American Journal of Plant Sciences
Vol.4 No.5A(2013), Article ID:32241,9 pages DOI:10.4236/ajps.2013.45A001
ABNORMAL SHOOT IN YOUTH, a Homolog of Molybdate Transporter Gene, Regulates Early Shoot Development in Rice
![]()
Graduate School of Agricultural and Life Sciences, University of Tokyo, Tokyo, Japan.
Email: #anagato@mail.ecc.u-tokyo.ac.jp
Copyright © 2013 Ken-Ichiro Hibara et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Received March 12th, 2013; revised April 15th, 2013; accepted May 1st, 2013
Keywords: ABNORMAL SHOOT IN YOUTH; Early Vegetative Stage; Shoot Meristem; Rice; Molybdenum; Transporter
ABSTRACT
We analyzed the abnormal shoot in youth (asy) mutant to understand the phase-specific regulation of shoot development. asy showed various shoot abnormalities, including small leaves due to the precocious termination of cell division, defects in leaf blade-sheath boundary formation, and abnormal shoot apical meristem maintenance at the early vegetative stage. These defects recovered with advanced development. ASY encodes a DUF791 domain protein, which is part of the major facilitator superfamily. Despite stage-specific phenotypes, the ASY expression level was roughly constant throughout development. A paralog of ASY, ASL, exists in the rice genome and is supposed to have redundant functions. ASL expression was relatively low in early-stage embryos but increased at later stages. Thus, asy phenotypes were limited to the stage when ASL expression was suppressed. A homology search revealed that ASY is a homolog of the Chlamydomonas CrMoT2 gene, which encodes a molybdate transporter. ASY was suggested to encode a molybdate transporter based on its sequence similarity with CrMoT2 and predicted transmembrane topology. This is the first report of a CrMOT2-type molybdate transporter in higher plants.
1. Introduction
The genetic regulatory mechanism of shoot development has been studied in many higher plants such as Arabidopsis, rice, and maize. CUP-SHAPED COTYLEDON 1 (CUC1), CUC2, and SHOOT MERISTEMLESS play important roles in differentiation of the shoot apical meristem (SAM) in Arabidopsis [1-3]. However, distinct genes related to small RNA metabolism in rice such as SHOOTLESS 1 (SHL1), SHL2, SHOOT ORGANIZATION 1 (SHO1), SHL4/SHO2, and WAVY LEAF have decisive roles in SAM formation [4,5]. The mechanism of SAM maintenance is another important aspect of shoot development. A large number and various classes of genes are involved in SAM maintenance, including CLAVATA, WUSCHEL, FAS, and FSM [6-9]. These genes function throughout the plant life cycle; however, plants show phases of development that include embryogenesis, and vegetative and reproductive phases. The vegetative phase is further divided into juvenile and adult phases. These phases indicate that plant development is regulated by phase-specific genes as well as by genes acting throughout the life cycle.
Recent advances in plant developmental biology have shown that the juvenile-adult phase change during vegetative development is an important event. Because a large number of traits differ between the juvenile and adult phases [10-12] and several genes associated with phase changes have been cloned [13-16], it is expected that juvenile or adult phase-specific regulatory mechanisms of shoot development are operating. Accordingly, another interesting aspect of plant vegetative development is the phase-specific regulation of gene expression.
In this study, we identified the abnormal shoot in youth (asy) mutant, which showed abnormal phenotypes during the early vegetative phase but near-normal later development. The asy causal gene encodes a molybdate transporter. Our findings indicate that a specific trace element plays a distinct role in plant development, including leaf morphogenesis, cell division, and shoot meristem maintenance.
2. Materials and Methods
2.1. Plant Materials
We identified a single recessive mutant from M2 population of rice (Oryza sativa L., ssp. japonica cv. Taichung 65) mutagenized with N-methyl-N-nitrosourea. Since the mutant developed weak plants at the early stage and recovered almost normal stature at the late vegetative stage, we named this mutant abnormal shoot in youth (asy). Taichung 65 was used as wild type. In the experiments on early vegetative seedlings, seeds were surface sterilized and grown on MS plates that contained 4.6 g/L Murashige and Skoog plant salts mixture [17], 30 g/L sucrose, 5 mg/L myo-inositol, 5 ng/L thiamine hydrochloride, 25 ng/L nicotinic acid, 25 ng/L pyridoxine hydrochloride, 0.1 mg glycine and 10 g/L agar (adjusted to pH 5.8 with KOH) under constant white light at 28˚C. Adult plants were grown in pots or paddy field. Transgenic plants were grown in biohazard phytotron.
2.2. Histological Analysis
For paraffin sectioning, samples were fixed in FAA (formaldehyde: glacial acetic acid: ethanol [1:1:18]) for 24 h at 4˚C, dehydrated in a graded ethanol series, and embedded in Paraplast plus (McCormick Scientific). Microtome sections (8 μm thick) were stained with Delafield’s hematoxylin, and then observed under a light microscope.
2.3. In Situ Hybridization
Shoot apices 7-day after germination (DAG) of wild type and asy were fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer for 48 h at 4˚C, and then dehydrated in a graded ethanol series. The dehydrated samples in 100% ethanol were replaced with butanol and embedded in Paraplast plus (McCormick Scientific). Paraffin sections (8 μm thick) were applied to microscope slides coated with MAS (Matsunami Glass). Digoxygenin-labeled antisense RNA probes of OSH1 and Histone H4 were prepared as described [18]. In situ hybridization and immunological detection with alkaline phosphatase were performed according to the methods of Kouchi and Hata (1993) [19].
