Crystal Structure Theory and Applications
Vol.1 No.2(2012), Article ID:23024,8 pages DOI:10.4236/csta.2012.12002
Homology Modelling and Structural Comparisons of Capsid-Associated Proteins from Circoviruses Reveal Important Virus-Specific Surface Antigens
1School of Animal and Veterinary Sciences, Charles Sturt University, Wagga Wagga, Australia
2School of Biomedical Sciences, Charles Sturt University, Wagga Wagga, Australia
Email: *shraidal@csu.edu.au
Received July 3, 2012; revised August 6, 2012; accepted August 15, 2012
Keywords: Circovirus; Capsid-Associated Protein; Structure Homology; Cap; Modelling
ABSTRACT
Circoviridae represent a growing family of small animal viruses. Some of these viruses have veterinary and medical importance, although, a vast amount of these newly discovered viruses have unknown effects on their hosts. The capsid-associated protein (Cap) of circoviruses is of interest because of its role in viral structure, immune evasion, host cell entry, and nuclear shuttling of viral components. The structure of the porcine circovirus 2 (PCV2) Cap has been solved and offered insight to these functions. Based on the crystallographic PCV2 Cap structure, models from circoviruses isolated from avian, fish, and mammalian hosts have been constructed and analyzed to better understand the roles of these proteins in the virus family. A high degree of conservation is observed in the models, however, the surface antigens differ among viruses. This is likely a reflection of the small genome harbored by circoviruses, and therefore the requirement of their few proteins to carry out specific vital functions, while maintaining enough variation to successfully infect their hosts. Here we describe the putative structures of a range of Cap proteins from circoviruses based on the crystallographic determination of porcine Cap, identifying key regions for function and inhibition of crystal formation.
1. Introduction
Members of the family Circoviridae all have small, circular, single-stranded DNA (ssDNA) genomes, surrounded by small, nonenveloped, icosahedral capsids [1]. Three genera and a recently proposed fourth genus exist in the Circoviridae family. The genus Circovirus officially contains porcine circovirus 1 (PCV1) and 2 (PCV2), beak and feather disease virus (BFDV), goose circovirus (GoCV), canary circovirus (CaCV), and columbid circovirus (CoCV) [2]. Although not officially classified, many circovirus-like viruses are known to infect several other avian species, barbell, catfish, bat, and dog [3-11] . New viral discoveries have led to a recently proposed fourth genus, Cyclovirus, which is closely related to Circovirus and is represented by small circular viruses isolated from feces of mammals, including chimpanzees, humans, and bats [10,12] . Some of these viruses have been known to have pathological effects on their host, while others have not been connected with any disease.
As few proteins are encoded in the viral genomes, these viruses are a model of efficiency and must use host cell machinery to replicate [13]. Two major proteins are characterized in circoviruses and circovirus-like viruses; these are the replication-associated protein (Rep), which is involved in the replication of the virus, and the capsid-associated protein (Cap), which is the structural component of the viral capsid. Cap is the main antigenic protein of circoviruses because the protein uses repeated subunits to compose the entire capsid structure of the virus. In PCV and BFDV, 60 repeating Cap subunits are used to make up the capsid [1].
The Cap sequence is highly variable, likely an important aspect to evasion of the host immune system. Amino acid identities in the Cap sequence of BFDV range from 73% - 99% [14,15]. The Cap protein is a single domain protein, but has several features that pertain to its functions. The N-terminus of the protein is positively charged and may contain several conspicuous nuclear localizing signals (NLS). A second significant function of Cap is its ability to bind DNA [16]. Affinity of Cap to DNA is a vital function because this interaction allows the viral genome to gain access to the host cell nucleus. Repbinding capability may also be an important role for Cap.
This interaction is specifically interesting because of discrepancies between circoviruses. Rep proteins of PCV possess NLSs and can enter the nucleus without Cap [17], however, in BFDV, Rep must bind to Cap for nuclear transport [16].
In 2011 [18] the crystal structure of PCV2 Cap was determined. To crystallize PCV2 Cap, 40 amino acids were omitted from the N-terminal, which contains the NLS, however, the structure placed the NLS inside the capsid [18]. The structural data provided important information with respect to the antigenic characteristics of this virus, where predictions about PCV epitopes had either been on the internalized surface and, therefore, not immunogenically relevant or were composed of consecutive amino acids [19,20]. The structure of Cap has illuminated epitopes for monoclonal antibody (MAb) binding and has been useful to compare variation in binding of MAbs to PCV2 and PCV1. Small mutations, sometimes involving only a single amino acid substitution, were attributed to specific differences in MAb binding between the two porcine viruses [18]. This highlights the selective pressure to employ mutations as a means to elude the host immune system, and can be related to the publication from Kundu et al. [21], showing that viruses infecting the same population may have diverse biological fitness due to genetic mutations that translate into slight amino acid variation. Specifically, Cap must successfully evade the host immune system, attach to a surface receptor on a cell that is appropriate for infection, bind to its own genome (and viral proteins) for localization into the nucleus where replication can occur, all while undergoing additional mutations, which select for optimal antigenicity and must not diminish critical function. These characteristics lead to the hypothesis that although variation among circoviruses and related viruses must be sufficiently different to infect their respective hosts and escape immune detection, they must have the same underlying structures, which allow them to carry out a core and critical set of functions.
Similarities between the amino acid sequences of circovirus Cap proteins and the solved PCV2 Cap also allow for models to be constructed. The differences among strains and between viruses may have different effects when comparing their structural attributes. The rapidly growing number of circoviruses and circovirus-like viruses being discovered in a diverse range of hosts begs the question of their structural relatedness [22]. Conserved regions among these viruses would be helpful for development of diagnostic and treatment strategies, and the recognition of new viral discoveries. Models for circoviruses infecting a variety of hosts were analyzed based on the structural data from PCV2 Cap. It was observed that changes in Cap sequences among the analyzed circovirus isolates do not translate into major structural differences of the proteins, and likely, no major structural changes in the viral capsid, however, key differences were observed in antigenic regions and are discussed within.
