American Journal of Plant Sciences
Vol.4 No.3(2013), Article ID:28683,15 pages DOI:10.4236/ajps.2013.43061
Pinus taeda cDNA Microarray as a Tool for Candidate Gene Identification for Local Red/Far-Red Light Adaptive Response in Pinus sylvestris
![]()
Umeå Plant Science Centre, Department of Forest Genetics and Plant Physiology, Swedish University of Agricultural Sciences, Umeå, Sweden.
Email: *sonalideo@yahoo.com
Received December 27th, 2012; revised February 4th, 2013; accepted February 14th, 2013
Keywords: Conifer; Cryptochrome1; Far-Red Light; Microarray; Red Light; Scots Pine
ABSTRACT
Light quality response is a vital environmental cue regulating plant development. Conifers, like angiosperms, respond to the changes in light quality including the level of red (R) and far-red (FR) light, which follows a latitudinal cline. R and FR wavelengths form a significant component of the entire plant life cycle, including the initial developmental stages such as seed germination, cotyledon expansion and hypocotyl elongation. With an aim to identify differentially expressed candidate genes, which would provide a clue regarding genes involved in the local adaptive response in Scots pine (Pinus sylvestris) with reference to red/far-red light; we performed a global expression analysis of Scots pine hypocotyls grown under two light treatments, continuous R (cR) and continuous FR (cFR) light; using Pinus taeda cDNA microarrays on bulked hypocotyl tissues from different individuals, which represented different genotypes. This experiment was performed with the seeds collected from northern part of Sweden (Ylinen, 68˚N). Interestingly, gene expression pattern with reference to cryptochrome1, a blue light photoreceptor, was relatively high under cFR as compared to cR light treatment. Additionally, the microarray data analysis also revealed expression of 405 genes which was enhanced under cR light treatment; while the expression of 239 genes was enhanced under the cFR light treatment. Differentially expressed genes were re-annotated using Blast2GO tool. These results indicated that cR light acts as promoting factor whereas cFR antagonises the effect in most of the processes like C/N metabolism, photosynthesis and cell wall metabolism which is in accordance with former findings in Arabidopsis. We propose cryptochrome1 as a strong candidate gene to study the adaptive cline response under R and FR light in Scots pine as it shows a differential expression under the two light conditions.
1. Introduction
Plants as sessile organisms, rely on their adaptive plasticity to respond to the changes in their local environmental conditions, to optimise growth and reproduction [1,2]. Light as a main environmental regulator, plays a central role in photosynthesis, photoperiodism, phototropism and photomorphogensis. The photomorphogenetic response includes several physiological mechanisms such as seed germination, hypocotyl elongation, neighbor detection, flowering and so on [3-5]. Photomorphogenesis is mediated by the perception of the light conditions by at least four families of photoreceptors: cryptochromes and phototropins, which monitor the blue and ultraviolet regions of the spectrum; the phytochromes, which mainly monitor the red (R) and far-red (FR) regions of the solar spectrum; and the ultraviolet B photoreceptor [3,6,7].
The phytochrome (phy) family consists of five members phyA through phyE in Arabidopsis [8]. PhyA is designated as phyO and phyB as phyP, in conifers [9]. Phytochromes exist as two interconvertible isoforms: a R-absorbing Pr form and a FR-absorbing Pfr form, of which Pfr is the physiologically active state [8]. On absorption of R light, Pr is converted to the Pfr form; and Pfr absorbs FR light, and gets converted to the Pr form. Two cryptochromes (cry) have been characterised in Arabidopsis—cry1 and cry2 encoded by CRY1 and CRY2 genes respectively [10].
Light regulated expression patterns in developing seedlings have been studied extensively in higher plants e.g. Arabidopsis [11,12], rice [13] and tomato [14]. Red and far-red light both form significant component of plant life cycle. Several investigations report the effect of these light wavelengths on growth and developmental patterns in plants [15]. R:FR ratio controls the seed germinationcotyledon expansion and hypocotyl elongation in the initial stages of plant development. In conifers, response to light quality has been extensively examined under prolonged dichromatic irradiation with R and FR light. Low R:FR ratio (e.g., due to absorption of R light by the chlorophyll in dense canopies) and removal of blue light is known to promote stem elongation, internode length and enhance apical dominance in order to facilitate the acquisition of the radiant energy for photosynthesis; so called shade avoidance mechanism [16-19]. The lateral root formation is also affected by R and FR lights. In Scots pine, lateral root formation was found to be decreased in seedlings grown under light enriched in the R part of the spectrum [20]. R:FR ratio also regulates processes like bud setting in conifers [21] and induces fall hardiness [22]. Seed germination is also largely regulated by light quality; R light enhances germination (especially in pine) [23] while FR inhibits it [24]. Trees also respond to light pulse treatments, e.g. in spruce, light enriched with FR or R prevented terminal bud formation [25] and female cone production [24] respectively. Earlier investigations have reported that the effect of R:FR ratio follows a latitudinal cline. Local adaptation to farred light has already been described in Norway spruce and in Scots pine. It has been demonstrated that Norway spruce (Picea abies) requires FR light to maintain growth, which increases clinally towards the northern latitude [21]. FR light is also necessary for maintaining seconddary needle extension growth in northern, but not in southern populations of Scots pine [26]. Latitudinal and altitudinal clines in response to UV light wavelength have also been reported in conifers [27,28]. Despite clinal adaptation in conifers has interested scientists in the field for many decades [29], there has been little success in identifying the genes responsible for the cline in conifers [30,31].
Scots pine (Pinus sylvestris) population has a wide and continuous distribution and consequently these populations are exposed to variable lengths of photoperiod and light quality. Scots pine being a shade intolerant species, R and FR light wavelengths influence plant development in a major way, which follows a latitudinal cline as discussed earlier. Scots pine lack the High Irradiance Response (HIR) for hypocotyl inhibition under continuous FR (cFR) light; which is the characteristic of angiosperms [e.g. 32-34]. As a result, the conifer hypocotyl developed under cFR or high intensity FR light does not differ from dark grown seedlings [34-36]. Furthermore, in contrast to most angiosperms, conifer seeds possess the ability to synthesize chlorophyll, develop chloroplastlike plastids and express photosynthesis-related genes in darkness [37-39].
Unlike angiosperms, the genome expression pattern under R and FR light has not been extensively studied in conifers; and despite the relevance of R and FR light qualities across all developmental stages of pine trees, the molecular mechanism involved is not clearly understood as yet. Moreover, in nature, most responses occur under continuous light, which can be triggered by short irradiation with high photon fluence rate or by continuous irradiation with low photon fluence rate [40]. In the current investigation we collected seeds from natural Scots pine populations from northern Sweden and compared the differential gene expression patterns between the seedlings grown under two light treatments—continuous R (cR) and cFR light, which are known to be the most essential wavelengths of the light spectrum in photomorphogenetic regulation. We also know that Scots pine populations are locally adapted to light quality. Hence, the main focus of this study aims at identifying candidate genes, which are potentially involved in local adaptation to red/far-red light, based on the differential gene expression pattern under these two light treatments; therefore, a dark control was not included in the experimental design as per standard procedure, which allows a baseline for categorising the regulation of gene expression e.g. upor down-regulation, instead we discuss in terms of differential expression across the discussion section of the manuscript. Moreover, the seedlings were not viable for such a long period until the hypocotyls were fully grown, under continuous darkness. However, we have taken into consideration the genetic variation, and therefore, we have performed the study on bulked hypocotyl tissues from different individuals representing different genotypes. Based on the candidate genes thus identified, further investigations on whether those genes follow a latitudinal cline could be carried out, as very little is known about genes responsible adaptive cline in conifers [31]. Moreover, in the phytochrome family Ingvarsson et al. [41], reported clinal variation in a mutation at phyB2 gene linked to day-length-induced growth cessation and bud set in Aspen (Populus tremula), but similar finding has not been supported in Scots pine [9].
Hypocotyl is known to be an excellent system to study the regulation of plant development due to its morphological simplicity and its high responsiveness to growth regulators such as light quality, temperature and hormones e.g. auxin, cytokinins, ethylene and gibberellins [12]. Hypocotyl as model system possesses another advantage in terms of number of genotypes that can be assayed at a given instance, which is especially relevant in large sized plants like conifers. Both cR and cFR light equally inhibits hypocotyl elongation in Betula pendula, but inhibition is higher when seedlings are irradiated by cR plus cFR light [42]. Similarly, in wild type Arabidopsis, hypocotyl elongation is inhibited by R and FR light [12].
Microarray technology allows transcriptional profiling for large scale experiments. For instance, microarray analysis was applied for the identification of genes involved in light quality regulation in Arabidopsis seedlings grown either in darkness, white, R or FR light [13,43]. In conifers, microarray studies have been focused on two different processes, wood formation [44-46] and stress response to pathogens [47-49]. However, genome-wide expression studies with respect to any aspect of light have not yet been investigated within conifers. Instead, most of the expression studies in response to light have been limited to barely few genes [39,50-52]. Van Zyl et al. demonstrated that it is possible to use a cDNA array from P. taeda hybridised with another species from Pinus or Picea with just a minor loss of sensitivity [53]. In this experiment we have used cDNA array from P. taeda, for expression analysis.