2.4. Map-Based Cloning of the ABNORMAL SHOOT IN YOUTH Gene
To map the ASY1 locus, asy homozygous plants (O. sativa L. ssp. japonica) were crossed with cv. Kasalath (ssp. indica), and the F2 and F3 populations were examined for recombination between the mutation and PCR-based polymorphic markers. The ASY locus was mapped in the 58.9 to 61.4 cM region on chromosome 10 using STS and CAPS markers obtained from the rice genome database (http://rgp.dna.affrc.go.jp/E/publicdata/caps/index.html). By finding recombination break points among 420 asy plants, the ASY locus was limited within a 100-kb region, including 20 candidate genes annotated in the RAP-DB database, and covered by three overlapping BAC clones. A single nucleotide substitution was identified in a gene (RAP Os ID; Os10g0519600 and MSU Os ID; LOC_ Os10g37520) in asy mutant. Accession numbers for ASY and ASL proteins are AB807476 and AB807477, respectively.
2.5. Complementation Test
The 6.8-kb ASY genomic DNA including 3-kb upstream and 1-kb downstream was used for complementation test. This fragment was introduced into Agrobacterium tumefaciens strain EHA101 and transformed into asy homozygous plants by the Agrobacterium mediated transformation method [20].
2.6. Transient Expression Assay
For construction of fluorescence fusion protein, the coding region of ASY was amplified with primers that included appropriate restriction sites. It was translationally fused to 5’ terminus of the sGFP gene driven by 35S promoter of the cauliflower mosaic virus (p35S::ASYsGFP). Then it was introduced into onion epidermal cells using a particle bombardment with gold particles according to the manufacturer’s instructions (PDS-1000/He; Bio-Rad, Hercules, CA, USA). The cells were observed using a fluorescence microscope (BZ-8000; Keyence Co.).
2.7. Expression Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For RT-PCR, 1 μg of RNA after DNase I digestion was reverse transcribed with the SuperScript III First Strand Synthesis System and oligo (dT) primer according to the manufacturer’s instructions (Invitrogen). Resulting cDNA was used for RT-PCR with gene specific primers; ASYReTiF: TTCAAGGTTGCAGCCAAAGC and ASYReTiR: AAGCAGGAGTCCACAGGAAA (for amplification of ASY cDNA), ASLReTiF: GGAGCACAACAAGAGAGGTT and ASLReTiR: CGGCAACGACCTTTCGAAAT (for amplification of ASL cDNA). The primers for an internal control, UBIQUITIN, were prepared as described [21].
For real-time PCR, 500 ng of RNA after DNase I digestion was reverse transcribed with High Capacity RNAto-cDNA Master Mix (Applied Biosystems, Foster City, CA, USA). Real-time PCR was performed with SYBR Green Master mix (Applied Biosystems) following the manufacturer’s instructions. Amplification was monitored in real time by Step One Plus real-time RT-PCR system (Applied Biosystems). The primers for ASY and ASL were the same as those for RT-PCR. The primers for ACT1 were ACT1F: TCCATCTTGGCATCTCTCAG and ACT1R: GTACCCCCATCAGGCATCTG. The expression levels of ASY and ASL were normalized to that of an internal control, ACT1.
2.8. Measurement of Molybdenum
Mature dry seeds of wild type and asy mutant were cut into embryo and endosperm. Mature dry seeds, endosperms, and embryos were digested with concentrated HNO3 at 110˚C. After complete digestion, the concentrations of molybdenum in the samples were determined by inductively coupled plasma mass spectrometry (ICP-MS) (model SPQ9700; SII, Chiba, Japan). Measurements were done for four (whole seed) or three (embryo and endosperm) independent samples.
3. Results
3.1. asy Phenotypes at the Early Vegetative Stage
asy plants showed a dwarf phenotype at 7 days after germination (DAG) (Figure 1(a)). Concomitantly, the second and third leaves of asy were shorter than those of wild type (Figure 1(b)). Extreme dwarf asy plants frequently died at about 7 DAG. Despite growth depression at the early stage, the surviving asy plants gradually recovered a normal phenotype; leaf length (Figure 1(b)) and plant height (Figure 1(c)) were similar between asy and wild type after the fourth leaf stage, or 14 DAG. The small leaves of asy indicate that the mutant had defects in cell division and/or cell growth. Thus, we measured cell size in the second leaf blade, but no difference was observed in cell size between wild type and asy (data not shown). Accordingly, asy was speculated to have a defect in cell division (see 3.3).
Other morphological aberrations were also detected in the leaves. The asy second leaf frequently lacked a ligule, which forms at the boundary between the leaf sheath and blade (Figures 1(d) and (e)). Moreover, the bulliform cells were enlarged and increased in number in the asy leaf blade (Figures 1(f) and (g)). In addition, the asy leaf blade tissues were partly transformed into those of the leaf sheath (Figure 1(e)). Transverse sections of the leaf blade revealed that air spaces had formed in the second leaf blades and infrequently in the third leaf blades of asy but not in subsequent leaf blades (Figure 1(g)). Because air spaces are characteristic of wild-type leaf sheaths, this result suggests that leaf blade and sheath identities are mixed in asy leaves.
