thaliana. Bioscience, Biotechnology, and Biochemistry, 71, 1557-1560.
http://dx.doi.org/10.1271/bbb.60692 - 468. Zheng, Z.L. (2009) Carbon and Nitrogen Nutrient Balance Signaling in Plants. Plant Signaling & Behavior, 4, 584-591.
- 469. Palenchar, P.M., Kouranov, A., Lejay, L.V. and Coruzzi, G.M. (2004) Genome-Wide Patterns of Carbon and Nitrogen Regulation of Gene Expression Validate the Combined Carbon and Nitrogen (CN)-Signaling Hypothesis in Plants. Genome Biology, 5, R91.
http://dx.doi.org/10.1186/gb-2004-5-11-r91 - 470. Egli, D.B., Leggett, J.E. and Duncan, W.G. (1976) Influence of N Stress on Leaf Senescence and N Redistribution in Soybeans. Agronomy Journal, 70, 43-47.
http://dx.doi.org/10.2134/agronj1978.00021962007000010011x - 471. Schildhauer, J., Wiedemuth, K. and Humbeck, K. (2008) Supply of Nitrogen Can Reverse Senescence Processes and Affect Expression of Genes Coding for Plastidic Glutamine Synthetase and Lysine-Ketoglutarate Reductase/Saccharopine Dehydrogenase. Plant Biology, 10, 76-84.
http://dx.doi.org/10.1111/j.1438-8677.2008.00075.x - 472. Castaings, L., Marchive, C., Meyer, C. and Krapp, A. (2011) Nitrogen Signaling in Arabidopsis: How to Obtain Insights into a Complex Signaling Network. Journal of Experimental Botany, 62, 1391-1397.
http://dx.doi.org/10.1093/jxb/erq375 - 473. Gutiérrez, R.A. (2012) Systems Biology for Enhanced Plant Nitrogen Nutrition. Science, 336, 1673-1675.
http://dx.doi.org/10.1126/Science.1217620 - 474. Balazadeh, S., Schildhauer, J., Araújo, W.L., Munné-Bosch, S., Fernie, A.R. and Proost, S. (2014) Reversal of Senescence by N Resupply to N-Starved Arabidopsis thaliana: Transcriptomic and Metabolomic Consequences. Journal of Experimental Botany, 65, 3975-3992.
http://dx.doi.org/10.1093/jxb/eru119 - 475. Breeze, E., Harrison, E., Mchattie, S., Hughes, L., Hickman, R. and Hill, C. (2011) High-Resolution Temporal Profiling of Transcripts during Arabidopsis Leaf Senescence Reveals a Distinct Chronology of Processes and Regulation. The Plant Cell, 23, 873-894.
http://dx.doi.org/10.1105/tpc.111.083345 - 476. Monfils, L.S., Bi, Y.M., Downs, G., Wu, W., Signorelli, T. and Lu, G. (2013) Nitrogen Transporter and Assimilation Genes Exhibit Developmental Stage-Selective Expression in Maize (Zea mays L.) Associated with Distinct Cis-Acting Promoter Motifs. Plant Signaling and Behavior Biosience, 8, Article ID: e26056.
http://dx.doi.org/10.4161/psb.26056 - 477. Kechid, M., Desbrosses, G., Rokhsi, W., Varoquaux, F., Djekoun, A. and Touraine, B. (2013) The NRT2.5 and NRT2.6 Genes Are Involved in Growth Promotion of Arabidopsis by the Plant Growth-Promoting Rhizobacterium (PGPR) Strain Phyllobacterium brassicacearum STM196. New Phytologist, 198, 514-524.
http://dx.doi.org/10.1111/nph.12158 - 478. Dechorgnat, J., Patrit, O., Krapp, A., Fagard, M. and Daniel-Vedele, F. (2012) Characterization of the Nrt2.6 Gene in Arabidopsis thaliana: A Link with Plant Response to Bioti and Abiotic Stress. PLoS ONE, 7, E42491.
http://dx.doi.org/10.1371/journal.pone.0042491 - 479. David, L.C., Dechorgnat, J., Berquin, P., Routaboul, J.M., Debeaujon, I. and Daniel-Vedele, F. (2014) Proanthocyanidin Oxidation of Arabidopsis Seeds Is Altered in Mutant of the High-Affinity Nitrate Transporter NRT2.7. Journal of Experimental Botany, 65, 885-893.
http://dx.doi.org/10.1093/jxb/ert481 - 480. Reddy, M.M. and Ulaganthan, K. (2015) RNA-Seq Analysis of Urea Nutrition Responsive Transcriptome of Oryza sativa Elite Indica Cultivar RP Bio 226. Genomics Data, 6, 112-113.
http://dx.doi.org/10.1016/j.gdata.2015.08.025
Abbreviations
ALMT1: aluminium activated malate transporter-1;
AS: asparagine synthase;
CLC: chloride channel proteins;
Dof1: DNA binding with one finger;
GACC: General amino acid control;
GS/GOGAT: Glutamine synthetase/glutamate synthase;
HATS: High affinity transport system;
LATS: Low affinity transport system;
LBD: Lateral organ boundary domain;
MFS: Major facilitator super family;
NLP: NIN like protein;
NNP: Nitrate-nitrite porter;
NPF: Nitrate protein family;
NUE: Nitrogen use efficiency;
SLAC1: Slow anion channel-like;
SLAH: Slow anion channel-like homologue;
SPL9: Squamosa promoter binding protein.