2. Methods
2.1. Plant Material and Growth Conditions
Cones were collected from unrelated trees in a natural population of Scots pine from northern Sweden (Ylinen, 68˚N). A total of 72 seeds were soaked in water and maintained overnight at 4˚C. Soaked seeds were sown in a randomised complete two-block design in trays containing vermiculite, in a growth chamber. The climate chamber was supplied with light from Halogen-metal lamp (Ostram; Powerstar HQI-TS, 400 W/D, daylight). Seeds were grown under continuous light conditions under red and far-red filters (Filters used: red (026-M) + blue (195-M) for FR and red for R light; Lee filters, Hampshire, UK) (refer to additional file—Light Spectrum). We applied high irradiance light treatment, which consisted of low intensity and continuous irradiation of R (620 nm) or FR (720 nm) light. We applied low light intensity for both light treatments (<1 W/m2·nm) to avoid saturating the photsyntetic capacity of the Scots pine seedlings under the long time light treatments. Further, lower intensity was chosen for this study to compensate for the long continuous light treatment. The growth chamber was maintained under constant 85% humidity and at 22˚C. Some of the earlier investigations reported that hourly light pulses providing the same total fluence were significantly less effective in causing response to light [32,34]; therefore we used continuous light for our experiment.
The relative intensity between cR and cFR was set following the experimental design of two essential publications on light quality research in conifers [34] and Arabidopsis [15], where higher intensity was applied under the R compared to FR light treatments. We are aware that with our experimental design, it is not possible to distinguish between the effects caused by intensity and light wavelength. However, it allowed us to reach our main goal, which was to reproduce the lack of response to high irradiance under FR, typical of Scots pine; and compare it to hypocotyl growth response under R light, where it is well known that R light inhibits hypocotyl growth as described by Fernbach and Mohr [34].
As the germination synchronisation was not efficient enough, the seedlings were individually harvested when the hypocotyls were fully grown i.e. after the hypocotyls growth ceased with the opening of the cotyledons for every seedling [35]. This was carried out in order to avoid measuring growth speed, as we were interested in the total growth. This methodology also allowed us to capture the hypocotyls at the same developmental stage, which was more appropriate for the subsequent global expression profile study. The length of the hypocotyls was determined as the distance between the crown of the root (usually prominent as the seat of a brownish veil) and the needle base. Seedlings were then snap frozen in liquid nitrogen and stored in −80˚C till further processing.
2.2. Microarray Procedure
For the global expression profile characterisation, we prepared an R-bulk and FR-bulk containing ten hypocotyls (representing different genotypes) treated with cR and cFR light, respectively. We used dye swap experimental design to eliminate the dye bias, therefore the array was performed twice and instead of having biological replicates, we prepared bulk samples of ten hypocotyls under each light condition. For the bulk preparation, equal amounts of hypocotyl tissue were weighed (roots and cotyledons were removed prior to weighing). Pinus taeda spotted cDNA microarrays (representing 12,523 genes) from The J. Craig Venter Institute, USA (formerly The Institute for Genomic Research, USA) were used to perform the expression analysis in an automated slide processor (ASP; Amersham Bioscience, Little Chalfont, UK). As this array was built on the RNA extracted from seedlings, it was very appropriate for our expression study in hypocotyls. Microarray was performed using the procedure. Total RNA from each bulk was extracted using commercially available kit (Qiagen). Total RNA was amplified with MessageAmp™ II aRNA Amplification Kit from Ambion. 7 µg of amplified RNA was used for cDNA synthesis (amino allyl-labelling). The cDNA was purified using Microcon 30 concentrators, eluted in water and dried in Speed-vac. Samples were re-suspended in 10 µL of 0.1 M NaHCO3, pH 9 and mixed with the appropriate Cy-dye, dissolved in 10 µL DMSO. The samples were coupled for 120 minutes and cleaned using a Cyscribe GFX purification kit (GE Healthcare). The cDNA was eluted in 100 µL of elution buffer and then the volume was reduced in a Speed-vac to 41 µL. Appropriate incorporated samples where pooled and mixed with 45 µL SSC (20X), 45 µL Formamide (100%), 4 µL SDS (10%), 1 µL tRNA (25 µg/µL) and 1 µL Oligo-dA (10 µg/µL). The slides were exposed to pre-hybridisation buffer (5X SSC, 50% Formamide, 2.5 X Denhardt’s solution, 0.1% BSA) for 30 minutes. The samples where denatured at 95˚C for 3 minutes and then injected into the ASP chamber, using a syringe. The slides where hybridised for 14 - 16 hours at 42˚C then washed with three buffers of sequentially decreasing concentration (1X SSC, 0.05% SDS followed by 0.3X SSC and finally 0.05X SSC). All kits were used according to the manufacturer’s instructions.
2.3. Data Analysis
Microarray slides were scanned with predetermined increasing laser power and phototube multiplier (PMT) settings using a Scanarray scanner (PerkinElmer AB, Sweden). The resulting images were analysed in Genepix 5.0 (Axon Instruments, CA, USA). The raw data was stored in the public microarray database UPSC-BASE [54]. The different scan levels for each slide were merged with Restricted Linear Scaling (RLS) [55] followed by print-tip lowess normalisation [56,57], using tools implemented in UPSC-BASE, before further analysis. The log ratios of the intensity measurements were computed. Statistical analysis was carried out by calculating the t-test statistic and the corresponding p-value from the M-value for each gene in the dye swap experiment [58].
The p-values were adjusted by false discovery ratio (FDR) method using the R-statistical package, R Development Core Team, 2006 [59]. We used a p-value cut-off of 0.05 and log2 expression ratios >0.95 as selection criteria for significantly expressed genes. Computational annotation of the statistically significant, differentially expressed genes was done using Blast2GO tool (Version: January 2010 release) [60].
3. Results
The focus of our study was to identify candidate genes for studying adaptive cline in Scots pine, based on differential gene expression pattern in Scots pine seedlings grown under cR and cFR light treatments; therefore, throughout the result and discussion chapter we have referred to the changes in gene expression under cR with reference to the expression pattern under cFR light and vice versa. Thus the genes which are enhanced under cR are suppressed under cFR and vice-versa.
3.1. Hypocotyl Elongation Response
The Kolmogorov-Smirnov test showed that there was a significant effect of light quality on the length of the hypocotyls (p-value = 4.12e−9) (Figure 1). Higher hypocotyl length under cFR light has also been reported by Fernbach and Mohr [34]. The difference in hypocotyl length between cR and cFR light that we observed, once more supports the well-known phenomenon of lack of high irradiance response (HIR) in Scots pine [34,61]. However, lack of HIR may not be general to all conifers; for example, long exposure to FR light caused inhibition of hypocotyl elongation to the same extent (or more) as compared to R in Picea abies [23,35]. This might be explained by the shadow-tolerant nature of Picea abies compared to Scots pine [62]. For example, under light environments enriched in FR light (e.g., under dense canopies) enhancement of internode growth has been described in seedlings and adult trees of Scots pine, but the same response was not observed in Picea abies [17, 19].
3.2. Microarray Analysis
Higher expression of cryptochrome1 was found under cFR with reference to cR light, suggesting that this can be regarded as a putative candidate gene involved in adaptive response with reference to red/far-red light. The microarray analysis also revealed 405 differentially expressed genes under cR light treatment, and 239 genes were found to be differentially expressed under cFR light treatment. Figure 2 represents the MA-plot of the loga-
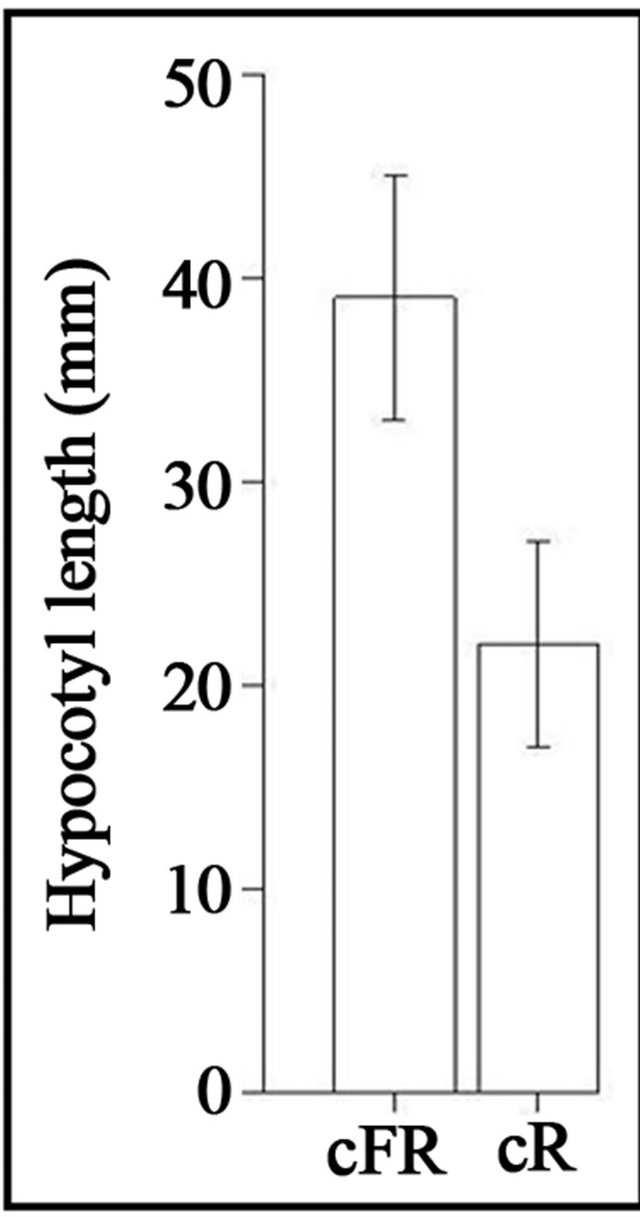
Figure 1. Histogram of the average hypocotyl length under cR and cFR light.