2. Materials and Methods
2.1. Modelling of Circovirus Capsid-Associated Proteins
The structure of PCV2 Cap monomer and viral capsid were retrieved from the RCSB Protein Data Bank (PDB ID: 3R0R). The recombinant PCV2 Cap protein that was crystallized did not contain the NLS; rather it lacked the first 40 N-terminal residues, and contained 193 PCV2 Cap residues [18]. Several circovirus Cap sequences from a range of hosts were retrieved from the National Center for Biotechnology Information (NCBI, U.S. National Library for Medicine). These sequences are outlined in Table 1. Models were created using SWISSMODEL (Swiss Institute of Bioinformatics) [23] and viewed with The PyMOL Molecular Graphics System, Version 1.3 (Schrodinger, LLC.). Alignment of the circovirus Cap sequences was performed with Clustal W2 (EMBL-EBI) [24]. A phylogenetic tree was made using MAFFT (CBRC) [25] and viewed using Archaeopteryx (phylosoft) [26].
3. Results
3.1. Molecular Replacement Models of Circovirus Caps and Using PCV2 Capsid Structure
The Cap sequences of related circoviruses varied in length from 214 to 273 amino acids, and shared between 23% - 32% identity and 35% - 46% similarity with the PCV2 Cap sequence (Table 1). The canine circovirus (CaCV-1), the most recent circovirus sequence to be discovered, is the least similar to PCV2 Cap, a strange diversity when considering the mammalian hosts of PCV2 and CaCV-1. The barbel (BaCV) and duck circovirus (DuCV) Cap sequences share the closest similarity to PCV2 Cap. Interestingly, the two circovirus sequences isolated from fish inhabit the opposite ends of the spectrum; the Silurus glanis circovirus (CfCV) being marginally more similar to PCV2 than CaCV-1. The isolates from mammalian hosts, CaCV-1 and Chimpanzee Stool avian-like circovirus-chimp17 (CsaCV-chimp17), also have major differences, although the CsaCV-chimp17 sequence has been likened to the raven circovirus (RaCV) so it is not surprising that its similarity to PCV2 fits appropriately with other avian isolates. All avian circoviruses compared in this study have over 40% similarity with PCV2 Cap.
The internal jelly roll structure described in PCV2 Cap is conserved among all models constructed of related circoviruses (Figure 1). This jelly roll is formed by seven to eight b-sheets, depending on the model. Although some surface characteristics are altered in each data set, there are still several b-sheets making up the core of all the selected viral Caps. All N-termini appear to be facing internally, and therefore, the NLS portions of all Cap structures likely remain inside the viral capsid.
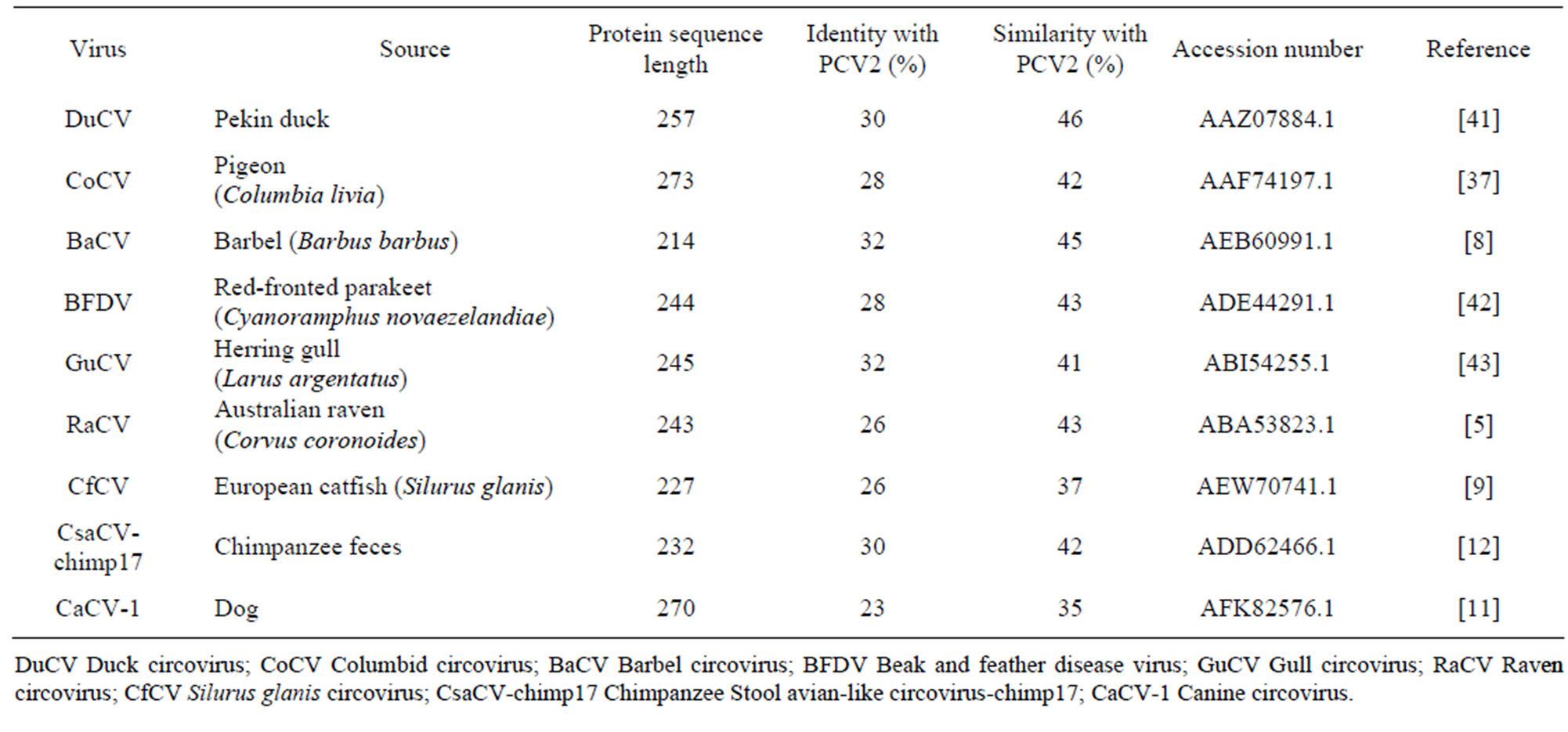
Table 1. Summary of the Circoviruses viewed and modelled, sources of virus extraction, Cap amino acid sequence length, and amino acid identity and similarity to PCV2 Cap.
