Advances in Microbiology
Vol.4 No.3(2014), Article ID:42785,8 pages DOI:10.4236/aim.2014.43019
The Optimization of Aspergillus sp. GM4 Tannase Production under Submerged Fermentation
1Department of Biology, Sector of Microbiology, Federal University of Lavras, Lavras, Brazil
2Department of Biology, University of São Paulo, Ribeirão Preto, Brazil
3Department of Food Science, Federal University of Lavras, Lavras, Brazil
4Department of Phytopathology, Federal University of Lavras, Lavras, Brazil
Email: *patricia@dbi.ufla.br
Copyright © 2014 Alessandra Gonçalves de Melo et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In accordance of the Creative Commons Attribution License all Copyrights © 2014 are reserved for SCIRP and the owner of the intellectual property Alessandra Gonçalves de Melo et al. All Copyright © 2014 are guarded by law and by SCIRP as a guardian.
Received December 17, 2013; revised January 17, 2014; accepted January 25, 2014
KEYWORDS
Aspergillus sp.; Response Surface Methodology; Submerged Fermentation; Tannase; Tannic Acid
ABSTRACT
Tannase is a hydrolytic enzyme that is involved in the biodegradation of tannins and it has biotechnological potential in the pharmaceutical, chemical, food and beverage industries. Microorganisms, especially filamentous fungi, are important tannase producers. The aims of this work were to find a potential tannase producer and to improve the cultivation conditions. Three Aspergillus species (A. japonicus 246A, A. tamarii 3 and Aspergillus sp. GM4) were investigated in different culture media (Adams, Czapeck, Khanna, M5 and Vogel) and inducers (1% and 2% tannic acid; 1% green tea; 1% methyl gallate; 1% gallic acid). Aspergillus sp. GM4 and Adams medium were selected. The tannase production by Aspergillus sp. GM4 in Adams medium was induced in the presence of 2% (w/v) tannic acid and gallic acid as carbon sources, while green tea was not able to induce tannase production. The Plackett-Burman screening design was performed with the variables MgSO4, KH2PO4, yeast extract, tannic acid, agitation rate and salt solution. The variables MgSO4 and agitation rate were selected for the optimization of tannase production using a Central Composite Rotatable Design. Under optimized conditions, a 2.66-fold increase in the enzyme production was observed with small modifications in the medium composition.
1. Introduction
Tannase (EC 3.1.1.20) is a hydrolytic enzyme involved in the biodegradation of tannins. This enzyme catalyzes the hydrolysis of the ester and depside bonds present in hydrolysable tannins, releasing gallic acid and glucose [1]. Tannase also has many applications in food, beverage, pharmaceutical and chemical industries, and even in bioremediation [2,3]. Industrial bioconversion of tannic acid is generally accomplished by tannase for the production of gallic acid, which is mostly used in the pharmaceutical industry to produce the anti-bacterial drug Trimethoprim [4]. Gallic acid is also an important substrate for the synthesis of propyl gallate, an antioxidant used in the food industry [1].
Microorganisms can continuously produce tannase in large quantities, resulting in an increased yield with the establishment of optimized fermentation methods. Tannase is produced in the presence of tannic acid by several filamentous fungi, mainly Aspergillus and Penicillium species [4]. Industrial production of microbial tannase is obtained by submerged culture because this simplifies sterilization and process control [5]. Studies evaluating the cultivation of tannase-producing strains are important for choosing strains and enzymes with desirable characteristics for industrial applications. The optimization of enzyme production is an important step that can be performed by individually optimizing each parameter (temperature, cultivation time and pH, among others), as has been done for Aspergillus flavus tannase production [6], or by using the factorial design methodology, as has been done for Bacillus massiliensis tannase production [7]. This article describes the optimization of tannase production under submerged fermentation by Aspergillus sp. GM4, a new strain producer, using the Response Surface Methodology (RSM).
2. Material and Methods
2.1. The Selection of the Microorganism and Culture Medium
Three Aspergillus species (A. japonicus 246A, A. tamarii 3 and Aspergillus sp. GM4) belonging to the culture collection of the Laboratory of Prospection and Genetics of Fungi (Department of Biology, Federal University of Lavras, Lavras, MG, Brazil) were previously selected due to their ability to produce tannase in tannic acid agar. The submerged fermentations for tannase production were conducted using 25 mL of Khanna [8], Czapeck [9], M5 [10], Adams [11] or Vogel [12] media in 125 mL Erlenmeyer flasks containing 2% (w/v) tannic acid as the main carbon source, pH 6.0. The media were previously autoclaved at 120˚C and 1.5 atm for 20 min and were inoculated with a spore suspension at 1 × 105 conidia/mL of medium. Erlenmeyer flasks containing the media were incubated at 30˚C for 72 h on a rotary shaker (100 rpm). Then, the biomass was separated by vacuum filtration through Whatman N˚ 1 filter paper, and the cell-free culture broth was used to measure the extracellular tannase activity.