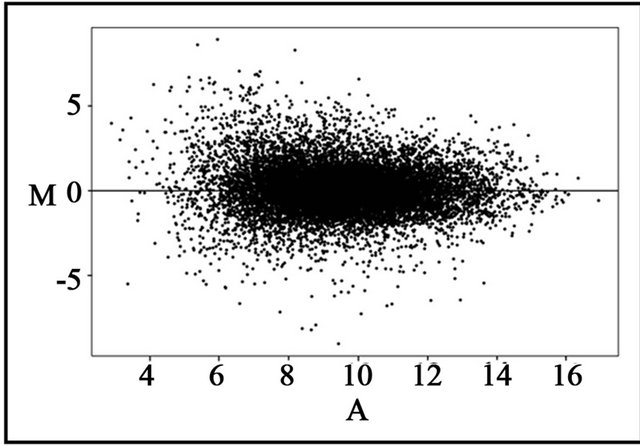
Figure 2. MA-plot of the gene intensity values under cR and cFR light.
rithmic values of the intensities, which shows a linear trend and the log ratios are centred around zero.
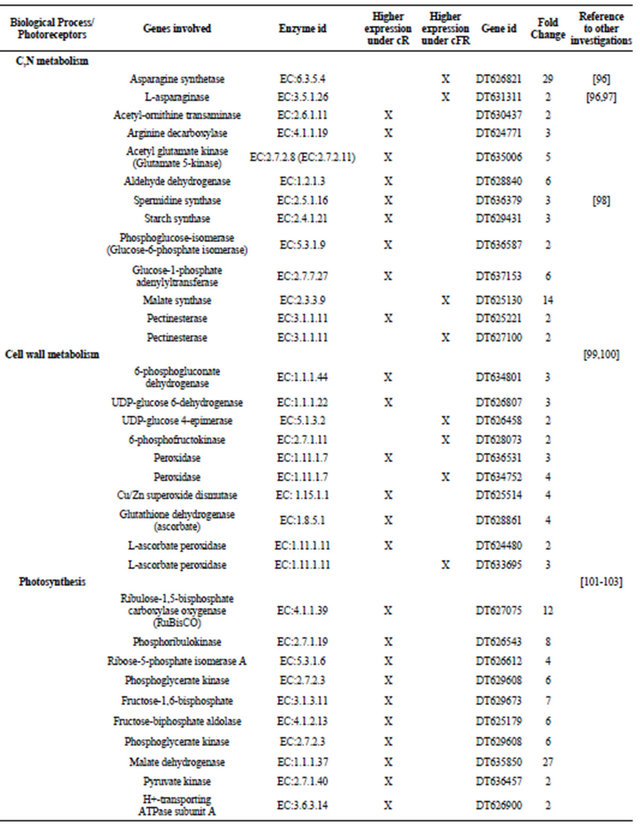
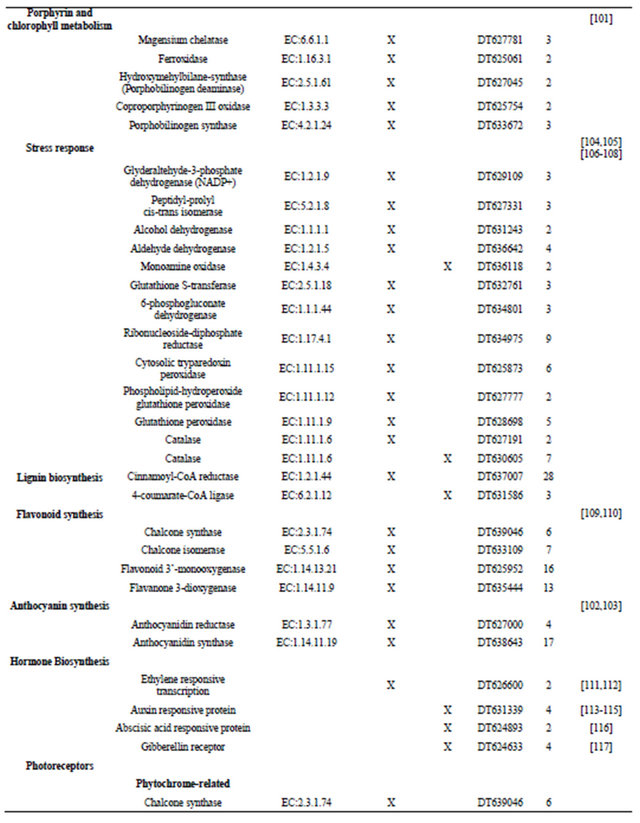
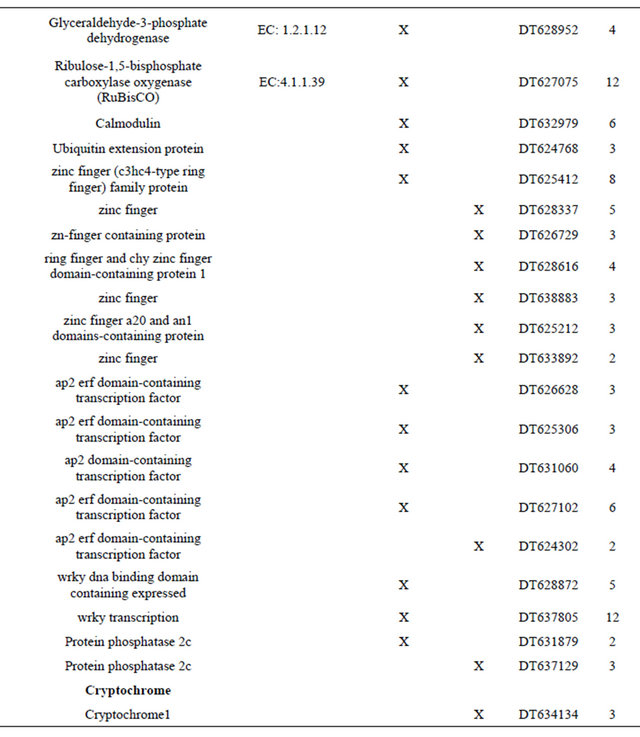
Higher expression of a larger number of genes involved in processes such as C,N metabolism (e.g. arginine decarboxylase, acetyl glutamate kinase, starch synthase, malate synthase), photosynthesis (e.g. phosphorribulokinase, pyruvate kinase, porphobilinogen synthase), lignin biosynthesis (e.g. cinnamoyl-CoA reductase), flavonoid synthesis (e.g. chalcone synthase), porphyrin and chlorophyll metabolism (e.g. porphobilinogen synthase), and anthocyanin synthesis (e.g. anthocyanidin synthase) was found under cR as compared to the cFR light. Table 1 gives the overview of the metabolic pathway and fold regulation of the gene expression under cR and cFR light.
The Blast2GO analysis of the percentage of secondlevel GO terms for three categories: biological process, cellular component and molecular function, is represented in Figures 3-5, respectively. With reference to biological processes under cR light, highest participation was of the metabolic process (~33%, Figure 3(a)), which was more than that of cFR light (~23%, Figure 3(b)). Cellular process contributed the highest among the various biological processes under cFR (~31%, Figure 3(b)), which was about the same, as observed under cR light (Figure 3(a)). A process such as localisation was observed to be almost doubled under cFR than under cR; whereas elevated expression of genes involved in multiorganism process and developmental process, was found under cR as compared to cFR light. The processes such as response to stimulus and immune system process contributed equally under cR and cFR light conditions.
Regarding the gene expression under the cellular component category (Figure 4), cell occupied the largest percentage under cR and cFR light; percentage being higher under cFR (54%, Figure 4(b)) as compared to cR light (40%, Figure 4(a)). The activity in envelop was almost four times more under cR (4.5%, Figure 4(a)) than under cFR light (~1%, Figure 4(b)). The percentage with regards to macromolecular complex was observed to be doubled under cR, whereas it was approximately the same for both light treatments with reference to extracellular region.
Under the category of molecular function (Figure 5), binding contributed the most among the other functions under both cR and cFR light conditions, followed very closely by the catalytic activity. In this aspect, binding occupied larger percentage under cFR (~48%, Figure 5(b)) than cR light (~37%, Figure 5(a)); whereas the catalytic activity contributed about 38% under cR (Figure 5(a)), which was higher than that of the cFR light condition (~33%, Figure 5(b)). Transport activity, translation regulator activity, transcription regulator activity and antioxidant activity was approximately the same under cR and cFR light. Molecular transducer activity, nutrient reservoir activity and enzyme regulator activity was observed to be comparatively higher under cFR while structural molecule activity and electron carrier activity was observed to be higher under cR light.
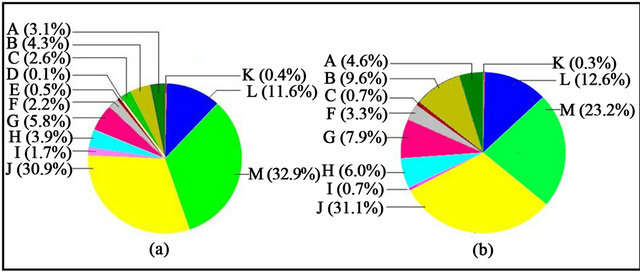
Figure 3. Level 2 pie charts of Biological Process—(a) Higher expression under cR light; (b) Higher expression under cFR light. A: Multicellular organism process; B: Localisation; C: Multi-organism process; D: Locomotion; E: Growth; F: Reproduction; G: Biological regulation; H: Developmental process; I: Anatomical structure formation; J: Cellular process; K: Immune system process; L: Response to stimulus; M: Metabolic process.