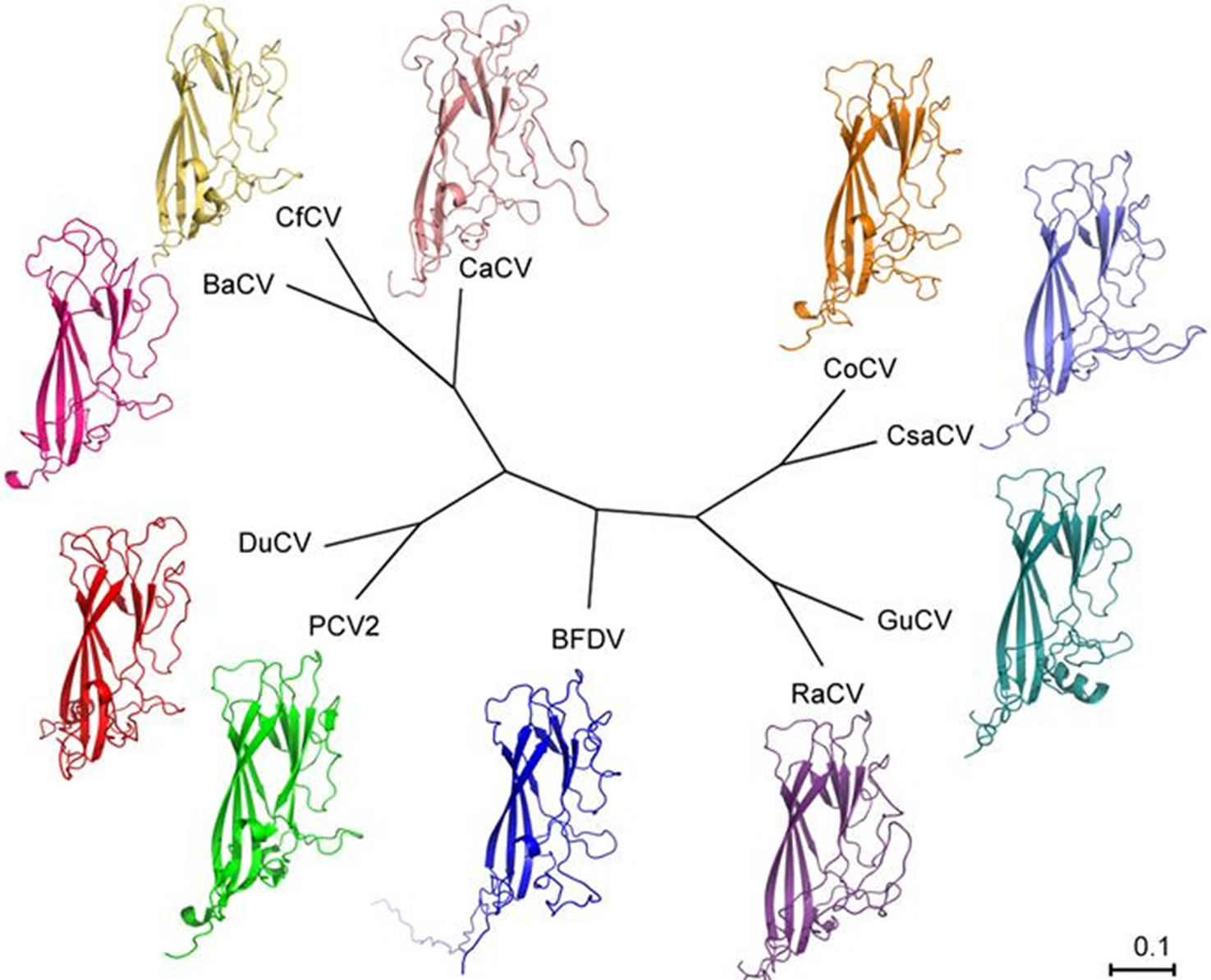
Figure 1. Unrooted phylogenetic tree showing the relationship between the amino acid sequences of the analyzed circovirus Caps. The accession numbers for each sequence are: CoCV AAF74197.1, Chimpanzee Stool avail-like circovirus-chimp17 (referred to as CsaCV in the figure) ADD62466.1, GuCV ABI54255.1, RaCV ABA53823.1, BFDV ADE44291.1, PCV2 3ROR (PDB ID number), DuCV AAZ07884.1, BaCV AEB60991.1, CfCV AEW70741.1, and canine circovirus (referred to as CaCV in the figure) AFK82576.1.
3.2. Beak and Feather Disease Virus
A full-length BFDV Cap sequence isolated from a redfronted parakeet (Cyanoramphus novaezelandiae) was superimposed on the PCV2 Cap structure to view the putative positioning of the NLS in the virus. The model of the full-length BFDV Cap structure contained one α-helix, and eight β-sheets. Of the four epitopes, B and C showed very little change in external positioning. Epitope A shows differences as the PCV2 Cap structure has a small β-sheet while the putative full-length BFDV Cap has another turn on its strand. Epitope E shows major changes with many additional amino acids prior to the PCV2 Cap epitope causing the strand to protrude. The NLS was expected to extend into the inside of the viral capsid (Figure 2).
3.3. Duck Circovirus
The DuCV Cap model has few deviations from the PCV2 Cap structure, with even less structural variability on the external surface (Figure 3). The model contains two ahelices and eight b-sheets. In the region homologous to PCV2 Cap residues between surface epitopes A and B the most striking difference appears. Several additional residues on the DuCV Cap sequence are organized into a a-helix. This landmark protrudes from the structure, however, it corresponds to the internal surface of the capsid, negating its antigenic relevance. Other differences attain similarities from the BFDV Cap model. These include the lack of a b-sheet corresponding to the latter portion of epitope A on PCV2 Cap, and a more modest protrusion prior to epitope E, consisting of seven additional residues, rather than the 13 seen in the BFDV Cap sequence. The DuCV Cap is the only avian-related virus among those analyzed to not contain a significant portion of sequential identical residues in epitope E. A pore, due to a tunnel in the DuCV Cap protein, would extend from the external surface to the internal surface of the viral capsid.