3.2. Defects in the asy Shoot Meristem and Embryo
The leaf abnormalities and seedling lethality we observed suggested abnormalities in the asy SAM. Longitudinal sections of SAMs revealed that although the width of the asy SAM was comparable to that of wild type, the height was smaller than that in wild type (Figures 2(a)-(d)). We also found a SAM in which many cells were vacuolated in an extreme dwarf asy plant (Figure 2(d)). As in the case of leaf phenotypes, SAM defects in asy were also recovered at 21 DAG (Figures 2(e) and (f)). Thus, ASY was proposed to have a significant role in the SAM at the early (juvenile) stage of vegetative development.
Because the first three leaves are formed in rice during seed development, we examined asy embryos at 10 days after pollination (DAP). Although all organs were formed in the embryos, morphological abnormalities were observed in the scutellum, coleoptile, and shoot apex (Fig-
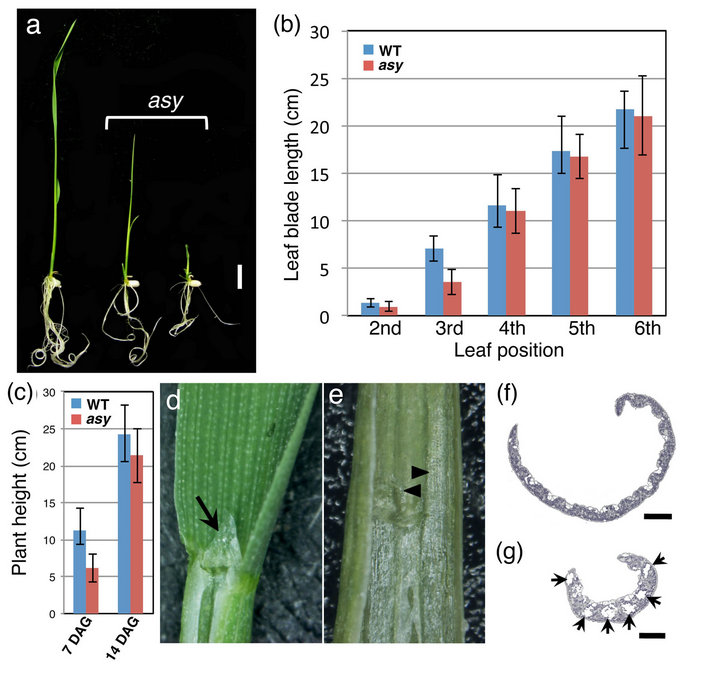
Figure 1. Phenotypes of asy at the early vegetative stage. (a) 7 DAG plants; (b) Change of leaf blade length during development; (c) Comparison of plant height between wild type and asy at 7 DAG; (d) Leaf blade-sheath boundary of wild type. Arrow points developing ligule (arrow); (e) Leaf blade-sheath boundary of asy lacking ligule. Arrowheads show leaf sheath tissue ectopically invading into blade region; (f), (g) Cross section of wild-type (f) and asy (g) second leaf blade. Arrows point airspaces. Bar, 1 cm in (a) and 250 μm in (f) and (g).
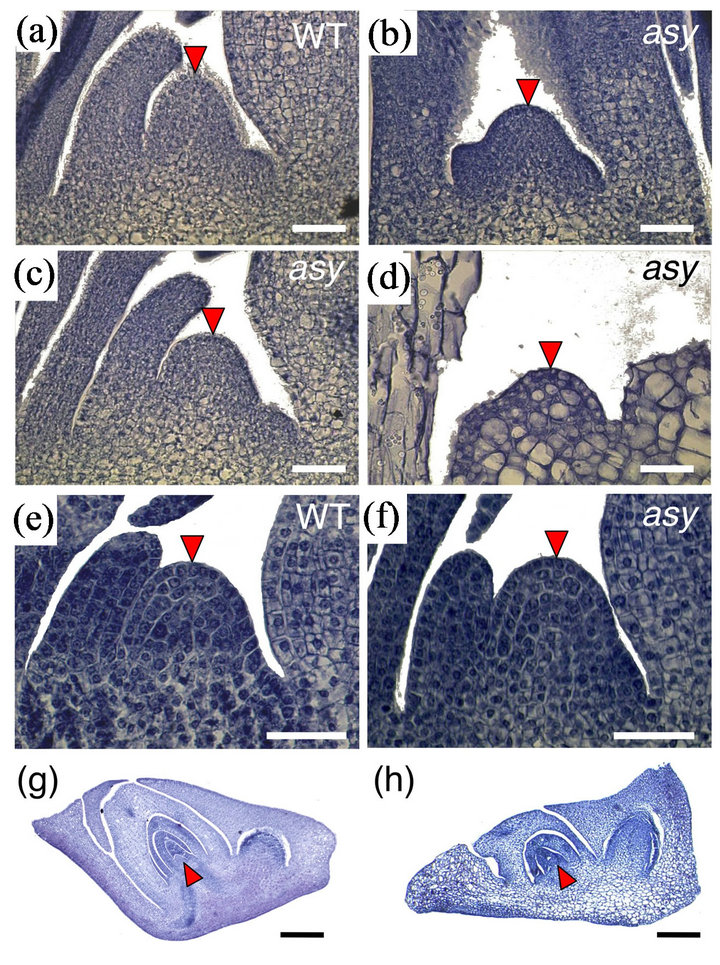
Figure 2. Shoot apex and embryo phenotype of asy. (a) 7 DAG wild-type shoot apex. (b)-(d) 7 DAG asy shoot apex. In (d), many cells in shoot meristem are vacuolated; (e) 21 DAG wild-type shoot apex; (f) 21 DAG asy shoot apex; (g, h) 10 DAP embryos of wild-type (g) and asy (h). Arrowheads point SAM. Bars, 25 μm in (a)-(f) and 100 μm in (g) and (h).
ures 2(j) and (k)). Vacuolization was detected in cells of the scutellum, coleoptile, and first leaf primordium. These results show that ASY acted as early as the embryonic stage.