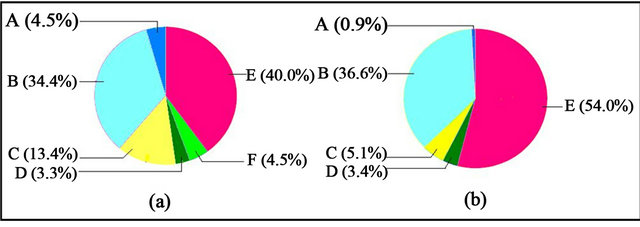
Figure 4. Level 2 pie charts of Cellular Component—(a) Higher expression under cR light; (b) Higher expression under cFR light. A: Envelop; B: Organelle; C: Macromolecular Complex; D: Extracellular region; E: Cell; F: Membrane-enclosed lumen.
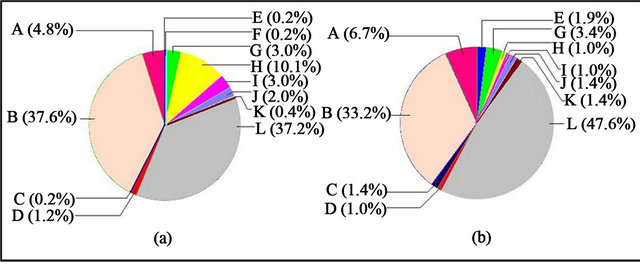
Figure 5. Level 2 pie charts of Molecular Function—(a) Higher expression under cR light; (b) Higher expression under cFR light. A: Transporter activity; B: Catalytic activity; C: Nutrient reservoir activity; D: Translation regulator activity; E: Molecular transducer activity; F: Metallochaperone activity; G: Transcription regulator activity; H: Structural molecule activity; I: Electron carrier activity; J: Antioxidant activity; K: Enzyme regulator activity; L: Binding.
Table 1. Metabolic pathway and fold regulation of the gene expression under cR and cFR light.
Continued
Continued
4. Discussion
A comprehensive discussion on all the photomorphogenetic changes that occur in response to light was beyond the scope of this article, instead, we focused on light response pathways, which have been better characterised in conifers.
Microarray data analysis reveals that expression pattern of most of the genes under R and FR light is in accordance with the earlier findings in Arabidopsis and other plant systems (refer Table 1). Plants respond to light signals through a complex network of photoreceptors [63]. Phytochromes are involved in the control of hypocotyl lengthening [64] and are the most well studied class of photoreceptors, which perceive R and FR light [8]. Cryptochromes perceive blue light; they mediate various light induced responses in plants and are demonstrated to be involved in inhibition of hypocotyl elongation under blue light [65].
Direct regulation of phytochrome was not detected in this study, as the array did not include phytochrome; but the results show that transcripts of the phytochromeregulated genes were observed under both cR and cFR light, although more number of the transcripts were found under cR light than under cFR, which suggests that there was a difference in phytochrome signalling between the conditions tested (refer Table 1). Higher expression of chalcone synthase [66,67], glyceraldehyde-3- phosphate dehydrogenase [68,69], ribulose-1,5-bisphosphate carboxylase/oxygenase [70,71], which are positively regulated by phytochrome, was observed under cR light. Calmodulin and ubiquitin extension protein expression was also found to be elevated under cR compared to cFR light; calmodulin being one of the early components in phytochrome signalling, and ubiquitin mediated protein degradation is a part of phytochrome signalling mechanism [3]. However, expression of isoforms of protein-phosphatase2c, which is indentified recently as phytochrome-interacting protein [72], was found under both cR and cFR light treatment. PhyA induced transcriptional cascade includes zinc-finger proteins, AP2-domain and WRKY factors; zinc-finger proteins being the largest class in this regard [73]. Regarding this aspect, we observed expression of various forms of zinc-finger and AP2-domain being expressed under both light conditions; more forms of zinc-finger were expressed under cFR, while more forms of AP2-domain were expressed under cR light. Expression of WRKY factors was elevated under cR light.
Cry1 is involved in inhibition of hypocotyl elongation under blue light [74,75]. Cry1 is blue light dependent, but it also plays a role in blue light independent regulation of circadian clock, cotyledon folding and inhibition of hypocotyl elongation in Arabidopsis [76]. Cry1-cry2 mutant exhibit reduced changes of mRNA expression to not only blue, but also R light. Transgenic Arabidopsis, over-expressing cry1 photoreceptor is hypersensitive to continuous blue light, exhibiting unusually short hypocotyls and high levels of anthocyanin [77,78]. In the present study, we found higher expression of cry1 transcripts under cFR light as compared to cR. Further, studies indicated that blue light acts through cry1, which affects the expression of many genes, a subset of which suppresses stem growth by repressing gibberellins and auxin levels and/or sensitivity [79]. We observed elevated expression of cry1 under cFR light in pine, but molecular and physiological mechanisms behind the response of cry1 under cFR light has not been investigated as yet in any species. One hypothesis is that, cry1 has altered function in pine due to sequence variability, therefore it might have promoted the elongation of hypocotyls in the current investigation. In fact, variation in cryptochrome sequence has been described with new altered biochemical properties [80]. Here, we also take into account that cry1 can act independently of phyA and phyB, as reported by an earlier investigation in Arabidopsis [81].
It is well known that phyA regulates seed germination in response to cFR whereas phyB controls germination under cR light [82]. PhyA [83-85] and phyB [86,87] predominantly mediates HIR of photoinhibition of hypocotyl elongation in Arabidopsis induced by cFR and cR light respectively. Cryptochromes play role in control of gene expression mediated by phytochromes [10]. Cry1 physically interacts with phyA [88] and it also shows functional dependence on phyA or phyB in Arabidopsis for the blue light induced inhibition of hypocotyl growth and anthocyanin accumulation [89]. We found indirect evidences of differential expression of phyA between cFR and cR light treatments. Therefore, another hypothesis is that Cry1-phyA interaction may function in a different way in pine species compared to Arabidopsis allowing for the elongation of hypocotyls and suppression of anthocyanin accumulation under cFR light, which may be due to sequence variation in cry1, as discussed earlier. Moreover, changes in the coding sequence of phyA has been shown to alter its biochemical properties, which in turn causes an altered response to light namely reduced far-red sensitivity [15,84,85,90,91]. The significant increase in the length of the pine hypocotyls in response to cFR light may be due to increased auxin production under effect of cry1 or cry1-phyA (phyO in pine) interaction (excess of auxin promotes elongation in Arabidopsis) [92]. This is based on our findings with reference to auxin-responsive protein, although we have not measured the auxin levels in the seedlings. Here, we would also like to point out that rapid high-level synthesis of auxin may be deployed by plants via alternative pathway to combat shade avoidance [93].
Based on our results from the microaaray data analysis, we propose the blue-light receptor/cryptochrome1 as the putative candidate gene involved in local adaptive response to red/far-red light, which may be used to study adaptive cline in Scots pine with reference to red/far-red light and it also deserves further investigation to help dissecting the molecular mechanisms behind lack of HIR response in pines and other conifers.
Under natural conditions, plant response to light is confounded with stress response processes related to temperature and decline in water potential [94]. In our experiment, we minimised the effect of other environmental factors, other than light quality, by keeping humidity and temperature constant. No tissue necrosis or seedling decay was observed, supporting that even if response to light stress was activated, it probably did not reach saturation. Our study indicates that gene expression is enhanced by cR light treatment, namely more number of genes show higher expression under cR compared to cFR light (Table 1). This result is in accordance with the findings in Arabidopsis, where most metabolic pathways have been seen more sensitive to R and blue light than to FR [43]; except in case of photoreceptors, where we found higher expression of the blue light receptor cryptochrome1 under cFR. Our expression analysis revealed the probable role of cry1 gene in the adaptive response to high irradiance under cFR and cR lights in Scots pine, which is considered as a blue-light sensor; indicating that the roles and interactions of phytochrome and cryptochrome gene families are not fully resolved.
We propose cryptochrome1 as putative candidate gene involved in local adaptive response in Scots pine with reference to red/far-red light. This study thus demonstrates that global expression analysis using microarray as an efficient tool to predict candidate genes potentially involved in local adaptation. The data represented here could be used as a clue to study adaptive cline in Scots pine. We also suggest, the light dependent AS, RuBisCO and cinnamoyl-CoA reductase genes as putative candidates to study the adaptive cline; as the fold regulation of these genes was significant under the two light treatments studied (Table 1). Our next step would be to carry out RT-PCR with our most relevant candidate gene cry1 and other genes in the light response pathway; but we foresee difficulties in this regard due to the insufficient genomic resources (number of mRNA and EST sequences in GenBank (2011): 3,431 and 666 respectively), and presence of pseudogenes and the complexity of the Scots pine genome [95]; which explains the reason behind why so little work has been done in conifers, with reference to the current topic of investigation. Future research in other high-throughput omics and micro RNAs (miRNA) would add further depth to our understanding of physiological process and gene regulation in conifers in response to light quality. If much remains to be learned in order to understand the underlying network that controls angiosperm photomorphogenesis, then our knowledge with reference to conifer species is still in its infancy.
5. Acknowledgements
The authors want to thank Skogforsk and specially, Bengt Andersson, Torgny Persson T and Monica Lundström for providing Scots pine seeds and Thomas Hiltonen from UPSC, Umeå for performing the microarray hybridisations. We also want to thank to Ross Whetten for suggestions on the experimental part of the work and Andreas Sjödin for providing help to normalise the microarray data. This work was supported by the Kempe foundation through the Research school of Forest Genetics, SLU, Umeå.
