Open Journal of Applied Biosensor
Vol.1 No.2(2012), Article ID:21880,11 pages DOI:10.4236/ojab.2012.12002
Aptasensors in Health, Environment and Food Safety Monitoring*
1Department of Biotechnology, Indian Institute of Technology Guwahati, Guwahati, India
2Centre for the Environment, Indian Institute of Technology Guwahati, Guwahati, India
Email: #ubora@iitg.ernet.in, ubora@rediffmail.com
Received July 10, 2012; revised August 8, 2012; accepted August 15, 2012
Keywords: Aptamer; Biosensor; Clinical Diagnosis; Food Safety; Environmental Pollution Control
ABSTRACT
Biosensors have been developed using various types of sensing elements like biomacromolecules (viz. enzymes, antibodies, receptors, nucleic acids, etc.) organelles, tissues, intact cells of both microorganisms and higher organisms. A recent trend is the emergence of aptamers as sensing elements that has the potential to replace all the above ligands. This is possible due to the unique features of aptamers (sensitivity, specificity, reusability, stability, non-immunogenicity), which can be easily exploited in biosensor technology. Aptasensors are thus basically biosensors based on aptamers as ligand molecules. Here we review the various applications of aptasensors in health (specifically in diagnostics), food industry and environmental monitoring.
1. Introduction
Aptamers are small oligonucleotides, peptides and peptide nucleic acids that can bind with high affinity and selectivity to diverse targets like small and macromolecules, from organic, inorganic and biological origin [1]. Aptamers can fold into distinct secondary and tertiary structures, bind to their targets with high affinity (dissociation constants on the order of nanoto picomolar) and recognize their targets with a specificity that challenges antibodies and other biological ligands. Nucleic acid aptamers are selected from a random pool by an iterative process called SELEX, who’s principle is similar to that of natural selection in evolution proposed by Charles Darwin over 150 years ago. The enriched pool shows high specificity and sensitivity to their target molecules. Due to their versatility aptamers are also called as “magic bullets” and are excellent example of functional biological molecules which are selected in vitro. Since its discovery aptamer based technologies have been drawn immense attention in various research communities [2]. Apart from its high specificity and sensitivity, aptamers offer a wide range of advantages over other existing molecules viz stability, design flexibility and cost-effectiveness. These features favour their use as bio recognition elements in biosensor development. Aptasensors are biosensors where aptamers are used in place of biological ligands to sense targeted analytes.
Oligonucleotide (both DNA, RNA), peptide and peptide-nucleic acid (PNA) based aptasensors offer high reproducibility against a wide variety of targets (e.g. proteins, peptides, drugs, small molecules, metal ions, and even whole cells) and are quickly emerging as desirable candidates for high throughput analytical methods that use minute amount of analytes (nano-microlitre). Availability of in-depth knowledge of nucleic acid aptamers in terms of their conformational and ligand binding mechanisms has evoked deep interest among researchers for developing aptamer based bioassays as reflected in the exponential increase of published articles related to aptasensors (Figure 1). Here we attempt to review the recent advancements of aptasensors based on different sensing technologies in diverse fields like, diagnostics, food safety, environmental toxicity studies etc (Figure 2).
Aptasensor based analysis is continuously evolving with various detection schemes ranging from label-free methods such as surface plasmon resonance (SPR) [3] and quartz crystal microbalance (QCM) measurements [4] to label dependant methods such as electrochemistry, fluorescence, chemiluminescence, field effect transistors [5] etc. However currently electrochemical and optical aptasensors [6] constitute the two predominant types under development. These can even distinguish between chiral molecules and are able to recognize a distinct epitope of a target molecule as well [7].
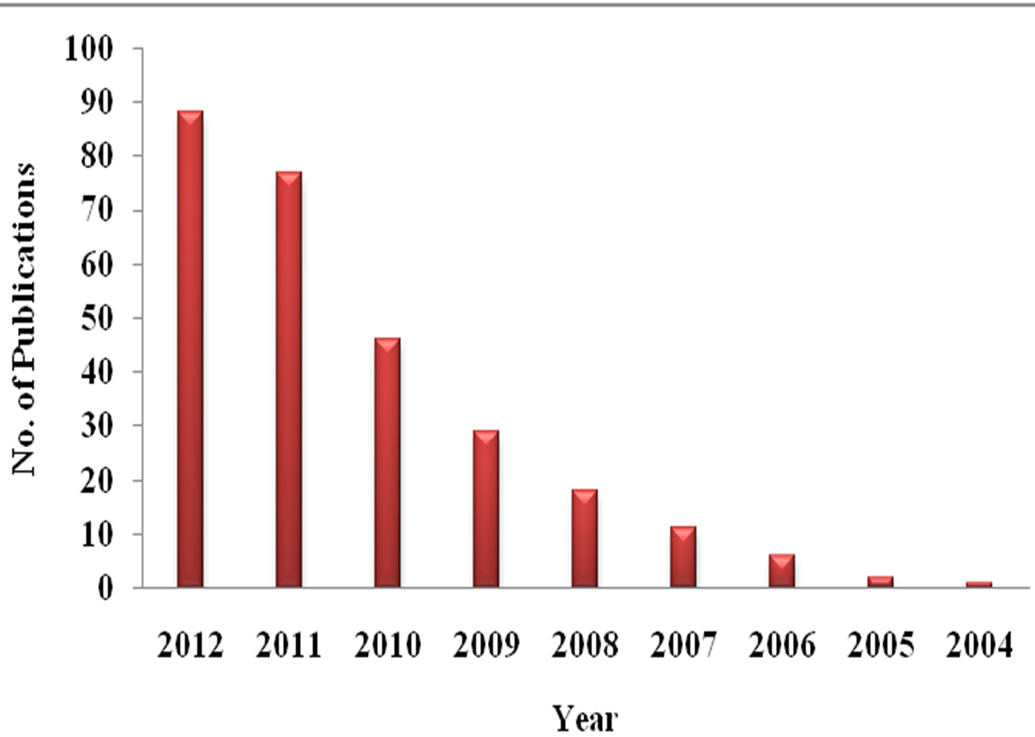
Figure 1. Publication trend in the field of aptasensors (Data obtained from Scopus; Search strings: “Aptasensor”).
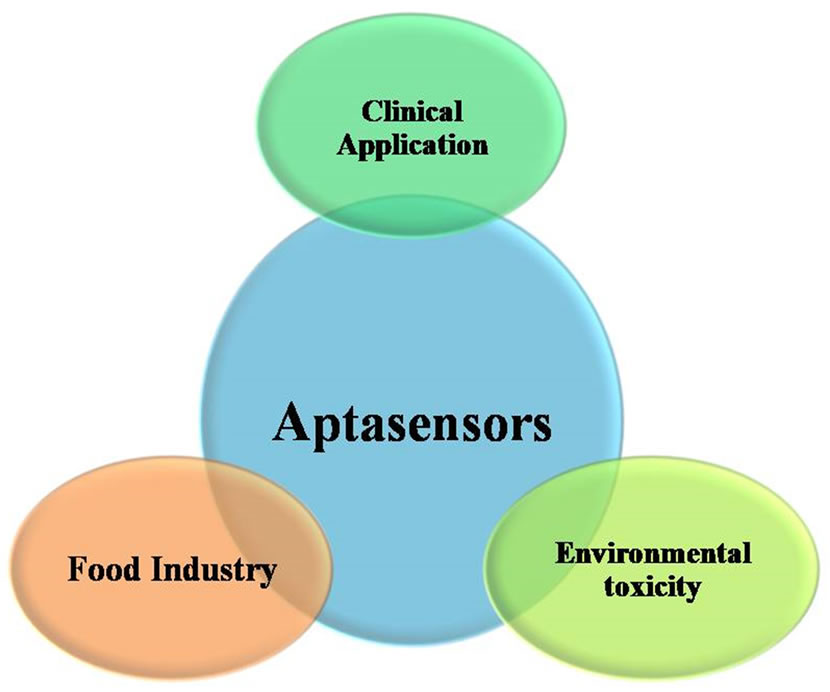
Figure 2. Applications of aptasensors.