3.4. Columbid Circovirus
The CoCV Cap model displays structures similar to an average compromise between PCV2 Cap and BFDV Cap. The similarity to PCV2 Cap leaves very little of the CoCV Cap to extend beyond the surface of the porcine virus structure. Not surprisingly, the differences are the most pronounced prior to the region homologous to epitope E. Here, the additional residues are split into two protrusions, with the median portion returning to similarity with the PCV2 Cap structure. Three sequences which contain several consecutive amino acids that are present in most of the avian-related circoviruses in this study are present in this area, WIPL183-186, which is part of the me dian portion that rejoins the PCV2 Cap strand’s trajectory, and HYGLAFS200-206 and PQP210-212, which are
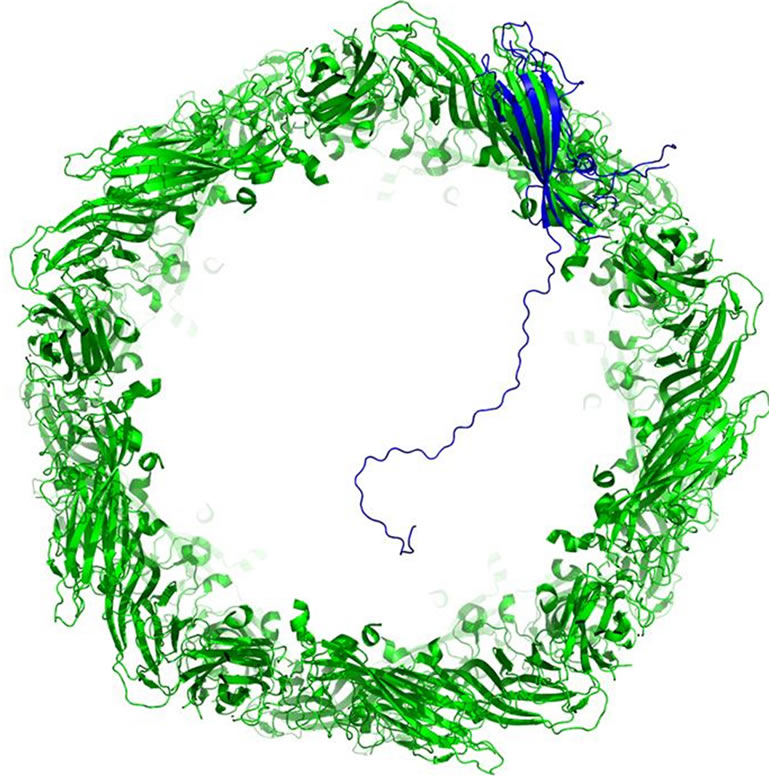
Figure 2. Cross-section of viral capsid of PCV2 composed of 60 units of PCV2 Cap (green). Full-length BFDV Cap is overlain one of the PCV2 Cap molecules to view the position of the NLS inside the viral capsid. The NLS extends well into the capsid cavity, likely interacting with the viral genome.
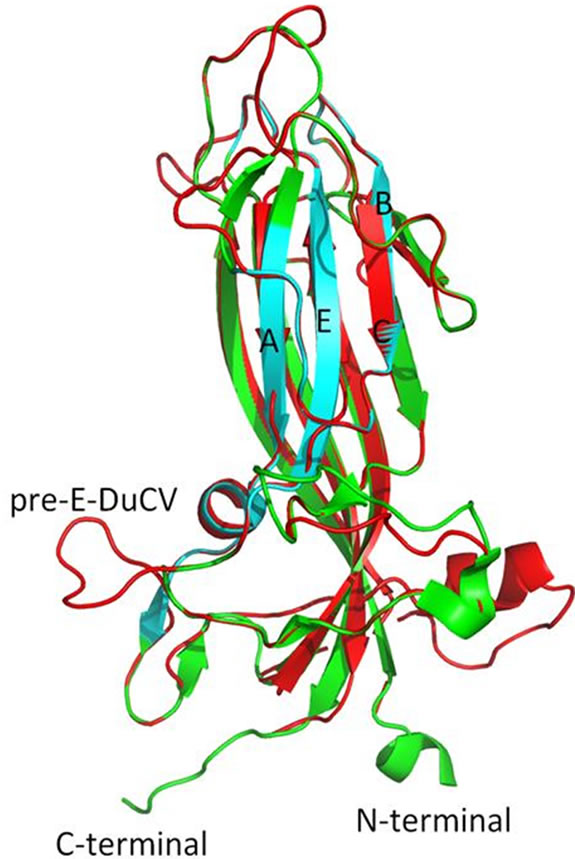
Figure 3. PCV2 Cap (green and cyan) overlain with DuCV (red). The cyan portions on the PCV2 Cap indicate amino acids presumed to be epitopes on the external capsid surface for MAb binding and are labelled A, B, C, and E according to Khayat et al. [18]. The protrusion on DuCV that includes residues prior to epitope E has been labeled pre-E-DuCV, and appears on the surface of the protein. The DuCV Cap sequence shares the highest amount of similarity to the PCV2 Cap sequence. The jelly roll structure and several other features remain conserved.
within epitope E. Interestingly, the WIPL sequence in the other models remains as part of the protrusion, not following the PCV2 Cap structure. The b-sheet corresponding to epitope A is also not present in CoCV Cap, along with other minor b-sheets seen in PCV2 Cap. CoCV Cap has a total of two a-helices and eight b-sheets. The model shows some small transprotein tunnels, although it is not clear whether these would allow the passage of particles into and out of the viral capsid.