REFERENCES
- A. D. Bradshaw, “Evolutionary Significance of Phenotypic Plasticity in Plants,” Advances in Genetics, Vol. 13, 1965, pp. 115-155.
- J. Thompson, “Phenotypic Plasticity as a Component of Evolutionary Change,” Trends in Ecology & Evolution, Vol. 6, No. 8, 1991, pp. 246-249. doi:10.1016/0169-5347(91)90070-E
- M. Chen, J. Chory and C. Fankhauser, “Light Signal Transduction in Higher Plants,” Annual Review of Genetics, Vol. 38, 2004, pp. 87-117. doi:10.1146/annurev.genet.38.072902.092259
- C. Kami, S. Lorrain, P. Hornitschek and C. Fankhauser, “Light-Regulated Plant Growth and Development,” Plant Development, Vol. 91, 2010, pp. 29-66. doi:10.1016/S0070-2153(10)91002-8
- R. E. Kendrick and G. H. M. Kronenberg, “Photomorphogenesis in Plants,” 2nd Edition, Kluwer Academic Publishers, Dordrecht, 1994, pp. 1-828.
- W. Briggs and J. Spudich, “Handbook of Photosensory Receptors,” Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, 2005. doi:10.1002/352760510X
- W. R. Briggs and J. M. Christie, “Phototropins 1 and 2: versatile Plant Blue-Light Receptors,” Trends in Plant Science, Vol. 7, No. 5, 2002, pp. 204-210.
- P. H. Quail, “Phytochrome Photosensory Signalling Networks,” Nature Reviews Molecular Cell Biology, Vol. 3, No. 2, 2002, pp. 85-93. doi:10.1038/nrm728
- M. R. Garcia-Gil, M. Mikkonen and O. Savolainen, “Nucleotide Diversity at Two Phytochrome Loci along a Latitudinal Cline in Pinus sylvestris,” Molecular Ecology, Vol. 12, No. 5, 2003, pp. 1195-1206. doi:10.1046/j.1365-294X.2003.01826.x
- X. H. Yu, H. L. John Klejnot and C. T. Lin, “The Cryptochrome Blue Light Receptors. The Arabidopsis Book,” The American Society of Plant Biologists, 2010.
- X. Li, Y. Yang, Y. Li, J. Wang, X. Xiao, X. Guo, D. Tang and X. Liu, “Protein Identification and mRNA Analysis of Phytochrome-Regulated Genes in Arabidopsis under Red Light,” Science in China Series C-Life Sciences, Vol. 52, No. 4, 2009, pp. 371-380. doi:10.1007/s11427-009-0045-0
- F. Vandenbussche, J. P. Verbelen and D. Van der Straeten, “Of Light and Length: Regulation of Hypocotyl Growth in Arabidopsis,” Bioessays, Vol. 27, No. 3, 2005, pp. 275- 284. doi:10.1002/bies.20199
- Y. L. Jiao, L. G. Ma, E. Strickland and X. W. Deng, “Conservation and Divergence of Light-Regulated Genome Expression Patterns during Seedling Development in Rice and Arabidopsis,” Plant Cell, Vol. 17, No. 12, 2005, pp. 3239-3256. doi:10.1105/tpc.105.035840
- G. A. Auge, S. Perelman, C. D. Crocco, R. A. Sanchez and J. F. Botto, “Gene Expression Analysis of LightModulated Germination in Tomato Seeds,” New Phytologist, Vol. 183, No. 2, 2009, pp. 301-314. doi:10.1111/j.1469-8137.2009.02867.x
- J. N. Maloof, J. O. Borevitz, T. Dabi, J. Lutes, R. B. Nehring, J. L. Redfern, G. T. Trainer, J. M. Wilson, T. Asami, C. C. Berry, D. Weigel and J. Chory, “Natural Variation in Light Sensitivity of Arabidopsis,” Nature Genetics, Vol. 29, No. 4, 2001, pp. 441-446. doi:10.1038/ng777
- S. Asakawa, S. Sasaki and Y. Morikawa, “Growth of Tree Seedlings under the Lights of Different Spectral Compositions,” Journal of the Japanese Forestry Society, Vol. 56, No. 12, 1974, pp. 441-447.
- J. Hoddinott and R. Scott, “The Influence of Light Quality and Carbon Dioxide Enrichment on the Growth and Physiology of Seedlings of Three Conifer Species. 1. Growth Responses,” Canadian Journal of Botany-Revue Canadienne De Botanique, Vol. 74, No. 3, 1996, pp. 383- 390. doi:10.1139/b96-048
- M. Sarala, E. Taulavuori, J. Karhu, E. M. Savonen, K. Laine, E. Kubin and K. Taulavuori, “Improved Elongation of Scots Pine Seedlings under Blue Light Depletion Is Not Dependent on Resource Acquisition,” Functional Plant Biology, Vol. 36, No. 8, 2009, pp. 742-751. doi:10.1071/FP09012
- I. J. Warrington, D. A. Rook, D. C. Morgan and H. L. Turnbull, “The Influence of Simulated Shadelight and Daylight on Growth, Development and Photosynthesis of Pinus radiata, Agathis australis and Dacrydium cupressinum,” Plant Cell and Environment, Vol. 12, No. 4, 1989, pp. 343-356. doi:10.1111/j.1365-3040.1989.tb01951.x
- T. M. de la Rosa, P. J. Aphalo and T. Lehto, “Effects of Far-Red Light on the Growth, Mycorrhizas and Mineral Nutrition of Scots Pine Seedlings,” Plant and Soil, Vol. 201, No. 1, 1998, pp. 17-25. doi:10.1023/A:1004383526878
- D. H. Clapham, I. Dormling, I. Ekberg, G. Eriksson, M. Qamaruddin and D. Vince-Prue, “Latitudinal Cline of Requirement for Far-Red Light for the Photoperiodic Control of Budset and Extension Growth in Picea abies (Norway Spruce),” Physiologia Plantarum, Vol. 102, No. 1, 1998, pp. 71-78. doi:10.1034/j.1399-3054.1998.1020110.x
- E. H. Beck, R. Heim and J. Hansen, “Plant Resistance to Cold Stress: Mechanisms and Environmental Signals Triggering Frost Hardening and Dehardening,” Journal of Biosciences, Vol. 29, No. 4, 2004, pp. 449-459. doi:10.1007/BF02712118
- H. Kvaalen and M. Appelgren, “Light Quality Influences Germination, Root Growth and Hypocotyl Elongation in Somatic Embryos But Not in Seedlings of Norway Spruce,” In Vitro Cellular & Developmental Biology— Plant, Vol. 35, No. 6, 1999, pp. 437-441. doi:10.1007/s11627-999-0064-3
- D. Durzan, R. Campbell and A. Wilson, “Inhibition of Female Cone Production in White Spruce by Red Light Treatment during Night Under Field Conditions,” Environmental and Experimental Botany, Vol. 19, No. 3, 1979, pp. 133-135. doi:10.1016/0098-8472(79)90042-X
- I. S. Fløystad and G. G. Patil, “Growth and Terminal Bud Formation in Picea abies Seedlings Grown with Alternating Diurnal Temperature and Different Light Qualities,” Scandinavian Journal of Forest Research, Vol. 17, No. 1, 2002, pp. 15-27. doi:10.1080/028275802317221046
- D. H. Clapham, I. Ekberg, G. Eriksson, L. Norell and D. Vince-Prue, “Requirement for Far-Red Light to Maintain Secondary Needle Extension Growth in Northern But Not Southern Populations of Pinus sylvestris (Scots Pine),” Physiologia Plantarum, Vol. 114, No. 2, 2002, pp. 207- 212. doi:10.1034/j.1399-3054.2002.1140206.x
- J. H. Sullivan and A. H. Teramura, “Effects of Ultraviolet-B Irradiation on Seedling Growth in the Pinaceae,” American Journal of Botany, Vol. 75, No. 2, 1988, pp. 225-230. doi:10.2307/2443888
- J. H. Sullivan and A. H. Teramura, “The Effects of Ultraviolet-B Radiation on Loblolly-Pine. 2. Growth of FieldGrown Seedlings,” Trees-Structure and Function, Vol. 6, No. 3, 1992, pp. 115-120. doi:10.1007/BF00202426
- O. Savolainen, T. Pyhajarvi and T. Knurr, “Gene Flow and Local Adaptation in Trees,” Annual Review of Ecology Evolution and Systematics, Vol. 38, 2007, pp. 595- 619. doi:10.1146/annurev.ecolsys.38.091206.095646
- A. E. Palme, M. Wright and O. Savolainen, “Patterns of Divergence among Conifer ESTs and Polymorphism in Pinus sylvestris Identify Putative Selective Sweeps,” Molecular Biology and Evolution, Vol. 25, No. 12, 2008, pp. 2567-2577. doi:10.1093/molbev/msn194
- W. Wachowiak, P. A. Balk and O. Savolainen, “Search for Nucleotide Diversity Patterns of Local Adaptation in Dehydrins and Other Cold-Related Candidate Genes in Scots Pine (Pinus sylvestris L.),” Tree Genetics & Genomes, Vol. 5, No. 1, 2009, pp. 117-132. doi:10.1007/s11295-008-0188-3
- J. J. Casal and M. A. Mazzella, “Conditional Synergism between Cryptochrome 1 and Phytochrome B Is Shown by the Analysis of phyA, phyB, and hy4 Simple, Double, and Triple Mutants in Arabidopsis,” Plant Physiology, Vol. 118, No. 1, 1998, pp. 19-25. doi:10.1104/pp.118.1.19
- M. M. Neff, S. M. Nguyen, E. J. Malancharuvil, S. Fujioka, T. Noguchi, H. Seto, M. Tsubuki, T. Honda, S. Takatsuto, S. Yoshida and J. Chory, “BAS1: A Gene Regulating Brassinosteroid Levels and Light Responsiveness in Arabidopsis,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 96, No. 26, 1999, pp. 15316-15323. doi:10.1073/pnas.96.26.15316
- E. Fernbach and H. Mohr, “Coaction of Blue Ultraviolet— A Light and Light Absorbed by Phytochrome in Controlling Growth of Pine (Pinus sylvestris L.) Seedlings,” Planta, Vol. 180, No. 2, 1990, pp. 212-216. doi:10.1007/BF00193998
- O. Scharff, “Effects of Red and Far-Red Light on Hypocotyl of Picea abies,” Physiologia Plantarum, Vol. 15, No. 4, 1962, pp. 804-814. doi:10.1111/j.1399-3054.1962.tb08129.x
- Y. Morikawa, S. Asakawa and S. Sasaki, “Growth of Pine and Birch Seedlings under Lights with Different Spectral Compositions and Intensities,” Journal of the Japanese Forestry Society, Vol. 58, No. 5, 1976, pp. 174-178.