2. Aptasensors in Health Monitoring
Aptamers can be modified and reused for both diagnostic and therapeutic purposes. Aptamers have more potential than antibodies and aptasensors can be used in detection of various biomarkers, in cancer diagnosis and even detection of pathogenic microorganisms.
2.1. Aptasensors for Biomarker Detection
The detection of biomarkers presents in blood, urine and other body fluids help in the early diagnosis of any disease in general. Aptasensors have been widely used for detection of biomarkers, such as thrombin, Immunoglobulin E, C-reactive protein, IFN gamma or platelet-derived growth factor (PDGF) in body fluids.
Similar to antigen-antibody interaction, binding properties of aptamers depends on the conformational plasticity and three-dimensional folding of the nucleic acid sequence that results in a 3D-structure “aptatope”, the equivalent of an “epitope”.
Aptasensor assays can be exploited for detection of biological molecules with various working principles. The sensitivity and longevity of a aptasensor was significantly increased by altering the size of nucleotide bases and modification of the capping to prevent nuclease activity. Pu et al. developed a smart polymeric transducer and aptamer/intercalating dye system that allows the label-free detection of thrombin with high sensitivity and specificity [8]. Further the methodology didn’t require any chemical modification of the probes or the analytes. The minimum amount of thrombin that could be measured by the fluorescence intensity changes was estimated to be 0.1 nM. This offers a new strategy to detect a wide spectrum of analytes and would be highly beneficial in future clinical applications.
A method based on combination of ex situ polarization modulation FTIR measurements of the RNA monolayer and in situ Surface Plasmon Resonance Imaging (SPRI) measurements of DNA hybridization adsorption onto the surface have been developed to detect protein factor IXa .This system can detect single stranded RNA having surface density of 4.0 × 1012 molecules/cm2 and a surface ligation efficiency of 85% ± 10% [9].
Single-walled carbon nanotubes typically functionalized with gold nanoparticles increases the surface area to capture a large amount of primary aptamers and amplify the detection response. Multi-labeled Platinum-nano particles (PtNPs)-redox probes-reduced glutathione-S nanocomposites display satisfying electrochemical redox activity and high electrocatalytic activity of PtNPs and bienzyme, which exhibit high sensitivity for detection of proteins like platelet derived growth factor (PDGF) and thrombin. The linear range of PDGF was found to be 0.01e35 nM with a detection limit of 8 pM, while the linear range was 0.02 to 45 nM with a detection limit of 11 pM for thrombin are obtained [10]. Numerous aptasensors available for biomarker detection are summarized in Table 1.
2.2. Aptasensors in Cancer Diagnosis
Early diagnosis is critical in cancer for prevention, improvement of patient comma missing between survival and disease disease prognosis etc., which in many cases is currently restricted by non availability of sensitive and specific methods of diagnostics.
Detection and identification of cancerous cells rely on the identification of specific markers that appear in case of lymphoma and leukemias. Recognition of cancer markers with specific probes help in identifying the potential risk factor. In blood plasma proteins or free DNA molecules are treated as tumor prognosis markers. A number of recent studies have successfully used aptasensors for detecting tumour markers which are summarized in Table 2. Feng et al. reported label-free cancer cell detection with an electrochemical sensor based on the first clinical oncology trial II used aptamer AS1411
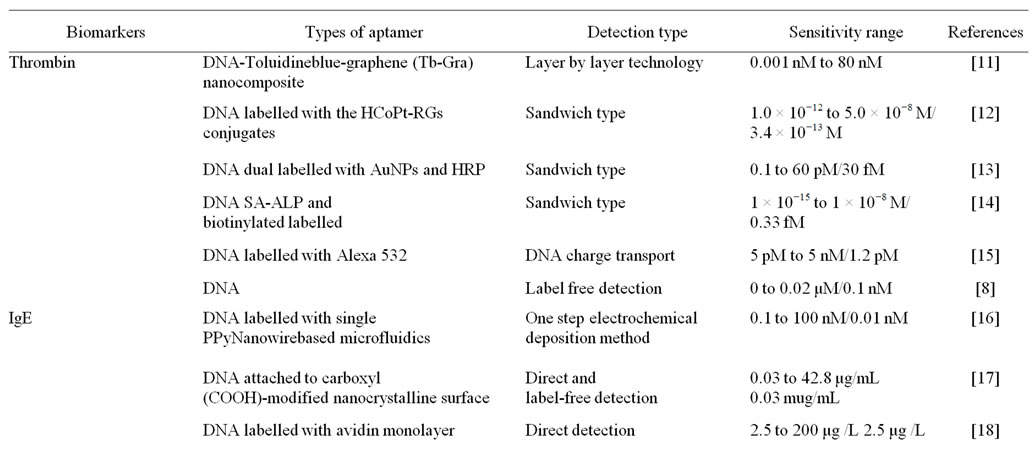
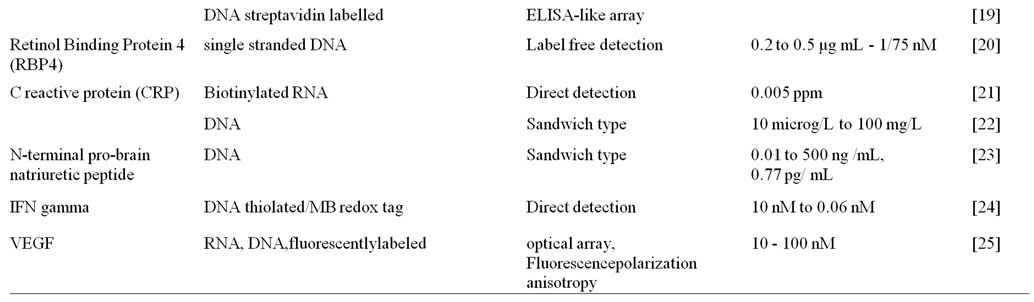
Table 1. Example of aptasensors in various biomarker detection.
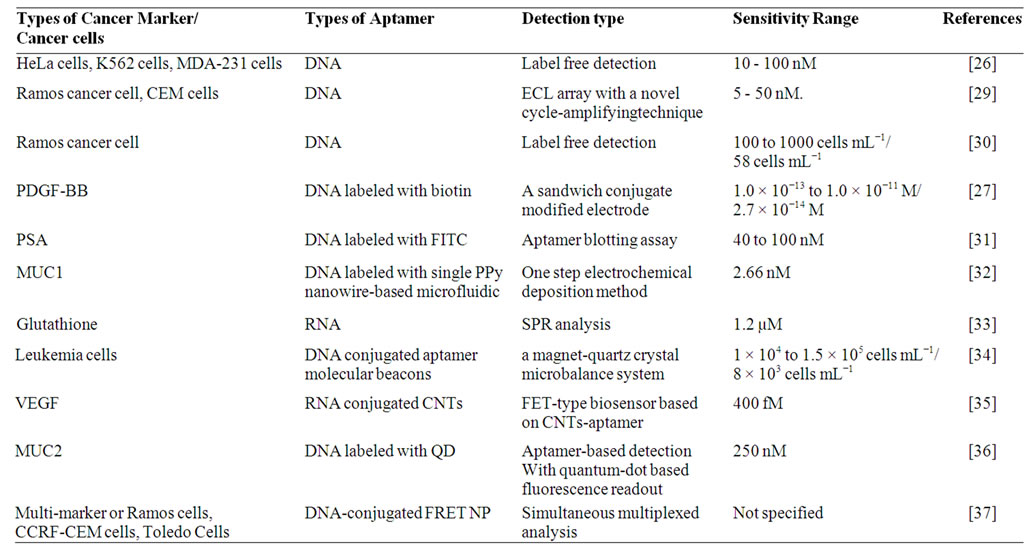
Table 2. Example of aptasensors used in cancer detection.
and graphene-modified electrode [26]. An aptamerperylenetetracarboxylic acid (PTCA) nanocomposite was utilized as nanoscale anchorage ligand to effectively capture cells on electrode surface through the specific binding between cell surface nucleolin and the aptamer AS1411. The aptasensor could distinguish cancer cells from normal cells and detect as low as 1000 cells/ml and can be regenerated and reused with ease.