3.3. Characterization of asy by Marker Gene Expression
Phenotypic abnormalities such as small leaves and an aberrant SAM suggest that related gene expression is disturbed in asy. First, we examined cell division activity by in situ hybridization probed with histone H4. At 4 DAG, when the fourth leaf primordium was being formed, a large number of histone H4 signals were uniformly observed in second (P3 stage), third (P2), and fourth (P1) leaf primordia in wild type (Figure 3(a)). In contrast, although the fourth leaf primordium (P1) showed many histone H4 signals comparable to those in wild type, histone H4 transcripts disappeared from the distal region of the asy third leaf primordium (P2) (Figure 3(b)). This tendency became more remarkable in the second leaf primordium; the expression domain was limited to a narrow basal region and the expression level decreased (Figure 3(b)). This result indicates that asy terminated cell division precociously, resulting in small leaves.
To examine SAM activity, we performed in situ hybridization probed with the OSH1 gene, which is expressed in indeterminate cells of the SAM. At 7 DAG, some asy SAMs showed a narrower OSH1 expression domain (Figures 3(c) and (d)), while other asy SAMs failed to express OSH1 (Figure 3(e)). Thus, asy is defective in SAM maintenance. Presumably, vacuolated and/or OSH1-non-expressing SAMs are inactive and the corresponding plants die at an early stage of development.
3.4. Identification of ASY Gene
asy homozygous plants were crossed with ssp. indica cv. Kasalath to identify the ASY gene using a map-based cloning method. The ASY locus was roughly mapped at 58.9 - 61.4 cM on chromosome 10 (Figure 4(a)). By fine mapping using 420 plants, candidates were confined to 20 genes from the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp/). Through sequencing, we identified a G-to-A substitution in Os10g0519600. The substitution occurred in the conserved AG dinucleotide of the splice acceptor site in the third intron (Figure 4(a)). We performed a complementation test by introducing a 6.8-kb genomic fragment containing the coding region of Os10g0519600 into the asy mutant. T2 lines carrying the 6.8-kb genomic fragment (PZP-gASY) fully complemented the asy mutant phenotype (Figure 4(b)). Thus, we concluded that Os10g0519600 is the asy causal gene. Reverse transcription-polymerase chain reaction (RTPCR) analysis showed that ASY transcripts of various sizes with unusual splicing or non-splicing in the third intron were produced in asy seedlings at 5 DAG (Figure 4(c)).
ASY is comprised of eight exons and seven introns,
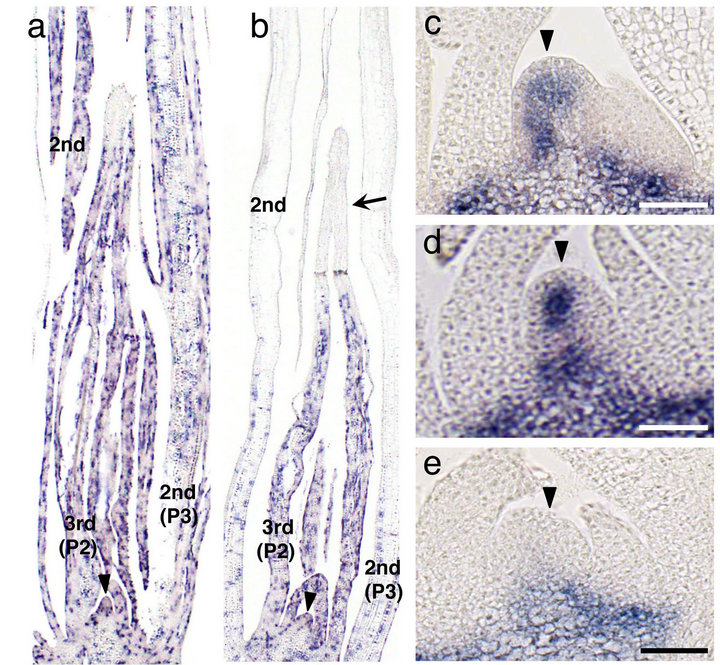
Figure 3. In situ localization of histone H4 and OSH1 transcripts in asy seedling. (a), (b) Expression pattern of histone H4 in wild-type (a) and asy (b) seedlings at 4 DAG. Leaf numbers and primordial stages (Plastochrone numbers) are indicated. Arrow indicates apical region of the third leaf primordium where the signals are absent. (c)-(e) Expression pattern of OSH1 in wild-type (b) and asy (c), (d) shoot apex at 7 DAG. Arrowheads in (c)-(e) point SAMs. Bars, 25 μm.