3.5. Barbel Circovirus
The small sequence length of the BaCV Cap leaves little extraneous residues to form protruding or additional structures beyond that seen in the PCV2 Cap. The a-helix appearing at the N-terminal of the PCV2 Cap structure is maintained in the BaCV Cap model, however, no other a-helices are present. Eight b-sheets are seen in the BaCV Cap model. The region corresponding to epitope A shows neither a-helix nor b-sheet. The most prominent protrusion from BaCV Cap appears to occur on the internal surface of the viral capsid, which takes place of another a-helix, although a divergence is also present in the region corresponding to epitopes B and C due to the inclusion of additional residues. Several tunnels exist in the protein, at least one of which would create a pore through to the internal environment of the capsid structure.
3.6. Gull Circovirus
A set of eight b-sheets make up the core of the gull circovirus (GuCV) Cap model. The single a-helix present in the model is an extension of an existing, shorter, a-helix from PCV2 Cap. This a-helix appears on the lateral surface of the protein, and would interact with an adjoining Cap protein in the viral capsid. Not surprisingly, the portion of GuCV Cap prior to epitope E from PCV2 Cap contains additional residues, causing a minor protrusion. The region within epitope E also contains a sequence which matches with several avain circoviruses analyzed, HYGLAFS180-186. The GuCV Cap contains transprotein tunnels, creating pores that would extend from the external surface to the internal surface of the viral capsid.
3.7. Raven Circovirus
The RaCV shares some striking similarities to the BFDV Cap structures and sequences. The model contains a a-helix and eight b-sheets. The surface structure homologous to epitope E from PCV2 Cap includes a preceding protrusion consisting of 17 additional residues. This protrusion possesses the WIPL185-188 sequence seen at the beginning of the additional region, and the VKHYGLA203-209 and PQP213-215 sequences in epitope E, both sequences that are identical or similar to the regions from the BFDV Cap sequence. The region homologous to epitope A also lacks the a-helix and b-sheet. The protein has pores, although these may not extend from the external to the internal surface of the viral capsid.
3.8. Silurus Glanis Circovirus
The CfCV Cap sequence, like the BaCV Cap sequence, is much shorter than PCV2 Cap at 227 amino acids. This allows the PCV2 Cap model to extend beyond the borders of the CfCV Cap model in several places. The first turn in the PCV2 Cap model extends for 12 residues longer than the CfCV Cap, immediately prior to epitope A. CfCV Cap contains two a-helices that lie adjacent to each other, one in epitope A and the other prior to epitope E, and seven b-sheets. The a-helix prior to epitope E is located on the surface and creates a small protrusion in the same position as the protrusions seen in the other models.
3.9. Chimpanzee Stool Avian-Like Circovirus-Chimp17
Although the CsaCV-chimp17 virus was isolated from a mammalian host and contains the closest amount of residues to the full-length PCV2 Cap sequence, its name asserts its likeness to the avian circoviruses. The relation to avian circoviruses is evident in the structure and sequence of the model. The overall structure maintains a shape consistent with the avian virus models, however, it is the only model not to contain a a-helix. The CsaCVchimp17 Cap model contains eight b-sheets, like the avian Cap models, and has a large protrusion prior to epitope E. The protrusion begins with the WIPL149-152 sequence seen in the BFDV, CoCV and RaCV Caps, as well as the sequence HYGLAFS166-172 as part of epitope E, which is seen in CoCV Cap, GuCV Cap and the BFDV Cap sequences that were analyzed.
3.10. Canine Circovirus
The CaCV-1 Cap is the least alike in sequence identity and similarity to PCV2 Cap; this is evident in the structure as well (Figure 4). The model includes one α-helix and seven β-sheets. Several stretches of non-homologous residues in CaCV-1 Cap result in additional features. The most conspicuous feature is a very large protrusion that extends from the external surface following the portion of the protein homologous with epitope C. This protrusion extends for 26 amino acids. Other protrusions from CaCV-1 Cap occur also occur on the external surface or on interfaces, which would articulate with other Cap proteins to form the viral capsid. No additional protrusions are present on the internal capsid surface. The model predicts the viral capsid structure to be porous, with multiple tunnels extending through the protein, which would reach from the external surface to the internal surface of the viral capsid.
4. Discussion
Models were made and evaluated for eight related cir-
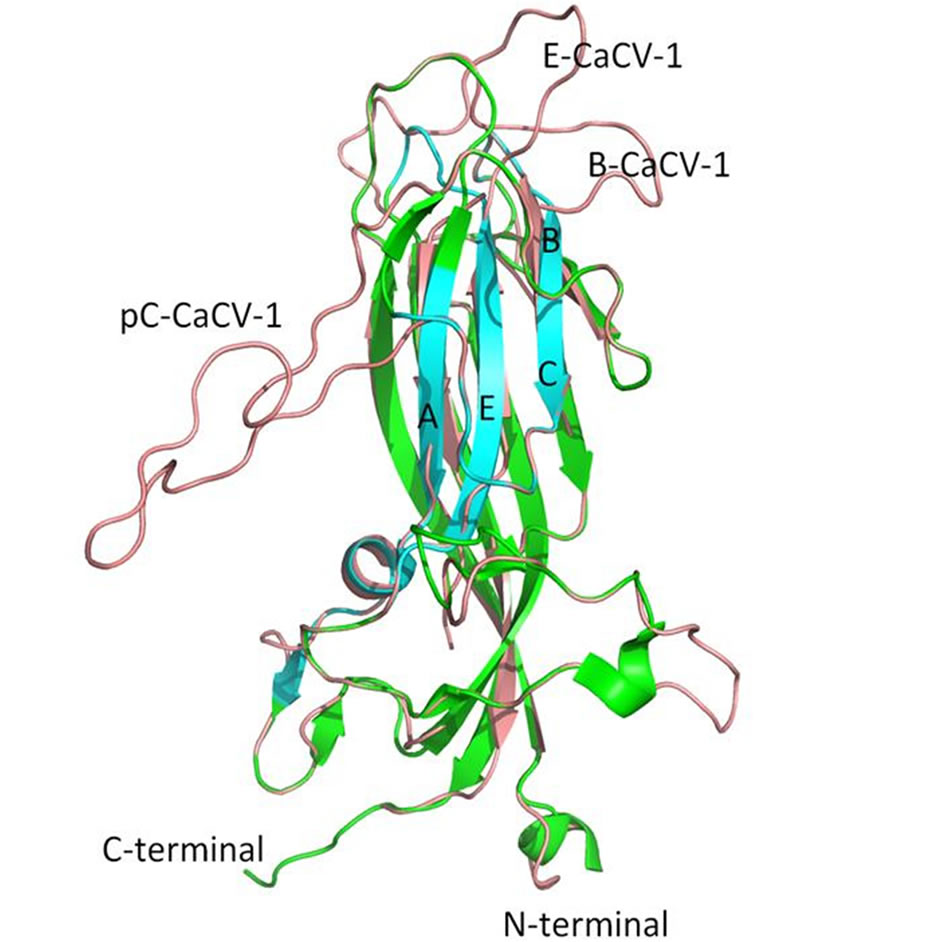
Figure 4. PCV2 Cap (green and cyan) overlain with CaCV-1 Cap model (pink). The CaCV-1 Cap model has the most discrepancies with PCV2 Cap. The regions pertaining to epitopes B and E from PCV2 Cap are labeled B-CaCV-1 and E-CaCV-1, respectively on the CaCV-1 Cap model. A large protrusion on the surface of CaCV-1 can also be seen on the post-epitope C region and is labeled pC-CaCV-1. The internal jelly roll structure, however, remains conserved.