Platelet-derived growth factor B chain (PDGF-BB) is a potential cancer biomarker and is known to be related to tumor growth and transformation. Recently, Chai et al. developed a electrochemiluminiscence (ECL) based aptasensor for PDGF-BB by assembling N-(aminobutyl)-N-ethylisoluminol functionalized gold nanoparticles (ABEI-AuNPs) with aptamers as nanoprobes which showed high sensitivity and specificity [27]. The biotinylated aptamer capture probes were first immobilized on a streptavidin coated gold nanoparticle electrode. The detection limit limit of the sensor was as low as 2.7 × 10−14 M.
In a recent study, Zhao et al. reported an electrochemical aptasensor for simultaneous determination of two tumor markers MUC1 and VEGF [28]. Since the cDNA immobilized on an electrode surface can hybridize with both MUC1 aptamer and VEGF165 aptamer to form a long double strand with ferrocene, the probe cannot give electrochemical signal. Nevertheless, the presence of the two markers inhibit the hybridization of cDNA with the aptamers, thus the distance between ferrocene and the electrode is changed, and a “signal-on” electrochemical method to detect two tumor markers is fabricated. It was proven experimentally that electrochemical aptasensors not only detect the two markers but also can discriminate their co-existence.
3. Aptasensors for Food Safety and Environmental Pollution Control
Food maybe contaminated by chemical compounds, toxin or pathogen leading to various food borne ailments. According to World Health Organization (WHO) food borne and waterborne diarrhoeal diseases kill about 2.2 million people annually out of which 1.9 million are children [38]. This growing public health problem demands the development of highly sensitive technologies for rapid detection of food contaminants. In this context, biosensors have a potential to emerge as a major tool in maintaining food safety. Traditional techniques are largely based on antibody-conjugated biosensors. However, many food contaminants are not conducive for antibody generation. Also antibodies suffer from batch to batch variation and short shelf lives necessitating the development of a simpler, consistent and cost effective approach for food safety testing.
Aptamers can address many of the challenges of a traditional biosensor without compromising on specificity and affinity. Based on the type of target, aptamers used in food safety testing and environmental pollution control can be broadly classified under two groups-aptamers against small molecule contaminants and those against pathogens.
3.1. Aptasensors against Small Molecules
Small molecule contaminants include antibiotics, toxins, pesticides and heavy metals that may be present in a variety of food products and environmental samples. For example, theophylline is a commonly used bronchodilator for asthma patients. However, its overdose leads to severe toxicity like, seizure, nausea etc. RNA based electrochemical aptasensors have been developed to monitor its level in serum [39]. Also aptasensors for detection of drugs like cocaine has been developed based on surface enhanced raman scattering spectroscopy (SERS) [40].
3.1.1. Aptasensors against Antibiotics
Antibiotics are often administered to farm animals along with their feed for prophylactic and therapeutic purposes. However, a large percentage of these antibiotics remain unmetabolized and accumulate in the tissues or are excreted out in the environment. The occurrence of antibiotics in the environment might lead to antibiotic resistance which may be transmitted to humans via the food chain [41].
3.1.1.1. Chloramphenicol (Cam)
Chloramphenicol is a popular bacteriostatic antimicrobial drug effective against a wide variety of gram positive and gram negative bacteria. It inhibits peptide bond formation by binding to peptidyl transferase loop of 23S ribosomal RNA. However it has lost its favour due to resistance and serious side effects like aplastic anemia. Burke et al. initially reported the use of RNA aptamers against chloramphenicol. These aptamers screened in vitro through SELEX resembled the Cam binding site in 23SrRNA. Various studies indicated that the aptamers structurally contained two symmetrically arranged A-rich bulges [42]. However, RNA aptamers are susceptible to nuclease attack and requires transcription and reverse transcription making it difficult to screen via SELEX. Recently Mehta et al., developed DNA aptamers to detect Cam having higher selectivity and affinity (Kd ~ 1 µM). Interestingly the two best aptamers screened by them had G-rich nucleotide regions distinguishing them from the rest [41]. These screened aptamers have now been used to develop label free electrochemical biosensors for detection of chloramphenicol. The aptamers were immobilized onto a gold electrode by self assembly approach. The developed electrochemical aptasensor was highly sensitive (detection limit of 1 nM) and specific to chloramphenicol over its other derivatives [43].
3.1.1.2. Tetracycline
Tetracyclines are a group of broad spectrum antibiotics which inhibits prokaryotic translation by interfering with the binding of amino-acyl tRNA to the ribosomal A-site [44]. Tetracyclines are often used as a veterinary drug to promote growth in animals. Thus residues of the antibiotic has been detected in meat [45], milk [46], honey [47], eggs [48] etc. On the other hand, tetracyclins, it has also been reported to be hepatotoxic to pregnant women [49]. Traditional detection methods for tetracycline like HPLC and capillary electrophoresis lack specificity and are often time consuming and expensive. Recently, aptamer based systems have been developed for detection of tetracyclines. Berens et al. first screened a RNA aptamer (cb28) against tetracycline which resembled its interaction with the small ribosomal subunit [50]. Later Xiao et al., improved upon the structure of the developed aptamer to obtain a very high affinity (Kd ~ 0.8 nM) [51]. A ssDNA electrochemical aptasensor was recently developed by Kim et al having high specifity for tetracycline over its other derivatives [52]. Zhang et al. reported another rapid electrochemical aptasensor for tetracycline using aptamers immobilized over glassy carbon electrodes. The aptasensor was able to rapidly detect tetracycline in milk with very high sensitivity (~1 ng/ml) [53]. A competitive enzyme linked aptamers assay (ELIAA) based aptasensor for tetracycline was developed by Jheong et al. using both DNA and RNA aptamers [54].
3.1.1.3. Aminoglycoside
Aminoglycosides are a class of antibiotics that bind with prokaryotic ribosomes leading to frameshift mutations which produce nonsense peptides eventually causing cell death [55]. Its clinical use has been restricted due to toxic side effects to the kidney and ear [56]. Aminoglycosides include antibiotics like tobramycin, kanamycin, neomycin etc, all having a common streptamine ring. Initially RNA aptamers were developed against tobramycin by Wang and Rando [57]. They later went on to develop RNA aptamers against all aminoglycosides based on the consensus sequence. [58]. RNA aptamers have often been reported to be unstable due to their vulnerability to endonucleases. In the quest to find the right balance between affinity and stability, Rowe et al., screened RNA aptamers to detect aminoglycoside antibiotics in human serum. Serum was separated from nucleases by ultrafiltration through a 3000 Da cut off spin column, so that the aptamers remain stable. This enabled them to develop a reagentless RNA based electrochemical aptasensor which could detect low levels of aminoglycoside antibiotics (2 - 6 µM) in human blood samples [59]. Recently, a ssDNA aptamer against kanamycin has also been screened by Song et al. The aptamers conjugated with gold nanoparticles showed dissociation constants in the nanomolar range and were able to detect kanamycin by colorimetric method [60]. Derbyshire et al., has been successful in screening RNA aptamers against the entire class of aminoglycosides. The aptamers were then coated with gold nanoparticle for a rapid and sensitive colorimetric assay based detection of antibiotics [61].