- M. Bogdanov, “Chlorophyll Formation in Dark. 1. Chlorophyll in Pine Seedlings,” Physiologia Plantarum, Vol. 29, No. 1, 1973, pp. 17-18. doi:10.1111/j.1399-3054.1973.tb04802.x
- F. Canovas, B. McLarney and J. Silverthorne, “LightIndependent Synthesis of LHC IIB Polypeptides and Assembly of the Major Pigmented Complexes during the Initial Stages of Pinus palustris Seedling Development,” Photosynthesis Research, Vol. 38, No. 1, 1993, pp. 89-97. doi:10.1007/BF00015065
- Y. Mukai, K. Tazaki, T. Fujii and N. Yamamoto, “LightIndependent Expression of Three Photosynthetic Genes, CAB, RBCS and RBCL, in Coniferous Plants,” Plant and Cell Physiology, Vol. 33, No. 7, 1992, pp. 859-866.
- E. Schafer, L. Fukshansky and W. Shropshire Jr., “Action Spectroscopy of Photoreversible Pigment Systems,” Encyclopedia of Plant Physiology, Vol. 16, 1983, pp. 39-68.
- P. K. Ingvarsson, M. V. Garcia, D. Hall, V. Luquez and S. Jansson, “Clinal Variation in phyB2, a Candidate Gene for Day-Length-Induced Growth Cessation and Bud Set, across a Latitudinal Gradient in European Aspen (Populus tremula),” Genetics, Vol. 172, No. 3, 2006, pp. 1845- 1853. doi:10.1534/genetics.105.047522
- B. A. Tsegay, J. E. Olsen and O. Juntttila, “Effect of Red and Far-Red Light on Inhibition of Hypocotyl Elongation in Ecotypes of Betula pendula Roth,” African Journal of Biotechnology, Vol. 4, No. 1, 2005, pp. 50-56.
- L. G. Ma, J. M. Li, L. J. Qu, J. Hager, Z. L. Chen, H. Y. Zhao and X. W. Deng, “Light Control of Arabidopsis Development Entails Coordinated Regulation of Genome Expression and Cellular Pathways,” Plant Cell, Vol. 13, No. 12, 2001, pp. 2589-2607. doi:10.2307/3871521
- R. Whetten, Y. H. Sun, Y. Zhang and R. Sederoff, “Functional Genomics and Cell Wall Biosynthesis in Loblolly Pine,” Plant Molecular Biology, Vol. 47, No. 1-2, 2001, pp. 275-291. doi:10.1023/A:1010652003395
- S. H. Yang, L. van Zyl, E. G. No and C. A. Loopstra, “Microarray Analysis of Genes Preferentially Expressed in Differentiating Xylem of Loblolly Pine (Pinus taeda),” Plant Science, Vol. 166, No. 5, 2004, pp. 1185-1195. doi:10.1016/j.plantsci.2003.12.030
- X. G. Li, H. X. Wu and S. G. Southerton, “Seasonal Reorganization of the Xylem Transcriptome at Different Tree Ages Reveals Novel Insights into Wood Formation in Pinus radiata,” New Phytologist, Vol. 187, No. 3, 2010, pp. 764-776. doi:10.1111/j.1469-8137.2010.03333.x
- A. Adomas, G. Heller, A. Olson, J. Osborne, M. Karlsson, J. Nahalkova, L. Van Zyl, R. Sederoff, J. Stenlid, R. Finlay and F. O. Asiegbu, “Comparative Analysis of Transcript Abundance in Pinus sylvestris after Challenge with a Saprotrophic, Pathogenic or Mutualistic Fungus,” Tree Physiology, Vol. 28, No. 6, 2008, pp. 885-897. doi:10.1093/treephys/28.6.885
- A. M. Morse, C. D. Nelson, S. F. Covert, A. G. Holliday, K. E. Smith and J. M. Davis, “Pine Genes Regulated by the Necrotrophic Pathogen Fusarium circinatum,” Theoretical and Applied Genetics, Vol. 109, No. 5, 2004, pp. 922-932. doi:10.1007/s00122-004-1719-4
- S. G. Ralph, S. Jancsik and J. Bohlmann, “Dirigent Proteins in Conifer Defense II: Extended Gene Discovery, Phylogeny, and Constitutive and Stress-Induced Gene Expression in Spruce (Picea spp.),” Phytochemistry, Vol. 68, No. 14, 2007, pp. 1975-1991. doi:10.1016/j.phytochem.2007.04.042
- M. C. Alosi, D. B. Neale and C. S. Kinlaw, “Expression of CAB Genes in Douglas-Fir Is Not Strongly Regulated by Light,” Plant Physiology, Vol. 93, No. 2, 1990, pp. 829- 832. doi:10.1104/pp.93.2.829
- Y. Mukai, N. Yamamoto and T. Koshiba, “Light-Independent and Tissue-Specific Accumulation of LightHarvesting Chlorophyll-A/B Binding-Protein and Ribulose Bisphosphate Carboxylase in Dark-Grown Pine-Seedlings,” Plant and Cell Physiology, Vol. 32, No. 8, 1991, pp. 1303-1306.
- N. Yamamoto, Y. Mukai, M. Matsuoka, Y. Kanomurakami, Y. Tanaka, Y. Ohashi, Y. Ozeki and K. Odani, “Light-Independent Expression of CAB and RBCS Genes in Dark-Grown Pine-Seedlings,” Plant Physiology, Vol. 95, No. 2, 1991, pp. 379-383. doi:10.1104/pp.95.2.379
- L. van Zyl, S. von Arnold, P. Bozhkov, Y. Z. Chen, U. Egertsdotter, J. MacKay, R. R. Sederoff, J. Shen, L. Zelena and D. H. Clapham, “Heterologous Array Analysis in Pinaceae: Hybridization of Pinus taeda cDNA Arrays with cDNA from Needles and Embryogenic Cultures of P. taeda, P. sylvestris or Picea abies,” Comparative and Functional Genomics, Vol. 3, No. 4, 2002, pp. 306-318. doi:10.1002/cfg.199
- A. Sjödin, M. Bylesjö, O. Skogström, D. Eriksson, P. Nilsson, P. Rydén, S. Jansson and J. Karlsson, “UPSCBASE—Populus Transcriptomics Online,” Plant Journal, Vol. 48, No. 5, 2006, pp. 806-817. doi:10.1111/j.1365-313X.2006.02920.x
- P. Ryden, H. Andersson, M. Landfors, L. Naslund, B. Hartmanova, L. Noppa and A. Sjostedt, “Evaluation of Microarray Data Normalization Procedures Using SpikeIn Experiments,” BMC Bioinformatics, Vol. 7, No. 300, 2006.
- G. K. Smyth, “Limma: Linear Models for Microarray Data,” In: R. Gentleman, V. Carey, S. Dudoit, R. Irizarry and W. Huber, Eds., Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Springer, New York, 2005, pp. 397-420.
- G. K. Smyth and T. Speed, “Normalization of cDNA Microarray Data,” Methods, Vol. 31, No. 4, 2003, pp. 265- 273. doi:10.1016/S1046-2023(03)00155-5
- M. Bremer, E. Himelblau and A. Madlung, “Introduction to the Statistical Analysis of Two-Color Microarray Data,” Methods in Molecular Biology, Vol. 620, 2010, pp. 287-313. doi:10.1007/978-1-60761-580-4_9
- R Development Core Team, “R: A Language and Environment for Statistical Computing,” R Foundation for Statistical Computing, Vienna, 2006.
- [61] A. Conesa, S. Gotees, J. M. García-Gómez, J. Terol, M. Talon and M. Robles, “Blast2GO: A Universal Annotation and Visualization Tool in Functional Genomics Research. Application Note,” Bioinformatics, Vol. 21, 2005, pp. 3674-3676. doi:10.1093/bioinformatics/bti610
- [62] M. J. Burgin, J. J. Casal, G. C. Whitelam and R. A. Sanchez, “A Light-Regulated Pool of Phytochrome and Rudimentary High-Irradiance Responses under Far-Red Light in Pinus elliottii and Pseudotsuga menziesii,” Journal of Experimental Botany, Vol. 50, No. 335, 1999, pp. 831- 836. doi:10.1093/jexbot/50.335.831
- [63] P. Jarvis and J. Leverenz, “Productivity of Temperate, Decidous, and Evergreen Forests, in Physiological Plant Ecology, IV. Ecosystem Processes: Mineral Cycling, Productivity and Man’s Influence,” Springer, New York, 1983, pp. 223-280.