coviruses, four from avian hosts, two from fishes, and two isolated from mammalian hosts. The structures for the models, along with sequence similarities show the relatedness of the avian circoviruses. Classifications between the other viruses, however, do not necessarily follow the same lines as their hosts. The publications reporting the viruses from non-avian hosts provide insight to the differences seen in these isolates. The discovery of BaCV and CfCV resulted in a new group of fish circoviruses, which are separated from circoviruses of other hosts [8,9]. Although BaCV is the closest related virus to CfCV, their Cap sequences only share 28% identity and 46% similarity so their structures need not be similar. Their discrepancy for similarity with PCV2 Cap is not surprising either, as phylogenetic analysis of amino acid sequence places BaCV closer than CfCV to PCV2 (Figure 1) [9]. The viruses isolated from mammalian hosts also differ greatly, however this is evident from their nomenclature. The CsaCV-chimp17 isolate was shown to relate to avian circoviruses more closely than other clusters (Figure 1) [12]. Meanwhile, the little data that exists on the newly discovered CaCV-1 classifies the canine virus as a separate species, as it shows minimal identity to other circoviruses [11]. Although both structures show distinct characteristics for both mammalian-isolated circoviruses, it can be seen that CsaCV-chimp17 resembles avian models with its eight b-sheets.
The avian circoviruses share many characteristics, including the previously mentioned eight b-sheets, and distinct surface amino acid sequences. The sequences WIPL, HYGLAFS, and PQP occur in several of the analyzed avian circovirus sequences, which corresponds to residues of epitope E of PCV2 Cap and the residues immediately preceding. Since these conserved regions are on the surface and are found in protrusions, the sequences may be a suitable antigen for MAbs that can be used to target a wide range of strains among a species of circoviruses, or possibly as a general diagnostic tool to cross-react with a range of viruses that infect different hosts. The ability of MAbs to cross-react with related viruses has been show to have neutralizing activity, and is of great interest as a diagnostic and characterization tool for many viral pathogens [27-29]. The location of the conserved sequences on the surface, as well as their close proximity may also select for MAbs that recognize linear epitopes, making them more robust for experimentation and diagnostics.
The overall structure of all circoviruses examined in this paper showed a high degree of similarity. Several studies have noted the high variation in circovirus sequences, especially those contributing to the capsid protein [14,15,30-37]. With the high diversity, even among isolates and strains from the same virus, structural changes may be anticipated. Many surface antigens will change, however, despite the diversity the configuration of the b-sheets at the centre of the structure was consistent throughout all models, creating a jelly roll formation as reported for PCV2 Cap and seen in other icosahedral viruses [18,38,39]. Even with PCV2 Cap exhibiting nearly twice as many b-sheets in the structure as CfCV and CaCV-1, the internal structure is still maintained. The presence of a-helices in the viruses may not be as significant due to their variation among closely related isolates and their generally small size. Long strands, containing the NLS would be seen on the N-terminal of all Cap proteins.
It was noted that all N-termini of the circoviruses viewed were directed internally into the viral capsid. The location of the NLS may also signify a function within the viral capsid. Citing proposed functions for similar Ntermini found in PCV and CAV which are used in packaging the viral genome into the capsid [13], the NLS on BFDV Cap was long assumed to be confined to the inside of the capsid in order to carry out this role [40]. Although the high concentration of positively charged arginines prevents a definitive structure from being determined without direct structural analysis of the full-length protein, PCV2 Cap structural data, even with a truncated NLS, is enough to show that the N-terminal of the protein is inward facing for all Cap models viewed. The NLS region was also observed binding DNA [16], giving credence to the claim that the N-terminal in Cap is responsible for viral DNA packaging in the capsid.
Since many changes occur on the external surface and do not affect the interaction with other Cap proteins, the viral capsid of all circoviruses likely share a high level of structural similarity. To continue to expand the structural knowledge of circovirus Caps, crystallographic or NMR data will need to be attained. It would be preferential to use the full-length protein, however, the NLS region has been shown to inhibit expression in E. coli [40]. Additionally, the long, positively charged NLS, characteristic to circovirus Caps, is predicted to extend outward from the tertiary structure of the protein creating a flexible region that would likely inhibit packing of a crystal for crystallographic determination. Although the size of these proteins is suitable for NMR determination difficulties arise with the high salt concentration required to purify and solubilize circovirus Caps, as well as its lack of solubility in the pH range compatible with NMR. Nevertheless, additional sequences of circovirus Cap, full-length and fragmented, should be expressed in an attempt to optimize for crystallization or NMR to continue to add knowledge to this rapidly growing family of viruses.