3.1.2. Aptasensors against Toxins
Mycotoxins are the major toxins that may be present in our food. They are a group of naturally occurring chemicals produced by moulds growing on a variety of different crops. Mycotoxins can cause adverse effects on humans from liver cancer, gastrointestinal diseases to kidney damage and immune suppression. A DNA aptamer was developed to detect ochratoxin A—a mycotoxin in food with high specificity [62]. This aptamer has been integrated into several biosensor detection systems including electrochemical, electrochemiluminescent, colorimetric and fluorescent platforms. Bacterial endotoxins (Lipopolysaccharides) are also one of the major contaminants present in commercially available proteins and pharmaceutical products. These can cause severe septic shock in humans and animals [63]. An electrochemical gold nanoparticle based aptasensor for detection of endotoxins from crude biological liquor was developed by Kim et al. [64]. The aptasensor showed excellent sensitivity and selectivity with a detection range of 0.01 to 1 ng/mL.
Seeds of the leguminous herb lupin have been widely used as a low cost protein source. However, there have been an increasing number of cases reporting severe allergic reactions to Lupin seeds. To meet this challenge and detect Lupin allergen levels in food a DNA aptamer based colorimetric sensing system was developed by Nadal et al. [65]. Many toxins are excreted by humans and animals which ultimately end up in water effluents. Endocrine disrupting compounds (EDC) form a major class of pollutants which causes severe health hazard by disrupting normal endocrine functions among human and aquatic organisms. 17β-estradiol is one such compound which has deleterious effects on the male reproductive system [66]. Recently, Yildrim et al. reported a highly selective and rapid fluorescence based DNA aptasensor for detection of low levels of 17β-estradiol in environmental water samples [67].
3.1.3. Aptasensors against Food Packaging Contaminants
Bisphenol A is used as a monomer compound in plastic polycarbonate products. However, it is potentially dangerous to humans and animals as it disrupts endocrine function by blocking binding of estrogen with its receptor. Using high affinity ssDNA aptamers Jo et al. developed a sol-gel biochip assay to detect Bisphenol A and measure its level in water samples [68].
3.1.4. Aptasensors against Heavy Metals
Meat, milk, egg, fish and other foodstuffs are often found contaminated by heavy metals like Mercury (Hg2+), Arsenic (As) etc. Hg2+ causes severe toxicity to the nervous and endocrine system whereas Arsenic (As) leads to serious ailments like heart problems, skin lesions and cancer. High arsenic contamination levels were reported in Mekong river delta in Vietnam. According to government sources the residual arsenic levels in water had exceeded the allowable limit by 10 µg/L leading to severe arsenic toxicity among the inhabitants. To address this crisis, Kim et al. screened DNA based aptamers showing highest affinity for Arsenate (V) and Arsenite (III) [69] which may be used to develop Arsenic based aptasensors in future.
Mercury (Hg2+) ions were successfully detected in water using gold nanoparticles based colorimetric aptasensors. The aptamer was rich in thymine regions which formed dimers in presence of Hg2+ ions. The gold nanoparticles were able to differentiate between dimers and short single stranded DNA and functioned as a colorimetric probe. The system had high sensitivity of around 1 nM [70]. Recently, Helwa et al., in their study, immobilized DNA aptamers on polyacrylamide gel. The setup provided rapid visual detection of Hg2+ and high sensitivity of around 10 nM [71].
Till date, a number of aptamer based sensors have been developed to detect very low levels of heavy metals in our environment. But not much research has been done on how to reduce the toxicity caused by these heavy metals. In an interesting work reported by Hu et al., aptamer-nanoparticle conjugate was used to reduce Hg2+ toxicity. The authors used selenomethionine (SeMet), having a higher binding affinity to mercury, as an antidote and conjugated it with PLGA naoparticle-aptamer complex. Rats fed with mercury contaminated food were treated with the conjugate and mercury accumulation was found to decrease appreciably in the brain and kidney [72].
3.1.5. Aptasensors against Pesticides
Atrazine is one of the most widely used pesticides to inhibit the growth of weeds. However, it can lead to severe reproductive damage in humans. Sinha et al. developed a novel strategy to engineer bacterial cells for detection and removal of atrazine from environmental samples. They first screened a series of aptamers against atrazine and then cloned them into E. coli cells. This construct then functioned as atrazine dependent riboswitches. [73].
Insecticides are also often applied to crops to protect them from insect attack. One such example is Acetamiprid, which is a neonicotinoid insecticide. However when leached into the environment it can cause toxicity in humans and animals. He et al. screened a series of aptamers via SELEX which bound to acetamiprid with a dissociation constant (Kd) of 4.98 µM [74]. Recently, Wang et al., successfully developed DNA aptamers which were able to detect upto four highly poisonous organo-phosphorous pesticides including phorate, profenofos, isocarbophos and omethoateas [75]. Although aptamers have been screened against various types of pesticides, its potential as a ligand molecule in a biosensor is largely unexplored.
3.2. Aptasensors against Pathogens
Biosensors capable of rapidly detecting pathogens with high sensitivity and specificity are essential to meet the ever increasing serious clinical and therapeutic challenges. Most importantly, aptasensors are able to target and specifically differentiate pathogens without prior information of their membrane molecules and structural genes. Various attempts to confront this challenge with aptasensors against pathogenic virus and bacteria are discussed herein.
3.2.1. Virus
The development of aptasensors against virus is in its infancy. Currently promising leads are available regarding the screening of aptamers against specific viruses which has the potential to be integrated with suitable biosensor platforms. Tang et al. reported the development of DNA aptamers against a specific protein of vaccinia virus. They infected a mammalian cell line (A549) with the virus and used the aptamers as probes for detection of the viral protein [76]. Those aptamers can be developed in aptasensors by modifying for immobilization purposes and labelled with reporter molecules, fluorophores without hampering their target specificity. Several aptasensors have been developed to detect viral proteins. Minunni et al. fabricated an aptasensor to detect HIV-1 Tat protein by immobilizing an RNA aptamer on a piezoelectric quartz crystal. Sensitivity, specificity, and reproducibility parameters were quantified. The aptasensor was also compared with the available immunosensor with immobilized anti-Tat antibodyies. Both the optimized aptasensor and the immunosensor showed a detection limit of 0.25 ppm [77]. The quartz crystal microbalance (QCM)-based aptasensor has also been compared with the corresponding surface plasmon resonance (SPR)-based aptasensor. The two aptasensors were developed by biotin-avidin linking onto the gold surface of the transducers (quartz crystals or chips) for the immobilization. Both platforms showed similar reproducibility, sensitivity and specificity. The linear range of SPR (1 - 2.5 ppm) was higher that of QCM (0 - 1.25 ppm) [78]. Another viral aptasensor developed against the heptatitis C virus (HCV) core antigen detector. Lee et al. experimented with the binding affinity of several aptamer sequences against the target candidate. After selection, the specific aptamer was immobilized in a 96-well plate, using the sol-gel-based immobilization method. Then, the immobilized aptamers on the chip were incubated with recombinant core antigens. The aptamer-core complexes were incubated with Cy3-labeled secondary antibodies. The platform was able to detect core-specific interaction with the aptamers, using pure recombinant protein as well as human sera matrixes [79].