- [64] K. A. Franklin, V. S. Larner and G. C. Whitelam, “The Signal Transducing Photoreceptors of Plants,” International Journal of Developmental Biology, Vol. 49, No. 5-6, 2005, pp. 653-664. doi:10.1387/ijdb.051989kf
- [65] H. Mohr, “Lectures on Photomorphogenesis,” SpringerVerlag, Berlin, 1972. doi:10.1007/978-3-642-65418-3
- [66] K. M. Folta and E. P. Spalding, “Unexpected Roles for Cryptochrome 2 and Phototropin Revealed by HighResolution Analysis of Blue Light-Mediated Hypocotyl Growth Inhibition,” Plant Journal, Vol. 26, No. 5, 2001, pp. 471-478. doi:10.1046/j.1365-313x.2001.01038.x
- [67] B. Ehmann, B. Ocker and E. Schafer, “DevelopmentDependent and Light-Dependent Regulation of the Expression of Two Different Chalcone Synthase Transcripts in Mustard Cotyledons,” Planta, Vol. 183, No. 3, 1991, pp. 416-422. doi:10.1007/BF00197741
- [68] E. M. Tobin and D. M. Kehoe, “Phytochrome Regulated Gene Expression,” Seminars in Cell Biology, Vol. 5, No. 5, 1994, pp. 335-346. doi:10.1006/scel.1994.1040
- [69] R. Cerff, “Glyceraldehyde 3-Phosphate Dehydrogenases and Glyoxylate Reductase: I. Their Regulation under Continuous Red and far Red Light in the Cotyledons of Sinapis alba L.,” Plant Physiology, Vol. 51, No. 1, 1973, p. 76. doi:10.1104/pp.51.1.76
- [70] J. Dewdney, T. R. Conley, M. C. Shih and H. M. Goodman, “Effects of Blue and Red-Light on Expression of Nuclear Genes Encoding Chloroplast Glyceraldehyd-3- Phospate Dehydrogenase of Aradopsis thaliana,” Plant Physiology, Vol. 103, No. 4, 1993, pp. 1115-1121. doi:10.1104/pp.103.4.1115
- [71] D. Ernst, F. Pfeiffer, K. Schefbeck, C. Weyrauch and D. Oesterhelt, “Phytochrome Regulation of mRNA Levels of Ribulose-1,5-Bisphosphate Carboxylase in Etiolated Rye Seedlings (Secale cereale),” Plant Molecular Biology, Vol. 10, No. 1, 1987, pp. 21-33. doi:10.1007/BF00014183
- [72] Y. Sasaki, T. Sakihama, T. Kamikubo and K. Shinozaki, “Phytochrome-Mediated Regulation of Two mRNAs, Encoded by Nuclei and Chloroplasts of Ribulose-1,5- Bisphosphate Carboxylase Oxygenase,” European Journal of Biochemistry, Vol. 133, No. 3, 1983, pp. 617-620. doi:10.1111/j.1432-1033.1983.tb07507.x
- [73] B. K. Phee, J. I. Kim, D. H. Shin, J. Yoo, K. J. Park, Y. J. Han, Y. K. Kwon, M. H. Cho, J. S. Jeon, S. H. Bhoo and T. R. Hahn, “A Novel Protein Phosphatase Indirectly Regulates Phytochrome-Interacting Factor 3 via Phytochrome,” Biochemical Journal, Vol. 415, 2008, pp. 247- 255. doi:10.1042/BJ20071555
- [74] J. M. Tepperman, T. Zhu, H. S. Chang, X. Wang and P. H. Quail, “Multiple Transcription-Factor Genes Are Early Targets of Phytochrome A Signaling,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 16, 2001, pp. 9437-9442. doi:10.1073/pnas.161300998
- [75] M. Ahmad and A. R. Cashmore, “Hy4 Gene of A. thaliana Encodes a Protein with Characteristics of a BlueLight Photoreceptor,” Nature, Vol. 366, No. 6451, 1993, pp. 162-166. doi:10.1038/366162a0
- [76] M. Koornneef, E. Rolff and C. J. P. Spruit, “GeneticControl of Light-Inhibited Hypocotyl Elongation in Arabidopsis thaliana (L) Heynh,” Zeitschrift Fur Pflanzenphysiologie, Vol. 100, No. 2, 1980, pp. 147-160.
- [77] Y. J. Yang, Z. C. Zuo, X. Y. Zhao, X. Li, J. Klejnot, Y. Li, P. Chen, S. P. Liang, X. H. Yu, X. M. Liu and C. T. Lin, “Blue-Light-Independent Activity of Arabidopsis Cryptochromes in the Regulation of Steady-State Levels of Protein and mRNA Expression,” Molecular Plant, Vol. 1, No. 1, 2008, pp. 167-177. doi:10.1093/mp/ssm018
- [78] C. Lin, M. Ahmad, D. Gordon and A. R. Cashmore, “Expression of an Arabidopsis Cryptochrome Gene in Transgenic Tobacco Results in Hypersensitivity to Blue, UV-A, and Green Light,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 92, No. 18, 1995, pp. 8423-8427. doi:10.1073/pnas.92.18.8423
- [79] C. T. Lin, M. Ahmad and A. R. Cashmore, “Arabidopsis Cryptochrome 1 Is a Soluble Protein Mediating Blue LightDependent Regulation of Plant Growth and Development,” Plant Journal, Vol. 10, No. 5, 1996, pp. 893-902. doi:10.1046/j.1365-313X.1996.10050893.x
- [80] K. M. Folta, M. A. Pontin, G. Karlin-Neumann, R. Bottini and E. P. Spalding, “Genomic and Physiological Studies of Early Cryptochrome 1 Action Demonstrate Roles for Auxin and Gibberellin in the Control of Hypocotyl Growth by Blue Light,” Plant Journal, Vol. 36, No. 2, 2003, pp. 203-214. doi:10.1046/j.1365-313X.2003.01870.x
- [81] S. E. D. El-Assal, C. Alonso-Blanco, A. J. M. Peeters, V. Raz and M. Koornneef, “A QTL for Flowering Time in Arabidopsis Reveals a Novel Allele of CRY2,” Nature Genetics, Vol. 29, No. 4, 2001, pp. 435-440. doi:10.1038/ng767
- [82] C. Poppe, U. Sweere, H. Drumm-Herrel and E. Schafer, “The Blue Light Receptor Cryptochrome 1 Can Act Independently of Phytochrome A and B in Arabidopsis thaliana,” Plant Journal, Vol. 16, No. 4, 1998, pp. 465- 471. doi:10.1046/j.1365-313x.1998.00322.x
- [83] Y. J. Han, P. S. Song and J. I. Kim, “Phytochrome-MeDiated Photomorphogenesis in Plants,” Journal of Plant Biology, Vol. 50, No. 3, 2007, pp. 230-240. doi:10.1007/BF03030650
- [84] A. Nagatani, J. W. Reed and J. Chory, “Isolation and Initial Characterization of Arabidopsis Mutants That Are Deficient in Phytochrome-A,” Plant Physiology, Vol. 102, No. 1, 1993, pp. 269-277.
- [85] B. M. Parks and P. H. Quail, “Hy8, a New Class of Arabidopsis Long Hypocotyl Mutants Deficient in Functional Phytochrome-A,” Plant Cell, Vol. 5, No. 1, 1993, pp. 39- 48. doi:10.2307/3869426
- [86] G. C. Whitelam, E. Johnson, J. R. Peng, P. Carol, M. L. Anderson, J. S. Cowl and N. P. Harberd, “PhytochromeA Null Mutants of Arabidopsis Display a Wild-Type Phenotype in White-Light,” Plant Cell, Vol. 5, No. 7, 1993, pp. 757-768. doi:10.2307/3869613
- [87] A. Nagatani, J. Chory and M. Furuya, “Phytochrome-B Is Not Detectable in the Hy3 Mutant of Arabidopsis, Which Is Deficient in Responding to End-Of-Day Far-Red Light Treatments,” Plant and Cell Physiology, Vol. 32, No. 7, 1991, pp. 1119-1122.