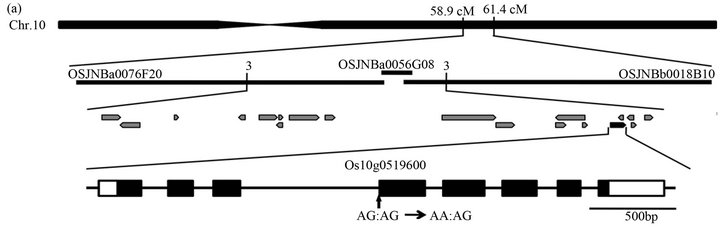
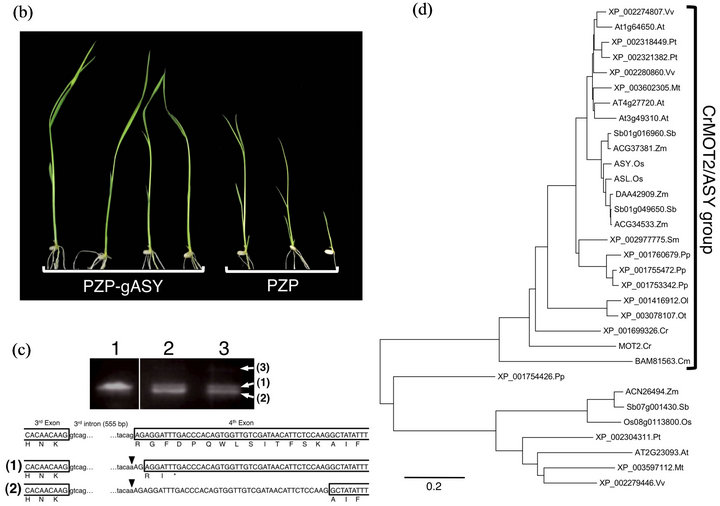
Figure 4. Molecular characterization of ASY gene. (a) Physical mapping of the ASY locus. The number shows recombination events. Black and white boxes indicate exons and untranslated regions, respectively. Arrow indicates mutation site in asy; (b) Complementation test. The left-side four plants are asy mutants transformed with 6.8-kb genomic ASY fragment (PZP-gASY). The right-side three plants are asy mutants transformed with empty vector (PZP); (c) Multiple splicing products in asy mutant. The upper panel shows RT-PCR analysis of ASY transcripts in wild-type (Lane 1) and asy (Lane 2, 3) seedlings at 5 DAG. Three different sizes of transcripts were detected. The lower illustration shows predicted mutant forms of ASY proteins corresponding to the transcripts in the upper panel. Arrowheads indicate mutation site in asy. The transcript as shown by (3) contains complete 3rd intron; (d) The phylogenetic tree of DUF791 domain-containing proteins from three monocots Oryza sativa (Os), Zea mays (Zm), Sorghum bicolor (Sb), four eudicots Arabidopsis thaliana (At), Vitis vinifera (Vv), Populus trichocarpa (Pt), Medicago truncatula (Mt), one fern Selaginella moellendorffii (Sm), one moss Physcomitrella patens (Pp), three green algae Chlamydomonas reinhardtii (Cr), Ostreococcus lucimarinus (Ol), Ostreococcus tauri (Ot) and red alga Cyanidioschyzon merolae (Cm). The alignment was obtained from ClustalW method and tree was build using MEGA5.1 software.
and encodes a protein composed of 456 amino acid residues (Figure 5(a)). The deduced amino acid sequence of ASY showed that it belongs to the DUF791 family, which is part of the major facilitator superfamily, with a domain of unknown function. The ASY protein contained a predicted 11 transmembrane domains and was proposed to act as a transporter [22] (Figure 5(b)).
BLAST searches against the rice protein database using the ASY amino acid sequence revealed that a paralogous DUF791 family gene exists in the rice genome, Os03g0114800, which had 93.2% identity with ASY. We named this paralog ASY-like (ASL) (Figures 4(d) and 5(a)).
A phylogenetic tree constructed from the DUF791 domain of ASY showed that DUF791 family proteins were divided into two clades. ASY group proteins, in which ASY and ASL are included, are well conserved not only in plants but also in algae (Figure 4(d)). In con-
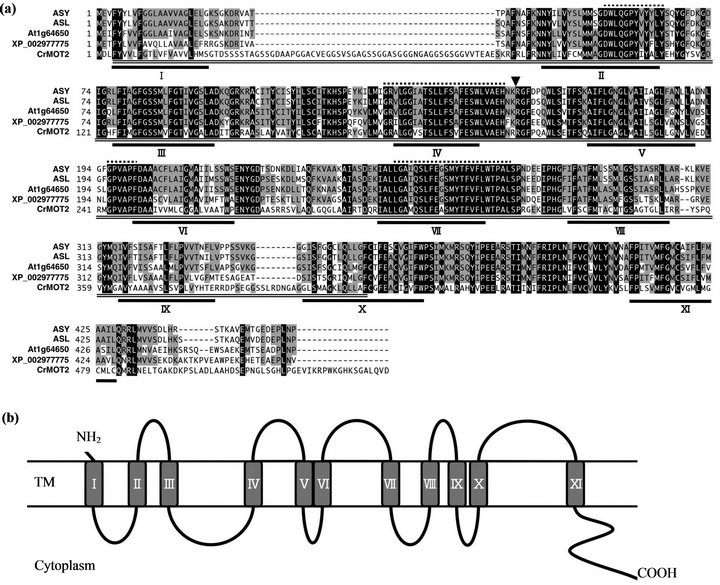
Figure 5. Alignment of amino acid sequences of ASY homologs and predicted topology. (a) Multiple alignment among ASY, ASL in Oryza sativa, ASY orthologs in Arabidopsis thaliana (At1g64650), Selaginella moellendorffii (XP_002977775) and CrMOT2 in Chlamydomonas reinhardtii. Amino acids shaded in black are conserved in all 5 proteins examined, and those in grey conserved in more than three proteins. Underlines indicate transmembrane regions predicted by TMpred software. Arrowhead shows the position of asy mutation causing splicing defects in asy. DUF791 domain is marked by double underline. Dushed overlines shows four conserved motifs reported by [23]; (b) Schematic illustration of predicted ASY transmembrane topology using TMpred software.