3.2.2. Bacteria
Detection of bacteria using aptasensors is a relatively new area. Two different strategy for whole-cell detection, using quantum dots (QDs) and carbon nanotubes (CNTs), have been proposed recently. Aptamer-functionalized QDs have been exploited to detect Bacillus thuringiensis spores [80]. In the study, zinc sulfide-capped cadmium selenide QDs were functionalized with a specific aptamer selected to detect the pathogen. After QD-aptamer incubation with the target, the spores were washed and collected for fluorescence measurement. Several controls with non-functionalized QDs and without spores were tested to measure the non-specific attachment of the QDs to the spores and the fluorescence background noise. The reported sensitivity was 103 CFU/ml. For specificity purposes, spores from B. globigii (B. subtilisvar. niger) were also tested. The system could differentiate B. thuringiensis from B. globigiiat concentrations above 105 CFU/ml. In another study, using aptamer-functionalized singlewalled carbon nanotube field-effect transistor (SWNTFET) arrays aptasensors was developed to detect E. coli DH5 α [81]. The binding between E. coli cells and the aptamer-functionalized FET resulted a difference in conductance (>50%) in culture samples with concentrations between 105 and 107 CFU/ml. Specificity assays were conducted with Salmonella typhimurium.
Recently, bioterrorism has become a major threat to national security. One of the leading examples is Anthrax caused by the spores of gram positive bacteria Bacillus anthracis. Cella et al. has developed an aptasensor for detection of protective antigen (PA) of anthrax. The aptasensor consisted of single stranded DNA aptamer functionalized to single walled carbon nanotubes having sensitivity in the nanomolar range (~1 nM) [82].
4. Conclusion
Aptamers can easily be integrated to the many existing biosensor schemes. The plethora of transduction platforms for affinity sensing can readily be adapted to the use of aptameric bio components. Additionally, catalytic aptamers (aptazymes) have also been reported, to offer modes of specific binding event detection. Moreover, it can be anticipated that the use of immobilisable molecular beacons can be exploited in the biosensor field (till date a handful of publications exist); a mode of detection highly suited to aptasensors facilitating reagentless, cost effective one step analysis. Overall, the potential of aptasensors is immense and this exciting and challenging area is on the brink of exponential growth. The ability to develop affinity based detection systems based on tailor made characteristics, like size, toxicity and matrix effects, offers the field of biosensing the opportunity to explore hitherto unexplored horizons in sensor development.
5. Acknowledgements
The authors thank the Department of Biotechnology, Govt. of India for supporting research and development activities through the Institutional Biotech Hub project (Sanction No: BT/04/NE/2009). AS and SD thank IITG & MHRD for the financial support in the form of fellowship.
REFERENCES
- E. N. Brody, M. C. Willis, J. D. Smith, S. Jayasena, D. Zichi and L. Gold, “The Use of Aptamers in Large Arrays for Molecular Diagnostics,” Molecular Diagnostics, Vol. 4, No. 4, 1999, pp. 381-388. doi:10.1016/S1084-8592(99)80014-9
- C. Tuerk and L. Gold, “Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase,” Science, Vol. 249, No. 4968, 1990, pp. 505-510. doi:10.1126/science.2200121
- Y. Li, H. J. Lee and R. M. Corn, “Fabrication and Characterization of RNA Aptamer Microarrays for the Study of Protein-Aptamer Interactions with SPR Imaging,” Nucleic Acids Research, Vol. 34, No. 22, 2006, pp. 6416- 6424. doi:10.1093/nar/gkl738
- Z. M. Dong and G. C. Zhao, “Quartz Crystal Microbalance Aptasensor for Sensitive Detection of Mercury(II) Based on Signal Amplification with Gold Nanoparticles,” Sensors, Vol. 12, No. 6, 2012, pp. 7080-7094. doi:10.3390/s120607080
- O. S. Kwon, S. J. Park, J. Y. Hong, A. R. Han, J. S. Lee, J. S. Lee, J. H. Oh and J. Jang, “Flexible FET-Type VEGF Aptasensor Based on Nitrogen-Doped Graphene Converted from Conducting Polymer,” ACS Nano, Vol. 6, No. 2, 2012, pp. 1486-1493. doi:10.1021/nn204395n
- A. Sassolas, L. J. Blum and B. D. Leca-Bouvier, “Optical Detection Systems Using Immobilized Aptamers,” Biosensors and Bioelectronics, Vol. 26, No. 9, 2011, pp. 3725-3736. doi:10.1016/j.bios.2011.02.031
- M. Michaud, E. Jourdan, A. Villet, A. Ravel, C. Grosset and E. Peyrin, “A DNA Aptamer as a New Target-Specific Chiral Selector for HPLC,” Journal of the American Chemical Society, Vol. 125, No. 28, 2003, pp. 8672-8679. doi:10.1021/ja034483t
- F. Pu, Z. Z. Huang, D. Hu, J. S. Ren, S. Wang and X. G. Qu, “Sensitive, Selective and Label-Free Protein Detection Using a Smart Polymeric Transducer and Aptamer/ Ligand System,” Chemical Communications, No. 47, 2009, pp. 7357-7359. doi:10.1039/b918241a
- Y. Li, H. J. Lee and R. M. Corn, “Fabrication and Characterization of RNA Aptamer Microarrays for the Study of Protein-Aptamer Interactions with SPR Imaging,” Nucleic Acids Research, Vol. 34, No. 22, 2006, pp. 6416- 6424. doi:10.1093/nar/gkl738
- L. J. Bai, R. Yuan, Y. Q. Chai, Y. Zhuo, Y. L. Yuan and Y. Wang, “Simultaneous Electrochemical Detection of Multiple Analytes Based on Dual Signal Amplification of Single-Walled Carbon Nanotubes and Multi-Labeled Graphene Sheets,” Biomaterials, Vol. 33, No. 4, 2012, pp. 1090-1096. doi:10.1016/j.biomaterials.2011.10.012
- S. B. Xie, R. Yuan, Y. Q. Chai, L. J. Bai, Y. L. Yuan and Y. Wang, “Label-Free Electrochemical Aptasensor for Sensitive Thrombin Detection Using Layer-by-Layer Self-Assembled Multilayers with Toluidine Blue-Graphene Composites and Gold Nanoparticles,” Talanta, Article in Press.