5. Acknowledgements
This research was supported by the Australian Research Council’s Discovery Projects funding scheme.
REFERENCES
- R. A. Crowther, et al., “Comparison of the Structures of Three Circoviruses: Chicken Anemia Virus, Porcine Circovirus Type 2, and Beak and Feather Disease Virus,” Journal of Virology, Vol. 77, No. 24, 2003, pp. 13036- 13041. doi:10.1128/JVI.77.24.13036-13041.2003
- C. M. Fauquet, et al., “Virus Taxonomy: The Eighth Report of the International Committee on Taxonomy of Viruses,” Academic Press, London, 2005.
- K. Hattermann, et al., “Cloning and Sequencing of Duck Circovirus (DuCV),” Archives of Virology, Vol. 148, No. 12, 2003, pp. 2471-2480. doi:10.1007/s00705-003-0181-y
- R. Johne, et al., “Genome of a Novel Circovirus of Starlings, Amplified by Randomly Primed Rolling-Circle Amplification,” Journal of General Virology, Vol. 87, No. 5, pp. 1189-1195. doi:10.1099/vir.0.81561-0
- M. Stewart, R. A. Perry and S. R. Raidal, “Identification of a Novel Circovirus in Australian Ravens (Corvus coronoides) with Feather Disease,” Avian Pathology, Vol. 35, No. 2, 2006, pp. 86-92. doi:10.1080/03079450600597345
- D. Todd, et al., “Molecular Characterization of Novel Circoviruses from Finch and Gull,” Avian Pathology, Vol. 36, No. 1, 2007, pp. 75-81. doi:10.1080/03079450601113654
- M. Y. Halami, et al., “Detection of a Novel Circovirus in Mute Swans (Cygnus olor) by Using Nested BroadSpectrum PCR,” Virus Research, Vol. 132, No. 1-2, 2008, pp. 208-212. doi:10.1016/j.virusres.2007.11.001
- M. Lorincz, et al., “First Detection and Analysis of a Fish Circovirus,” Journal of General Virology, Vol. 92, No. 8, 2011, pp. 1817-1821. doi:10.1099/vir.0.031344-0
- M. Lőrincz, et al., “Novel Circovirus in European Catfish (Silurus glanis),” Archives of Virology, Vol. 157, No. 6, 2012, pp. 1173-1176. doi:10.1007/s00705-012-1291-1
- X. Y. Ge, et al., “Genetic Diversity of Novel Circular Ssdna Viruses in Bats in China,” Journal of General Virology, Vol. 92, No. 11, 2011, pp. 2646-2653. doi:10.1099/vir.0.034108-0
- A. Kapoor, et al., “Complete Genome Sequence of the First Canine Circovirus,” Journal of Virology, Vol. 86, No. 12, 2012, pp. 7018-7018. doi:10.1128/JVI.00791-12
- L. Li, et al., “Multiple Diverse Circoviruses Infect Farm Animals and Are Commonly Found in Human and Chimpanzee Feces,” Journal of Virology, Vol. 84, No. 4, 2010, pp. 1674-1682. doi:10.1128/JVI.02109-09
- D. Todd, “Circoviruses: Immunosuppressive Threats to Avian Species: A Review,” Avian Pathology, Vol. 29, No. 5, 2000, pp. 373-394. doi:10.1080/030794500750047126
- M. R. Bassami, et al., “Genetic Diversity of Beak and Feather Disease Virus Detected in Psittacine Species in Australia,” Virology, Vol. 279, No. 2, 2001, pp. 392-400. doi:10.1006/viro.2000.0847
- R. Raue, et al., “Nucleotide Sequence Analysis of a C1 Gene Fragment of Psittacine Beak and Feather Disease Virus Amplified by Real-Time Polymerase Chain Reaction Indicates a Possible Existence of Genotypes,” Avian Pathology, Vol. 33, No. 1, 2004, pp. 41-50. doi:10.1080/03079450310001636219
- L. Heath, A. L. Williamson and E. P. Rybicki, “The Capsid Protein of Beak and Feather Disease Virus Binds to the Viral DNA and Is Responsible for Transporting the Replication-Associated Protein into the Nucleus,” Journal of Virology, Vol. 80, No. 14, 2006, pp. 7219-7225. doi:10.1128/JVI.02559-05
- T. Finsterbusch, et al., “Analysis of the Subcellular Localization of the Proteins Rep, Rep’ and Cap of Porcine Circovirus Type 1,” Virology, Vol. 343, No. 1, 2005, pp. 36-46. doi:10.1016/j.virol.2005.08.021
- R. Khayat, et al., “The 2.3-Angstrom Structure of Porcine Circovirus 2,” Journal of Virology, Vol. 85, No. 21, 2011, p. 11542. doi:10.1128/JVI.05863-11
- S. B. Shang, et al., “Fine Mapping of Antigenic Epitopes on Capsid Proteins of Porcine Circovirus, and Antigenic Phenotype of Porcine Circovirus Type 2,” Molecular Immunology, Vol. 46, No. 3, 2009, pp. 327-334. doi:10.1016/j.molimm.2008.10.028
- D. Mahe, et al., “Differential Recognition of ORF2 Protein from Type 1 and Type 2 Porcine Circoviruses and Identification of Immunorelevant Epitopes,” Journal of General Virology, Vol. 81, No. 7, 2000, pp. 1815-1824.