trast, the other group of proteins is found only in plants and has little significant conservation to ASY group proteins. For example, rice Os08g0113800, one of the three DUF791 domain-containing proteins in rice, has 23.2 and 22.8% identity with ASY and ASL, respectively.
The ASY group includes CrMOT2, which has been recently identified to be a molybdate transporter in Chlamydomonas [23]. ASY has 53.1% amino acid identity with CrMOT2. In addition, the deduced transmembrane topology was shared between ASY and CrMOT2 (Figure 5(b)). Next, we measured the molybdenum (Mo) content in mature seeds. The Mo contents (ng/mg dry weight) in whole mature seeds, mature embryos, and endosperm were 0.311 ± 0.010, 0.480 ± 0.028, and 0.287 ± 0.014 (mean ± standard deviation), respectively, in the asy mutant and 0.324 ± 0.009, 0.452 ± 0.023, and 0.308 ± 0.005, respectively, in wild type. We could not detect any differences in Mo content in mature embryos, endosperm, or whole seeds between wild type and asy.
3.5. ASY Expression Pattern
ASY was deduced to localize to the membrane based on its amino acid sequence. Therefore, we examined the subcellular localization of ASY using onion epidermal cells transformed with 35S::ASY-GFP. GFP signals were detected mainly in the plasma membrane (Figures 6(a) and (b)), suggesting that ASY acts as a transporter.
ASY expression in different organs was examined by RT-PCR. ASY was expressed in all organs examined (Figure 6(c), although the expression level fluctuated to some extent depending on the organ. ASL showed a similar expression pattern, but its expression level was relatively low in 5 DAP embryos and young inflorescences (Figure 6(c). Next, the expression of ASY and ASL during embryogenesis and at the early vegetative stage was

Figure 6. Expression of ASY and ASL. (a) Subcellular localization of GFP protein in onion epidermal cell; (b) Subcellular localization of ASY-GFP fusion protein in onion epidermal cell; (c) RT-PCR analysis of ASY and its paralog ASL in various organs. SA: shoot apex, IL: immature leaf, IF-1: 0.6 - 0.9 mm long young panicle, IF-2: 5 cm long young panicle, FO: floral organs, EM: immature embryo at 5 days after pollination, R: root; (d) Real-time PCR of ASY and ASL in 7 - 14 DAP seeds and in 4 - 14 DAG seedlings. Expression level was represented relative to that of ACTIN.
examined by real-time PCR. ASY was expressed rather constantly during seed development, whereas it was low until 10 DAP but largely increased at 14 DAP (Figure 6(d)). ASY expression was higher than that of ASL in 7 DAP seeds when the second leaf primordium was forming in the embryo (Figure 6(d)). After germination, ASY and ASL expression was comparable until 7 DAG, but the ASL expression level was much higher than that of ASY at 10 and 14 DAG (Figure 6(d)). These expression patterns suggest that the early stage-specific phenotypes of asy, despite nearly constant expression in all organs and at all stages, resulted from the expression of its paralog ASL.
4. Discussion
No rice mutants have been reported to show early development-specific phenotypes and recovery at later stages. Thus, asy is a unique mutant in that ASY plays important roles only during early development. Phenotypic defects in asy were observed in various traits of embryo and juvenile phase seedlings, including abnormal embryo formation, precocious termination of cell division in leaves associated with small leaves, aberrant leaf morphogenesis, and defective SAM maintenance. These abnormalities eventually caused seedling death. Interestingly, these stage-specific phenotypes were not caused by stage-specific ASY expression. A highly homologous gene, ASL, exists in the rice genome. ASL transcripts showed a similar expression profile to that of ASY. Notably, ASY expression was relatively higher than that of ASL in 7 DAP seed when the rice embryo developed second and third leaf primordia. Thus, the different expression levels of these genes during early embryogenesis might be responsible for the asy mutant defects. Although functional redundancy between ASY and ASL has not been confirmed, the high level of expression of ASL at the late stage explains the temporal specificity of asy phenotypes. A lossof-function ASL mutant is not available. To clarify the relationship between ASY and ASL, identification of the asl mutant and its detailed functional analysis are needed in the future.