- Y. Wang, R. Yuan, Y. Q. Chai, Y. L. Yuan, L. J. Bai and Y. H. Liao, “A Multi-Amplification Aptasensor for Highly Sensitive Detection of Thrombin Based on High-Quality Hollow CoPt Nanoparticles Decorated Graphene,” Biosensors and Bioelectronics, Vol. 30, No. 1, 2011, pp. 61- 66. doi:10.1016/j.bios.2011.08.027
- J. Zhao, Y. Y. Zhang, H. T. Li, Y. Q. Wen, X. Y. Fan, F. B. Lin, L. Tan and S. Z. Yao, “Ultrasensitive Electrochemical Aptasensor for Thrombin Based on the Amplification of Aptamer-AuNPs-HRP Conjugates,” Biosensors and Bioelectronics, Vol. 26, No. 5, 2011, 2297-2303. doi:10.1016/j.bios.2010.09.056
- Y. H. Liao, R. Yuan, Y. Q. Chai, Y. Zhuo, Y. L. Yuan, L. J. Bai, L. Mao and S. R. Yuan, “In-Situ Produced Ascorbic Acid as Coreactant for an Ultrasensitive Solid-State tris(2,2′-bipyridyl) Ruthenium(II) Electrochemiluminescence Aptasensor,” Biosensors and Bioelectronics, Vol. 26, No. 12, 2011, pp. 4815-4818. doi:10.1016/j.bios.2011.04.019
- X. Y. Zhang, Z. L. Zhao, H. C. Mei, Y. P. Qiao, Q. L. Liu, W. X. Luo, T. Xia and X. H. Fang, “Fluorescence Aptasensor Based on DNA Charge Transport for Sensitive Protein Detection in Serum,” Analyst, Vol. 136, No. 22, 2011, pp. 4764-4769. doi:10.1039/c1an15265c
- J. Y. Huang, X. L. Luo, I. Lee, Y. S. Hu, X. T. Cui and M. Yun, “Rapid Real-Time Electrochemical Detection of Proteins Using Single Conducting Polymer NanowireBased Microfluidic Aptasensor,” Biosensors and Bioelectronics, Vol. 30, No.1, 2011, pp. 306-309. doi:10.1016/j.bios.2011.08.016
- D. T. Tran, V. Vermeeren, L. Grieten, S. Wenmackers, P. Wagner, J. Pollet, K. P. Janssen, L. Michiels and J. Lammertyn, “Nanocrystalline Diamond Impedimetricaptasensor for the Label-Free Detection of Human IgE,” Biosensors and Bioelectronics, Vol. 26, No. 6, 2011, pp. 2987- 2993. doi:10.1016/j.bios.2010.11.053
- C. Y. Yao, Y. Z. Qi, Y. H. Zhao, Y. Xiang, Q. H. Chen and W. L. Fu, “Aptamer-Based Piezoelectric Quartz Crystal Microbalance Biosensor Array for the Quantification of IgE,” Biosensors and Bioelectronics, Vol. 24, No. 8, 2009, pp. 2499-2503. doi:10.1016/j.bios.2008.12.036
- J. L. Wang, R. J. Lv, J. J. Xu, D. K. Xu and H. Y. Chen, “Characterizing the Interaction between Aptamers and Human IgE by Use of Surface Plasmon Resonance,” Analytical and Bioanalytical Chemistry, Vol. 390, No. 4, 2008, pp. 1059-1065. doi:10.1007/s00216-007-1697-x
- S. Lee, Y. S. Kim, M. Jo, M. Jin, D. K. Lee and S. Kim, “Chip-Based Detection of Hepatitis C Virus Using RNA Aptamers That Specifically Bind to HCV Core Antigen,” Biochemical and Biophysical Research Communications, Vol. 358, No. 1, 2007, pp. 47-52. doi:10.1016/j.bbrc.2007.04.057
- A. Bini, S. Centi, S. Tombelli, M. Minunni and M. Mascini, “Development of an Optical RNA-Based Aptasensor for C-Reactive Protein,” Analytical and Bioanalytical Chemistry, Vol. 390, No. 4, 2008, pp. 1077-1086. doi:10.1007/s00216-007-1736-7
- J. Pultar, U. Sauer, P. Domnanich and C. Preininger, “Aptamer-Antibody On-Chip Sandwich Immunoassay for Detection of CRP in Spiked Serum,” Biosensors and Bioelectronics, Vol. 24, No. 5, 2009, pp. 1456-1461. doi:10.1016/j.bios.2008.08.052
- L. Mao, R. Yuan, Y. Q. Chai, Y. Zhuo and Y. Xiang, “Signal-Enhancer Molecules Encapsulated Liposome as a Valuable Sensing and Amplification Platform Combining the Aptasensor for Ultrasensitive ECL Immunoassay,” Biosensors and Bioelectronics, Vol. 26, No. 10, 2011, 4204-4208. doi:10.1016/j.bios.2011.02.035
- Y. Liu, N. Tuleouva, E. Ramanculov and A. Revzin, “Aptamer-Based Electrochemical Biosensor for Interferon Gamma Detection,” Analytical Chemistry, Vol. 82, No. 19, 2010, pp. 8131-8136. doi:10.1021/ac101409t
- T. G. McCauley, N. Hamaguchi and M. Stanton, “Aptamer-Based Biosensor Arrays for Detection and Quantification of Biological Macromolecules,” Analytical Biochemistry, Vol. 319, No. 2, 2003, pp. 244-250. doi:10.1016/S0003-2697(03)00297-5
- L. Y. Feng, Y. Chen, J. S. Ren and X. G. Qu, “A Graphene Functionalized Electrochemical Aptasensor for Selective Label-Free Detection of Cancer Cells,” Biomaterials, Vol. 32, No. 11, 2011, pp. 2930-2937. doi:10.1016/j.biomaterials.2011.01.002
- Y. Chai, D. Y. Tian, J. Gu and H. Cui, “A Novel Electrochemiluminescence Aptasensor for Protein Based on a Sensitive N-(aminobutyl)-N-Ethylisoluminol-Functionalized Gold Nanoprobe,” Analyst, Vol. 136, No. 16, pp. 3244- 3251. doi:10.1039/c1an15298j
- J, Zhao, X. L. He, B. Bo, X, J. Liu, Y. M. Yin, G. X. Li; “A ‘Signal-On’ Electrochemical Aptasensor for Simultaneous Detection of Two Tumor Markers,” Biosensors and Bioelectronics, Vol. 34, No. 14, 2012, pp. 249-252. doi:10.1016/j.bios.2012.02.016
- G. F. Jie, L. Wang, J. X. Yuan and S. S. Zhang, “Versatile Electrochemiluminescence Assays for Cancer Cells Based on Dendrimer/CdSe-ZnS-Quantum Dot Nanoclusters,” Analytical Chemistry, Vol. 83, No. 10, 2011, pp. 3873-3880. doi:10.1021/ac200383z
- X. Hun, H. C. Chen and W. Wang, “A Electrogenerated Chemiluminescence Biosensor for Ramos Cancer Cell Using DNA Encapsulated Ru(bpy)3Cl2 as Signal Probe,” Biosensors and Bioelectronics, Vol. 26, No. 9, 2011, pp. 3887-3893. doi:10.1016/j.bios.2011.03.004
- N. Savory, K. Abe, K. Sode and K. Ikebukuro, “Selection of DNA Aptamer against Prostate Specific Antigen Using a Genetic Algorithm and Application to Sensing,” Biosensors and Bioelectronics, Vol. 26, No. 4, 2010, pp. 1386-1391. doi:10.1016/j.bios.2010.07.057
- J. Huang, X. Luo, I. Lee, Y. Hu, X. T. Cui and M. Yun, “Rapid Real-Time Electrochemical Detection of Proteins Using Single Conducting Polymer Nanowire-Based Microfluidic Aptasensor,” Biosensors and Bioelectronics, Vol. 30, No. 1, 2011, pp. 306-309. doi:10.1016/j.bios.2011.08.016
- J. Bala, A. Bhaskar, A. Varshney, A. K. Singh, S. Dey and P. Yadava, “In Vitro Selected RNA Aptamer Recognizing Glutathione Induces ROS Mediated Apoptosis in the Human Breast Cancer Cellline MCF 7,” RNA Biology, Vol. 8, No. 1, 2011, pp. 101-111. doi:10.4161/rna.8.1.14116
- Y. Pan, M. Guo, Z. Nie, Y. Huang, C. Pan, K. Zeng, Y. Zhang and S. Yao, “Selective Collection and Detection of Leukemia Cells on a Magnet-Quartz Crystal Microbalance System Using Aptamer-Conjugated Magnetic Beads,” Biosensors and Bioelectronics, Vol. 25, No. 7, 2010, pp. 1609-1614. doi:10.1016/j.bios.2009.11.022
- O. S. Kwon, S. J. Park and J. Jang, “A High-Performance VEGF Aptamer Functionalized Polypyrrole Nanotube Biosensor,” Biomaterials, Vol. 31, No. 17, 2010, pp. 4740- 4747. doi:10.1016/j.biomaterials.2010.02.040