- S. Kundu, et al., “Tracking Viral Evolution during a Disease Outbreak: The Rapid and Complete Selective Sweep of a Circovirus in the Endangered Echo Parakeet,” Journal of Virology, Vol. 86, No. 9, 2012, pp. 5221-5229. doi:10.1128/JVI.06504-11
- E. Delwart and L. Li, “Rapidly Expanding Genetic Diversity and Host Range of the Circoviridae Viral Family and Other Rep Encoding Small Circular SsDNA Genomes,” Virus Research, Vol. 164, No. 1-2, 2012, pp. 114-121. doi:10.1016/j.virusres.2011.11.021
- K. Arnold, et al., “The SWISS-MODEL Workspace: A Web-Based Environment for Protein Structure Homology Modeling,” Bioinformatics, Vol. 22, No. 2, 2006, pp. 195-201. doi:10.1093/bioinformatics/bti770
- M. A. Larkin, et al., “Clustal W and Clustal X Version 2.0,” Bioinformatics, Vol. 23, No. 21, 2007, pp. 2947- 2948. doi:10.1093/bioinformatics/btm404
- K. Katoh and H. Toh, “Recent Developments in the MAFFT Multiple Sequence Alignment Program,” Briefings in Bioinformatics, Vol. 9, No. 4, 2008, pp. 286-298. doi:10.1093/bib/bbn013
- M. V. Han and C. M. Zmasek, “PhyloXML: XML for Evolutionary Biology and Comparative Genomics,” BMC Bioinformatics, Vol. 10, No. 356, 2009.
- N. Kitamoto, et al., “Cross-Reactivity among Several Recombinant Calicivirus Virus-Like Particles (VLPs) with Monoclonal Antibodies Obtained from Mice Immunized Orally with One Type of VLP,” Journal of Clinical Microbiology, Vol. 40, No. 7, 2002, pp. 2459-2465. doi:10.1128/JCM.40.7.2459-2465.2002
- N. Kitamoto, et al., “Cross-Reactivity among Cowpox, Ectromelia and Vaccinia Viruses with Monoclonal Antibodies Recognizing Distinct Antigenic Determinants in A-Type Inclusion-Bodies,” Archives of Virology, Vol. 91, No. 3-4, 1986, pp. 357-366. doi:10.1007/BF01314294
- R. Z. Rizk, et al., “Reactivity Pattern of 92 Monoclonal Antibodies with 15 Human Papillomavirus Types,” Journal of General Virology, Vol. 89, No. 1, 2008, pp. 117- 129. doi:10.1099/vir.0.83145-0
- R. Larochelle, R. Magar and S. D’Allaire, “Genetic Characterization and Phylogenetic Analysis of Porcine Circovirus Type 2 (PCV2) Strains from Cases Presenting Various Clinical Conditions,” Virus Research, Vol. 90, No. 1-2, 2002, pp. 101-112. doi:10.1016/S0168-1702(02)00141-7
- M. Hess, A. Scope and U. Heincz, “Comparitive Sensitivity of Polymerase Chain Reaction Diagnosis of Psittacine Beak and Feather Disease on Feather Samples, Cloacal Swabs and Blood from Budgerigars (Melopsittacus undulates, Shaw 18005),” Avian Pathology, Vol. 33, No. 5, 2004, pp. 477-481. doi:10.1080/03079450400003619
- B. Khalesi, et al., “A Comparison of Haemagglutination, Haemagglutination Inhibition and PCR for the Detection of Psittacine Beak and Feather Disease Virus Infection and a Comparison of Isolates Obtained from Loriids,” Journal of General Virology, Vol. 86, No. 11, 2005, pp. 3039-3046. doi:10.1099/vir.0.81275-0
- M. Fenaux, et al., “Genetic Characterization of Type 2 Porcine Circovirus (PCV-2) from Pigs with Postweaning Multisystemic Wasting Syndrome in Different Geographic Regions of North America and Development of a Differential PCR-Restriction Fragment Length Polymorphism Assay to Detect and Differentiate between Infections with PCV-1 and PCV-2,” Journal of Clinical Microbiology, Vol. 38, No. 7, 2000, pp. 2494-2503.
- G. H. Fu, et al., “Genetic Diversity and Genotype Analysis of Duck Circovirus,” Avian Diseases, Vol. 55, No. 2, 2011, pp. 311-318. doi:10.1637/9584-102010-ResNote.1
- D. Wang, et al., “Detection of Duck Circovirus in China: A Proposal on Genotype Classification,” Veterinary Microbiology, Vol. 147, No. 3-4, 2011, pp. 410-415. doi:10.1016/j.vetmic.2010.07.014
- Z. C. Zhang, et al., “Molecular Characterization and Epidemiological Investigation of Pigeon Circovirus Isolated in Eastern China,” Journal of Veterinary Diagnostic Investigation, Vol. 23, No. 4, 2011, pp. 665-672. doi:10.1177/1040638711407878
- A. Mankertz, et al., “Cloning and Sequencing of Columbid Circovirus (CoCV), a New Circovirus from Pigeons,” Archives of Virology, Vol. 145, No. 12, 2000, pp. 2469- 2479. doi:10.1007/s007050070002
- S. C. Harrison, et al., “Tomato Bushy Stunt Virus at 2.9-A Resolution,” Nature, Vol. 276, No. 5686, 1978, pp. 368-373. doi:10.1038/276368a0
- J. A. Speir and J. E. Johnson, “Virus Particle Structure: Nonenveloped Viruses,” In: B. W. J. Mahy and M. H. V. Van Regenmortel, Eds., Encyclopedia of Virology, Elsevier, Oxford, 2008, pp. 380-393. doi:10.1016/B978-012374410-4.00600-2
- R. Johne, et al., “Recombinant Expression of a Truncated Capsid Protein of Beak and Feather Disease Virus and Its Application in Serological Tests,” Avian Pathology, Vol. 33, No. 3, 2004, pp. 328-336. doi:10.1080/0307945042000220589
- A. Banda, et al., “Genetic Analysis of a Duck Circovirus Detected in Commercial Pekin Ducks in New York,” Avian Diseases, Vol. 51, No. 1, 2007, pp. 90-95. doi:10.1637/0005-2086(2007)051[0090:GAOADC]2.0.CO;2
- L. Ortiz-Catedral, et al., “A New Isolate of Beak and Feather Disease Virus from Endemic Wild Red-Fronted Parakeets (Cyanoramphus novaezelandiae) in New Zealand,” Archives of Virology, Vol. 155, No. 4, 2010, pp. 613-620. doi:10.1007/s00705-010-0607-2
- D. Todd, et al., “Molecular Characterization of Novel Circoviruses from Finch and Gull,” Avian Pathology, Vol. 36, No. 1, 2007, pp. 75-81. doi:10.1080/03079450601113654