ASY is a homolog of Chlamydomonas CrMoT2, which encodes a molybdate transporter, and is also found in humans [23]. ASY is the first homolog of CrMoT2 reported in higher plants. Our results suggest a significant role for the trace element Mo in rice development. The sequence similarity and subcellular localization of ASY suggest that it also acts as a molybdate transporter. Two genes, MOT1 and MOT2, encode molybdate transporters in Arabidopsis, but they are different from ASY/CrMOT2 [24-28]. In the presence of Mo, the Arabidopsis mot1 mutant does not show severe phenotypes [24,26,29]. In contrast, asy showed more severe phenotypes than Arabidopsis mot1 under appropriate soil conditions (seemingly in the presence of sufficient Mo), suggesting a functional difference between ASY and MOT1.
No significant differences were detected in the seed and embryo Mo contents between wild type and asy, although severe defects were observed in asy embryos. Because the Km value of Chlamydomonas CrMOT2 for molybdate is higher than that of Arabidopsis MOT1 and CrMOT1 [24,26], ASY may have a different function from the MOT1-type molybdate transporter. It is possible that ASY creates a local concentration gradient of Mo, which is required for organ development. It is also conceivable that a significant difference in Mo content can be detected when early stage seeds at 7 DAP are examined, because ASL is predominant at later stages. Questions about Mo uptake activity, Mo affinity, and the detailed subcellular localization of ASY in planta should be addressed in future studies.
5. Acknowledgements
This work is supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (202480 01 to YN and 23012006 to JI).
REFERENCES
- M. Aida, T. Ishida, H. Fukaki, H. Fujiwara and M. Tasaka, “Genes Involved in Organ Separation in Arabidopsis: An Analysis of the Cup-Shaped Cotyledon Mutant,” Plant Cell, Vol. 9, No. 6, 1997, pp. 841-857. doi:10.1105/tpc.9.6.841
- M. Aida, T. Ishida and M. Tasaka, “Shoot Apical Meristem and Cotyledon Formation during Arabidopsis Embryogenesis: Interaction among the Cup-Shaped Cotyledon and Shoot Meristemless Genes,” Development, Vol. 126, 1999, pp. 1563-1570.
- J. A. Long, E. I. Moan, J. I. Medford and M. K. Barton, “A Member of the Knotted Class Homeodomain Proteins Encoded by the STM Gene of Arabidopsis,” Nature, Vol. 379, No. 6560, 1996, pp. 66-69. doi:10.1038/379066a0
- H. Nagasaki, J.-I. Itoh, K. Hayashi, K. Hibara, N. SatohNagasawa, M. Nosaka, M. Mukouhata, M. Ashikari, K. Kitano, M. Matsuoka, Y. Nagato and Y. Sato, “Small Interfering RNA Production Pathway Is Required for Shoot Meristem Initiation in Rice,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 104, No. 37, 2007, pp. 14867-14871. doi:10.1073/pnas.0704339104
- M. Abe, T. Yoshikawa, M. Nosaka, H. Sakakibara, Y. Satoh, Y. Nagato and J.-I. Itoh, “Wavy Leaf1, an Ortholog of Arabidopsis HEN1, Regulates Shoot Development by Maintaining microRNA and Trans-Acting siRNA Accumulation in Rice,” Plant Physiology, Vol. 154, No. 3, 2010, pp. 1335-1346. doi:10.1104/pp.110.160234
- U. Brand, J. C. Fletcher, M. Hobe, E. M. Meyerowitz and R. Simon, “Dependence of Stem Cell Fate in Arabidopsis on a Feedback Loop Regulated by CLV3 Activity,” Science, Vol. 289, No. 5479, 2000, pp. 617-619. doi:10.1126/science.289.5479.617
- H. Schoof, M. Lenhard, A. Haecker, K. F. X. Mayer, G. Jurgens and T. Laux, “The Stem Cell Population of Arabidopsis Shoot Meristems Is Maintained by a Regulatory Loop between the Clavata and Wuschel Genes,” Cell, Vol. 100, No. 6, 2000, pp. 635-644. doi:10.1016/S0092-8674(00)80700-X
- H. Kaya, K. I. Shibahara, K. I. Taoka, M. Iwabuchi, B. Stillman and T. Araki, “Fasciata Genes for Chromatin Assembly Factor-1 in Arabidopsis Maintain the Cellularorganization of Apical Meristems,” Cell, Vol. 104, No. 1, 2001, pp. 131-142. doi:10.1016/S0092-8674(01)00197-0
- M. Abe, H. Kuroshita, M. Umeda, J.-I. Itoh and Y. Nagato, “The Rice Flattened Shoot Meristem, Encoding CAF-1 p150 Subunit, Is Required for Meristem Maintenance by Regulating the Cell-Cycle Period,” Developmental Biology, Vol. 319, 2008, pp. 384-393. doi:10.1016/j.ydbio.2008.04.040
- R. S. Poethig, “Phase Change and the Regulation of Shoot Morphogenesis in Plants,” Science, Vol. 250, No. 4983, 1990, pp. 923-930. doi:10.1126/science.250.4983.923
- E. J. Lawson and R. S. Poethig, “Shoot Development in Plants: Time for a Change,” Trends in Genetics, Vol. 11, No. 7, 1995, pp. 263-268. doi:10.1016/S0168-9525(00)89072-1
- K. Asai, N. Satoh, H. Sasaki, H. Satoh and Y. Nagato, “A Rice Heterochronic Mutant, mori1, Is Defective in the Juvenile-Adult Phase Change,” Development, Vol. 12, 2002, pp. 265-273.