- A. K. Cheng, H. Su, Y. A. Wang and H. Z. Yu, “Aptamer-Based Detection of Epithelial Tumor Marker Mucin 1 with Quantum Dot-Based Fluorescence Readout,” Analytical Chemistry, Vol. 81, No. 15, 2009, pp. 6130-6139. doi:10.1021/ac901223q
- X. Chen, M. C. Estevez, Z. Zhu, Y. F. Huang, Y. Chen, L. Wang and W. Tan, “Using Aptamer-Conjugated Fluorescence Resonance Energy Transfer Nanoparticles for Multiplexed Cancer Cell Monitoring,” Analytical Chemistry, Vol. 81, No. 16, 2009, pp. 7009-7014. doi:10.1021/ac9011073
- http://www.who.int/foodsafety/en/
- E. E. Ferapontova, E. M. Olsen and K. V. Gothelf, “An RNA Aptamer-Based Electrochemical Biosensor for Detection of Theophylline in Serum,” Journal of the American Chemical Society, Vol. 130, No. 13, 2008, pp. 4256- 4258. doi:10.1021/ja711326b
- J. Chen, J. Jiang, X. Gao, G. Liu, G. Shen and R. Yu, “A New Aptameric Biosensor for Cocaine Based on Surface-Enhanced Raman Scattering Spectroscopy,” Chemistry, Vol. 14, No. 27, 2008, pp. 8374-8382. doi:10.1002/chem.200701307
- J. Mehta, B. Van Dorst, E. Rouah-Martin, T. W. Herrebou, M. L. Scippo, R. Blust and J. Robbens, “In Vitro Selection and Characterization of DNA Aptamers Recognizing Chloramphenicol,” Journal of Biotechnology, Vol. 155, No. 4, 2011, pp. 361-369. doi:10.1016/j.jbiotec.2011.06.043
- D. H. Burke, D. C. Hoffman, A. Brown, M. Hansen, A. Pardi and L. Gold, “RNA Aptamers to the Peptidyl Transferase Inhibitor Chloramphenicol,” Chemistry & Biology, Vol. 4, No. 11, 1997, pp. 833-843. doi:10.1016/S1074-5521(97)90116-2
- S. Pilehvar, J. Mehta, F. Dardenne, J. Robbens, R. Blust and K. De Wael, “Aptasensing of Chloramphenicol in the Presence of Its Analogues: Reaching the Maximum Residue Limit,” Analytical Chemistry, Vol. 84, No. 15, 2012, pp. 6753-6758. doi:10.1021/ac3012522
- C. M. Spahn and C. D. Prescott, “Throwing a Spanner in the Works: Antibiotics and the Translation Apparatus,” Journal of Molecular Medicine, Vol. 74, No. 8, 1996, pp. 423-439. doi:10.1007/BF00217518
- F. K. Muriuki, W. O. Ogara, F. M. Njeruh and E. S. Mitema, “Tetracycline Residue Levels in Cattle Meat from Nairobi Salughter House in Kenya,” Journal of Veterinary Science, Vol. 2, No. 2, 2001, pp. 97-101.
- A. L. Pena, C. M. Lino and I. N. Silveira, “Determination of Oxytetracycline, Tetracycline, and Chlortetracycline in Milk by Liquid Chromatography with Postcolumn Derivatization and Fluorescence Detection,” Journal of AOAC International, Vol. 82, No. 1, 1999, pp. 55-60.
- M. Jeon and I. R. Paeng, “Quantitative Detection of Tetracycline Residues in Honey by a Simple Sensitive Immunoassaym,” Analytica Chimica Acta, Vol. 626, No. 2, 2008, pp. 180-185. doi:10.1016/j.aca.2008.08.003
- S. M. Croubels, K. E. Vanoosthuyze and C. H. Van Peteghem, “Use of Metal Chelate Affinity Chromatography and Membrane-Based Ion-Exchange as Clean-Up Procedure for Trace Residue Analysis of Tetracyclines in Animal Tissues and Egg,” Journal of Chromatography B: Biomedical Sciences and Applications, Vol. 690, No. 1-2, 1997, pp. 173-179. doi:10.1016/S0378-4347(96)00368-4
- M. C. Gwee, “Can Tetracycline-Induced Fatty Liver in Pregnancy Be Attributed to Choline Deficiency?” Medical Hypotheses, Vol. 9, No. 2, 1982, pp. 157-162. doi:10.1016/0306-9877(82)90131-1
- C. Berens, A. Thain and R. Schroeder, “A TetracyclineBinding RNA Aptamer,” Bioorganic & Medicinal Chemistry, Vol. 9, No. 10, 2001, pp. 2549-2556. doi:10.1016/S0968-0896(01)00063-3
- H. Xiao and T. E. Edwards and A. R. Ferré-D’Amaré, “Structural Basis for Specific, High-Affinity Tetracycline Binding by an in Vitro Evolved Aptamer and Artificial Riboswitch,” Chemistry & Biology, Vol. 15, No. 10, 2008, pp. 1125-1137. doi:10.1016/j.chembiol.2008.09.004
- Y. J. Kim, Y. S. Kim, J. H. Niazi and M. B. Gu, “Electrochemical Aptasensor for Tetracycline Detection,” Bioprocess and Biosystems Engineering, Vol. 33, No. 1, 2010, pp. 31-37. doi:10.1007/s00449-009-0371-4
- J. Zhang, B. Zhang, Y. Wu, S. Jia, T. Fan, Z. Zhang and C. Zhang, “Fast Determination of the Tetracyclines in Milk Samples by the Aptamer Biosensor,” Analyst, Vol. 135, No. 10, 2010, pp. 2706-2710. doi:10.1039/c0an00237b
- S Jeong and I. R. Paeng, “Sensitivity and Selectivity on Aptamer-Based Assay: The Determination of Tetracycline Residue in Bovine Milk,” Scientific World Journal, Vol. 2012, 2012, Article ID: 159456. doi:10.1100/2012/159456
- J. Wirmer and E. Westhof, “Molecular Contacts between Antibiotics and the 30S Ribosomal Particle,” Methods in Enzymology, Vol. 415, 2006, pp. 180-202. doi:10.1016/S0076-6879(06)15012-0
- M. P. Mingeot-Leclercq and P. M. Tulkens, “Aminoglycosides: Nephrotoxicity,” Antimicrobial Agents and Chemotherapy, Vol. 43, No. 5, 1999, pp. 1003-1012.
- Y. Wang and R. R. Rando, “Specific Binding of Aminoglycoside Antibiotics to RNA,” Chemical Biology, Vol. 2, No. 5, 1995, pp. 281-290. doi:10.1016/1074-5521(95)90047-0
- Y. Wang, J. Killian, K. Hamasaki and R. R. Rando, “RNA Molecules That Specifically and Stoichiometrically Bind Aminoglycoside Antibiotics with High Affinities,” Biochemistry, Vol. 35, No. 38, 1996, pp. 12338- 12346. doi:10.1021/bi960878w
- A. A. Rowe, E. A. Miller and K. W. Plaxco, “Reagentless Measurement of Aminoglycoside antIbiotics in Blood Serum via an Electrochemical, Ribonucleic Acid Aptamer-Based Biosensor,” Analytical Chemistry, Vol. 82, No. 17, 2010, pp. 7090-7095. doi:10.1021/ac101491d
- K. M. Song, M. Cho, H. Jo, K. Min, S. H. Jeon, T. Kim, M. S. Han, J. K. Ku and C. Ban, “Gold NanoparticleBased Colorimetric Detection of Kanamycin Using a DNA Aptamer,” Analytical Biochemistry, Vol. 415, No. 2, 2011, pp. 175-181. doi:10.1016/j.ab.2011.04.007
- N. Derbyshire, S. J. White, D. H. Bunka, L. Song, S. Stead, J. Tarbin, M. Sharman, D. Zhou and P. G. Stockley, “Toggled RNA Aptamers against Aminoglycosides Allowing Facile Detection of Antibiotics Using Gold Nanoparticle Assays,” Analytical Chemistry, Vol. 84, No. 15, 2012, pp. 6595-6602. doi:10.1021/ac300815c
- J. A. Cruz-Aguado and G. Penner, “Determination of Ochratoxin a with a DNA Aptamer,” Journal of Agricultural and Food Chemistry, Vol. 56, No. 22, 2008, pp. 10456-10461. doi:10.1021/jf801957h
- P. O. Magalhães, A. M. Lopes, P. G. Mazzola, C. RangelYagui, T. C. Penna and A. Pessoa Jr., “Methods of Endotoxin Removal from Biological Preparations: A Review,” Journal of Pharmacy & Pharmaceutical Sciences, Vol. 10, No. 3, 2007, pp. 388-404.