- [88] J. W. Reed, P. Nagpal, D. S. Poole, M. Furuya and J. Chory, “Mutations in the Gene for the Red Far-Red Light Receptor Phytochrome-B Alter Cell Elongation and Physiological-Responses throughout Arabidopsis Development,” Plant Cell, Vol. 5, No. 2, 1993, pp. 147-157. doi:10.2307/3869581
- [89] M. Ahmad, J. A. Jarillo, O. Smirnova and A. R. Cashmore, “The CRY1 Blue Light Photoreceptor of Arabidopsis Interacts with Phytochrome A in Vitro,” Molecular Cell, Vol. 1, No. 7, 1998, pp. 939-948. doi:10.1016/S1097-2765(00)80094-5
- [90] M. Ahmad and A. R. Cashmore, “The Blue-Light Receptor Cryptochrome 1 Shows Functional Dependence on Phytochrome A or Phytochrome B in Arabidopsis thaliana,” Plant Journal, Vol. 11, No. 3, 1997, pp. 421-427. doi:10.1046/j.1365-313X.1997.11030421.x
- [91] M. Boylan, N. Douglas and P. H. Quail, “DominantNegative Suppression of Arabidopsis Photoresponses by Mutant Phytochrome-A Sequences Identifies Spatially Discrete Regulatory Domains in the Photoreceptor,” Plant Cell, Vol. 6, No. 3, 1994, pp. 449-460. doi:10.2307/3869764
- [92] K. Dehesh, C. Franci, B. M. Parks, K. A. Seeley, T. W. Short, J. M. Tepperman and P. H. Quail, “Arabidopsis Hy8 Locus Encodes Phytochrome-A,” Plant Cell, Vol. 5, No. 9, 1993, pp. 1081-1088. doi:10.2307/3869628
- [93] C. P. Romano, P. R. H. Robson, H. Smith, M. Estelle and H. Klee, “Transgene-Mediated Auxin Overproduction in Arabidopsis-Hypocotyl Elongation Phenotype and Interactions with the Hy6-1 Hypocotyl Elongation and Axr1 Auxin-Resistant Mutants,” Plant Molecular Biology, Vol. 27, No. 6, 1995, pp. 1071-1083. doi:10.1007/BF00020881
- [94] Y. Tao, J. L. Ferrer, K. Ljung, F. Pojer, F. X. Hong, J. A. Long, L. Li, J. E. Moreno, M. E. Bowman, L. J. Ivans, Y. F. Cheng, J. Lim, Y. D. Zhao, C. L. Ballare, G. Sandberg, J. P. Noel and J. Chory, “Rapid Synthesis of Auxin via a New Tryptophan-Dependent Pathway Is Required for Shade Avoidance in Plants,” Cell, Vol. 133, No. 1, 2008, pp. 164-176. doi:10.1016/j.cell.2008.01.049
- [95] E. L. Singsaas, M. M. Laporte, J. Z. Shi, R. K. Monson, D. R. Bowling, K. Johnson, M. Lerdau, A. Jasentuliytana and T. D. Sharkey, “Kinetics of Leaf Temperature Fluctuation Affect Isoprene Emission from Red Oak (Quercus rubra) Leaves,” Tree Physiology, Vol. 19, No. 14, 1999, pp. 917-924. doi:10.1093/treephys/19.14.917
- [96] M. R. Garcia-Gil, “Evolutionary Aspects of Functional and Pseudogene Members of the Phytochrome Gene Family in Scots Pine,” Journal of Molecular Evolution, Vol. 67, No. 2, 2008, pp. 222-232. doi:10.1007/s00239-008-9135-z
- [97] R. A. Cañas, F. de la Torre, F. M. Canovas and F. R. Canton, “High Levels of Asparagine Synthetase in Hypocotyls of Pine Seedlings Suggest a Role of the Enzyme in Re-Allocation of Seed-Stored Nitrogen,” Planta, Vol. 224, No. 1, 2006, pp. 83-95. doi:10.1007/s00425-005-0196-6
- [98] R. A. Cañas, F. de la Torre, F. M. Canovas and F. R. Canton, “Coordination of PsAS1 and PsASPG Expression Controls Timing of Re-Allocated N Utilization in Hypocotyls of Pine Seedlings,” Planta, Vol. 225, No. 5, 2007, pp. 1205-1219. doi:10.1007/s00425-006-0431-9
- [99] A. W. Galston, “Polyamines as Modulators of Plant Development,” Bioscience, Vol. 33, No. 6, 1983, pp. 382- 388. doi:10.2307/1309107
- [100] E. P. Lorences and I. Zarra, “Hypocotyl Growth of Pinus pinaster Seedlings—Changes in Osmotic Potential and Cell-Wall Composition,” Physiologia Plantarum, Vol. 67, No. 3, 1986, pp. 377-382. doi:10.1111/j.1399-3054.1986.tb05751.x
- [101] J. Pedreira, N. Sanz, M. J. Pena, M. Sanchez, E. Queijeiro, G. Revilla and I. Zarra, “Role of Apoplastic Ascorbate and Hydrogen Peroxide in the Control of Cell Growth in Pine Hypocotyls,” Plant and Cell Physiology, Vol. 45, No. 5, 2004, pp. 530-534. doi:10.1093/pcp/pch059
- [102] K. Mitrakos, “The Participation of Red Far-Red Reaction System in Chlorophyll-Metabolism,” Physiologia Plantarum, Vol. 14, No. 3, 1961, pp. 497-503. doi:10.1111/j.1399-3054.1961.tb07908.x
- [103] C. Shichijo and T. Hashimoto, “A Red Light Signal Distinct from the Far-Red-Absorbing form of Phytochrome in Anthocyanin Induction of Sorghum Bicolor,” Journal of Photochemistry and Photobiology B-Biology, Vol. 38, No. 1, 1997, pp. 70-75. doi:10.1016/S1011-1344(96)07418-0
- [104] Y. Zhou and B. R. Singh, “Red Light Stimulates Flowering and Anthocyanin Biosynthesis in American Cranberry,” Plant Growth Regulation, Vol. 38, No. 2, 2002, pp. 165-171. doi:10.1023/A:1021322418740
- [105] U. Bechtold, N. R. Leyland and P. M. Mullineaux, “Does Elevated Foliar Glutathione Alter Responses to Biotic and Abiotic Stress?” Free Radical Research, Vol. 37, 2003, pp. 20-20.
- [106] M. Kimura, Y. Y. Yamamoto, M. Seki, T. Sakurai, M. Sato, T. Abe, S. Yoshida, K. Manabe, K. Shinozaki and M. Matsui, “Identification of Arabidopsis Genes Regulated by High Light-Stress Using cDNA Microarray,” Photochemistry and Photobiology, Vol. 77, No. 2, 2003, pp. 226-233. doi:10.1562/0031-8655(2003)0770226IOAGRB2.0.CO2
- [107] C. H. Foyer and G. Noctor, “Redox Sensing and Signalling Associated with Reactive Oxygen in Chloroplasts, Peroxisomes and Mitochondria,” Physiologia Plantarum, Vol. 119, No. 3, 2003, pp. 355-364. doi:10.1034/j.1399-3054.2003.00223.x
- [108] S. Karpinski, C. Escobar, B. Karpinska, G. Creissen and P. M. Mullineaux, “Photosynthetic Electron Transport Regulates the Expression of Cytosolic Ascorbate Peroxidase Genes in Arabidopsis during Excess Light Stress,” Plant Cell, Vol. 9, No. 4, 1997, pp. 627-640. doi:10.2307/3870512
- [109] T. Maruta, A. Tanouchi, M. Tamoi, Y. Yabuta, K. Yoshimura, T. Ishikawa and S. Shigeoka, “Arabidopsis Chloroplastic Ascorbate Peroxidase Isoenzymes Play a Dual Role in Photoprotection and Gene Regulation under Photo-Oxidative Stress,” Plant and Cell Physiology, Vol. 51, No. 2, 2010, pp. 190-200. doi:10.1093/pcp/pcp177
- [110] M. L. Molas, J. Z. Kiss and M. J. Correll, “Gene Profiling of the Red Light Signalling Pathways in Roots,” Journal of Experimental Botany, Vol. 57, No. 12, 2006, pp. 3217- 3229. doi:10.1093/jxb/erl086
- [111] D. W. Russell and A. W. Galston, “Flavonoid Complexes in Pisum sativum. 4. Effect of Red Light on Synthesis of Kaempferol Complexes and on Growth in Sub-Apical Internode Tissues,” Phytochemistry, Vol. 6, No. 6, 1967, pp. 791-797. doi:10.1016/S0031-9422(00)86024-0
- [112] B. Michalczuk and R. M. Rudnicki, “The Effect of Monochromatic Red-Light on Ethylene Production in Leaves of Impatiens balsamina L. and Other Species,” Plant Growth Regulation, Vol. 13, No. 2, 1993, pp. 125-131. doi:10.1007/BF00024254
- [113] C. L. Steed, L. K. Taylor and M. A. Harrison, “Red Fight Regulation of Ethylene Biosynthesis and Gravitropism in Etiolated Pea Stems,” Plant Growth Regulation, Vol. 43, No. 2, 2004, pp. 117-125. doi:10.1023/B:GROW.0000040116.10016.c3
- [114] W. R. Briggs, “Red Light, Auxin Relationships, and Phototropic Responses of Corn and Oat Coleoptiles,” American Journal of Botany, Vol. 50, No. 2, 1963, pp. 196-207. doi:10.2307/2439853
- [115] M. Iino, “Inhibitory-Action of Red-Light on the Growth of the Maize Mesocotyl—Evaluation of the Auxin Hypothesis,” Planta, Vol. 156, No. 5, 1982, pp. 388-395. doi:10.1007/BF00393308
- [116] A. M. Jones, D. S. Cochran, P. M. Lamerson, M. L. Evans and J. D. Cohen, “Red Light-Regulated Growth. 1. Changes in the Abundance of Indoleacetic-Acid and a 22-Kilodalton Auxin-Binding Protein in the Maize Mesocotyl,” Plant Physiology, Vol. 97, No. 1, 1991, pp. 352-358. doi:10.1104/pp.97.1.352
- [117] D. J. Tucker, “Effects of Far-Red Light on Lateral Bud Outgrowth in Decapitated Tomato Plants and Associated Changes in Levels of Auxin and Abscisic-Acid,” Plant Science Letters, Vol. 8, No. 4, 1977, pp. 339-344. doi:10.1016/0304-4211(77)90152-3
- [118] F. D. Beall, E. C. Yeung and R. P. Pharis, “Far-Red Light Stimulates Internode Elongation, Cell Division, Cell Elongation, and Gibberellin Levels in Bean,” Canadian Journal of Botany-Revue Canadienne De Botanique, Vol. 74, No. 5, 1996, pp. 743-752. doi:10.1139/b96-093
NOTES
*Corresponding author.