- C. Hunter, H. Sun and R. S. Poethig, “The Arabidopsis Heterochronic Gene Zippy Is an Argonaute Family Member,” Genes & Development, Vol. 18, 2003, pp. 2368- 2379.
- G. Chuck, A. M. Cigan, K. Saeteurn and S. Hake, “The Heterochronic Maize Mutant Corngrass1 Results from Over-Expression of a Tandem microRNA,” Nature Genetics, Vol. 39, 2007, pp. 544-549. doi:10.1038/ng2001
- G. Wu, M. Y. Park, S. R. Conway, J. W. Wang, D. Weigel and R. S. Poethig, “The Sequential Action of miR156 and miR172 Regulates Developmental Timing in Arabidopsis,” Cell, Vol. 138, 2009, pp. 750-759. doi:10.1016/j.cell.2009.06.031
- N. Tanaka, H. Ito, N. Sentoku, M. Kojima, H. Sakakibara, T. Izawa, J.-I. Itoh and Y. Nagato, “The COP1 Ortholog PPS Regulates the Juvenile-Adult and Vegetative-Reproductive Phase Changes in Rice,” Plant Cell, Vol. 23, 2011, pp. 2143-2154. doi:10.1105/tpc.111.083436
- T. Murashige and F. Skoog, “A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Cultures,” Physiologia Plantarum, Vol. 15, No. 3, 1962, pp. 473- 497. doi:10.1111/j.1399-3054.1962.tb08052.x
- J. I. Itoh, H. Kitano, M. Matsuoka and Y. Nagato, “Shoot Organization Genes Regulate Shoot Apical Meristem Organization and the Pattern of Leaf Primordium Initiation in Rice,” Plant Cell, Vol. 12, No. 11, 2000, pp. 2161- 2174.
- H. Kouchi and S. Hata, “Isolation and Characterization of Novel Nodulin cDNAs Representing Genes Expressed at Early Stages of Soybean Nodule Development,” Molecular Genetics and Genomics, Vol. 238, No. 1-2, 1993, pp. 106-119.
- Y. Hiei, S. Ohta, T. Komari and T. Kumashiro, “Efficient Transformation of Rice (Oryza sativa L.) Mediated by Agrobacterium and Sequence Analysis of the Boundaries of the T-DNA,” The Plant Journal, Vol. 6, No. 2, 1994, pp. 271-282. doi:10.1046/j.1365-313X.1994.6020271.x
- M. Jain, A. Nijhawan, A. K. Tyagi and J. P. Khurana, “Validation of Housekeeping Genes as Internal Control for Studying Gene Expression in Rice by Quantitative Real-Time PCR,” Biochemical and Biophysical Research Communications, Vol. 345, 2006, pp. 646-651. doi:10.1016/j.bbrc.2006.04.140
- S. S. Pao, I. T. Paulsen and M. H. Saier, “Major Facilitator Superfamily,” Microbiology and Molecular Biology Reviews, Vol. 12, 1998, pp. 1-34.
- M. Tejada-Jiménezl, A. Galván and E. Fernández, “Algae and Humans Share a Molybdate Transporter,” Proceedings of the National Academy of Sciences of the United States of America, Vol.108, No. 16, 2011, pp. 6420-6425. doi:10.1073/pnas.1100700108
- H. Tomatsu, J. Takano, H. Takahashi, A. Watanabe-Takahashi, N. Shibagaki and T. Fujiwara, “An Arabidopsis Thaliana High-Affinity Molybdate Transporter Required for Efficient Uptake of Molybdate from Soil,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 104, No. 47, 2007, pp. 18807- 18812. doi:10.1073/pnas.0706373104
- M. Tejada-Jimenez, A. Llamas, E. Sanz-Luque, A. Galvan and E. Fernandez, “A High-Affinity Molybdate Transporter in Eukaryotes,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 104, 2007, pp. 20126-20130. doi:10.1073/pnas.0704646104
- I. Baxter, B. Muthukumar, H. C. Park, P. Buchner, B. Lahner, J. Danku, K. Zhao, J. Lee, M. J. Hawkesford, M. L. Guerinot and D. E. Salt, “Variation in Molybdenum Content across Broadly Distributed Populations of Arabidopsis Thaliana Is Controlled by a Mitochondrial Molybdenum Transporter (MOT1),” PLoS Genetics, Vol. 4, 2008, Article ID: e1000004. doi:10.1371/journal.pgen.1000004
- R. R. Mendel, “Cell Biology of Molybdenum in Plants,” Plant Cell Reports, Vol. 30, Vol. 10, 2011, pp. 1787-1797. doi:10.1007/s00299-011-1100-4
- R. R. Mendel and T. Kruse, “Cell Biology of Molybdenum in Plants and Humans,” Biochimica et Biophysica Acta, Vol. 1823, No. 8, 2012, pp. 1568-1579. doi:10.1016/j.bbamcr.2012.02.007
- Y. Ide, M. Kusano, A. Oikawa, A. Fukushima, H. Tomatsu, K. Saito, M. Yokota Hirai and T. Fujiwara, “Effects of Molybdenum Deficiency and Defects in Molybdate Transporter MOT1 on Transcript Accumulation and Nitrogen/Sulphur Metabolism in Arabidopsis thaliana,” Journal of Experimental Botany, Vol. 62, 2010, pp. 1-15.
NOTES
*These authors contributed equally to this work.
#Corresponding author.