- S. E. Kim, W. Su, M. Cho, Y. Lee and W. S. Choe, “Harnessing Aptamers for Electrochemical Detection of Endotoxin,” Analytical Biochemistry, Vol. 424, No. 1, 2012, pp. 12-20. doi:10.1016/j.ab.2012.02.016
- P. Nadal, A. Pinto, M. Svobodova and N. Canela, “O’Sullivan CK. DNA Aptamers against the Lup an 1 Food Allergen,” PLoS One, 2012, Vol. 7, No. 4, Article ID: e35253. doi:10.1371/journal.pone.0035253
- H. S. Chang, K. H. Choo, B. Lee and S. J. Choi. “The Methods of Identification, Analysis, and Removal of Endocrine Disrupting Compounds (EDCs) in Water,” Journal of Hazardous Materials, Vol. 172, No. 1, 2009, pp. 1-12. doi:10.1016/j.jhazmat.2009.06.135
- N. Yildirim, F. Long, C. Gao, M. He, H. C. Shi and A. Z. Gu, “Aptamer-Based Optical Biosensor for Rapid and Sensitive Detection of 17β-Estradiol in Water Samples,” Environmental Science & Technology, 2012, Vol. 46, No. 6, pp. 3288-3294.
- M. Jo, J. Y. Ahn, J. Lee, S. Lee, S. W. Hong, J. W. Yoo, J. Kang, P. Dua, D. K. Lee, S. Hong and S. Kim, “Development of Single-Stranded DNA Aptamers for Specific Bisphenol a Detection,” Oligonucleotides, Vol. 21, No. 2, 2011, pp. 85-91. doi:10.1089/oli.2010.0267
- M. Kim, H. J. Um, S. Bang, S. H. Lee, S. J. Oh, J. H. Han, K. W. Kim, J. Min and Y. H. Kim, “Arsenic Removal from Vietnamese Groundwater Using the Arsenic-Binding DNA Aptamer,” Environmental Science & Technology, 2009, Vol. 43, No. 24, pp. 9335-9340. doi:10.1021/es902407g
- L. Li, B. Li, Y. Qi and Y. Jin, “Label-Free AptamerBased Colorimetric Detection of Mercury Ions in Aqueous Media Using Unmodified Gold Nanoparticles as Colorimetric Probe,” Analytical and Bioanalytical Chemistry, 2009, Vol. 393, No. 8, pp. 2051-2057. doi:10.1007/s00216-009-2640-0
- Y. Helwa, N. Dave, R. Froidevaux, A. Samadi and J. Liu, “Aptamer-Functionalized Hydrogel Microparticles for Fast Visual Detection of Mercury(II) and Adenosine,” ACS Applied Materials Interfaces, Vol. 4, No. 4, 2012, pp. 2228-2233. doi:10.1021/am300241j
- X. G. Hu, K. L. Tulsieram, Q. X. Zhou, L. Mu and J. P. Wen, “Polymeric Nanoparticle-Aptamer Bioconjugates Can Diminish the Toxicity of Mercury in Vivo,” Toxicology Letters, Vol. 208, No. 1, 2012, pp. 69-74. doi:10.1016/j.toxlet.2011.10.006
- J. Sinha, S. J. Reyes and J. P. Gallivan, “Reprogramming Bacteria to Seek and Destroy an Herbicide,” Nature Chemical Biology, Vol. 6, No. 6, 2010, pp. 464-470. doi:10.1038/nchembio.369
- J .He, Y. Liu, M. Fan and X. Liu, “Isolation and Identification of the DNA Aptamer Target to Acetamiprid,” Journal of Agricultural and Food Chemistry, Vol. 59, No. 5, 2011, pp. 1582-1586. doi:10.1021/jf104189g
- L. Wang, X. Liu, Q. Zhang, C. Zhang, Y. Liu, K. Tu and J. Tu, “Selection of DNA Aptamers That Bind to Four Organophosphorus Pesticides,” Biotechnology Letters, Vol. 34, No. 5, 2012, pp. 869-874. doi:10.1007/s10529-012-0850-6
- Z. Tang, P. Parekh, P. Turner, R. W. Moyer and W. Tan. “Generating Aptamers for Recognition of Virus-Infected cells,” Clinical Chemistry, Vol. 55, No. 4, 2009, pp. 813- 822. doi:10.1373/clinchem.2008.113514
- M. Minunni, S. Tombelli, A. Gullotto, E. Luzi and M. Mascini, “Development of Biosensors with Aptamers as Bio-Recognition Element: The Case of HIV-1 Tat Protein,” Biosensors and Bioelectronics, Vol. 20, No. 6, 2004, pp. 1149-1156. doi:10.1016/j.bios.2004.03.037
- S. Tombelli, M. Minunni, E. Luzi and M. Mascini, “Aptamer-Based Biosensors for the Detection of HIV-1 Tat Protein,” Bioelectrochemistry, Vol. 67, No. 2, 2005, pp. 135-141. doi:10.1016/j.bioelechem.2004.04.011
- S. Lee, Y. S. Kim, M. Jo, M. Jin, D. K. Lee and S. Kim, “Chip-Based Detection of Hepatitis C Virus Using RNA aptamers That Specifically Bind to HCV Core Antigen,” Biochemical and Biophysical Research Communications, Vol. 358, No. 1, 2007, pp. 47-52. doi:10.1016/j.bbrc.2007.04.057
- M. Ikanovic, W. E. Rudzinski, J. G. Bruno, A. Allman, M. P. Carrillo, S. Dwarakanath, S. Bhahdigadi, P. Rao, J. L. Kiel and C. J. Andrews, “Fluorescence Assay Based on Aptamer-Quantum Dot Binding to Bacillus thuringiensis Spores,” Journal of Fluorescence, Vol. 17, No. 2, 2007, pp. 193-199. doi:10.1007/s10895-007-0158-4
- H. M. So, D. W. Park, E. K. Jeon, Y. H. Kim, B. S. Kim, C. K. Lee, S. Y. Choi, S. C. Kim, H. Chang and J. O. Lee, “Detection and Titer Estimation of Escherichia Coli Using Aptamer-Functionalized Single-Walled Carbon-Nanotube Field-Effect Transistors,” Small, Vol. 4, No. 2, 2008, pp. 197-201. doi:10.1002/smll.200700664
- L. N. Cella, P. Sanchez, W. Zhong, N. V. Myung, W. Chen and A. Mulchandani, “Nano Aptasensor for Protective Antigen Toxin of Anthrax,” Analytical Chemistry, 2010, Vol. 82, No. 5, pp. 2042-2047. doi:10.1021/ac902791q
NOTES
*All authors have contributed equally.
#Corresponding author.