2.2. The Effect of the Incubation Time on Tannase Production
A spore suspension of Aspergillus sp. GM4 (1 × 105 conidia/mL) was inoculated in modified Adams medium (pH 6.0) with the following composition (g/L): yeast extract, 2.0; KH2PO4, 1.0; MgSO4·7H2O, 0.5; and tannic acid, 20.0. Erlenmeyer flasks were incubated at 30˚C and 100 rpm for 24, 48, 72, 96 and 120 h. After each incubation period, the culture filtrate, obtained as described above, was analyzed for tannase activity.
2.3. The Effect of Inducers on the Tannase Activity
The fungus Aspergillus sp. GM4 was initially grown at 30˚C for 48 h (100 rpm) in the modified Adams medium containing 2% (w/v) glucose as a carbon source. Then, the mycelia mass was separated by vacuum filtration, washed with sterilized distilled water, transferred in aseptic conditions to the modified Adams medium with the appropriate inducers (w/v) (1% and 2% tannic acid; 1% green tea; 1% methyl gallate; 1% gallic acid) and incubated at 30˚C for 48 h and 100 rpm. The mycelia mass was also transferred to a flask containing 1% (w/v) glucose and to a control flask without a carbon source. After the incubation time, the tannase activity was measured, and the induction ratio was calculated by dividing the induced tannase activity by the basal tannase activity from the medium containing glucose as the sole carbon source [13].
2.4. Enzyme Assay
The tannase activity was estimated using a method described by Deschamps [14]. One unit of tannase activity was defined as the amount of enzyme required to release one µmol of gallic acid per minute under the standard assay conditions.
2.5. Protein Estimation
The protein content in the culture filtrate was estimated using the Bradford assay [15] with BSA as a standard.
2.6. Optimization Procedure and Experimental Designs: Plackett-Burman Design, Central Composite Rotatable Design and Response Surface Analysis
To determine which variables significantly influence tannase production by Aspergillus sp. GM4 under submerged fermentation, a Plackett-Burman (PB) screening design was used [16,17]. Six independent variables (MgSO4·7H2O, KH2PO4, yeast extract, tannic acid, agitation rate and salt solution) were investigated, and each variable was tested at low (−1) and high (+1) levels, with three more repetitions at the central point in 15 experimental runs (Table 1). The initial concentrations of the salts were as follows (g/L): ZnSO4·7H2O, 0.0035; CuSO4·5H2O, 0.0031; MnSO4·H2O, 0.0069; FeSO4, 0.0033; and KCl, 0.049. A 1000-fold concentrate solution was prepared, and the appropriate dilution was performed to test the initial concentration and the double of this concentration. Fermentation was carried out by inoculating the media with a spore suspension at 1 × 105 conidia/mL medium at 30˚C for 72 hours at a specified agitation rate (80, 100 and 120 rpm). The medium conditions are shown in the matrix (Table 1). For each assay, the tannase production was measured and calculated in terms of U/mL. The software STATISTICA 8.0 (Stat Soft. Inc.®, Tulsa—Ok, USA) was used to analyze the experimental data. The variables with significant effects on tannase production were those with a significance level less than 10% (p-value < 0.1). These variables were selected for a Central Composite Rotatable Design.
The experiments were based on a Central Composite Rotatable Design (CCRD) with the two variables selected by the PB design. A CCRD with a total of 11 experiments was adopted, and four factorial points and three central points were included to verify the curvature of the experimental region and to provide additional degrees of freedom for error, which are important for testing the significance of effects [18]. The independent variables, their levels and real values are presented in Table 2. In addition, four axial points (at a distance of
±1.41 from the central point) were added to the matrix to fit this design to a second order model. The tannase activity was taken as the dependent variable (Y). The tannase production data were subjected to analysis of variance (ANOVA) to examine the adjustment of the model, and the deviation between the predicted and observed values was calculated. The proportion of variance explained by the model is given by the coefficient of determination (R2). The mathematical relationship between the independent and dependent variables was obtained using a second order polynomial Equation (1):
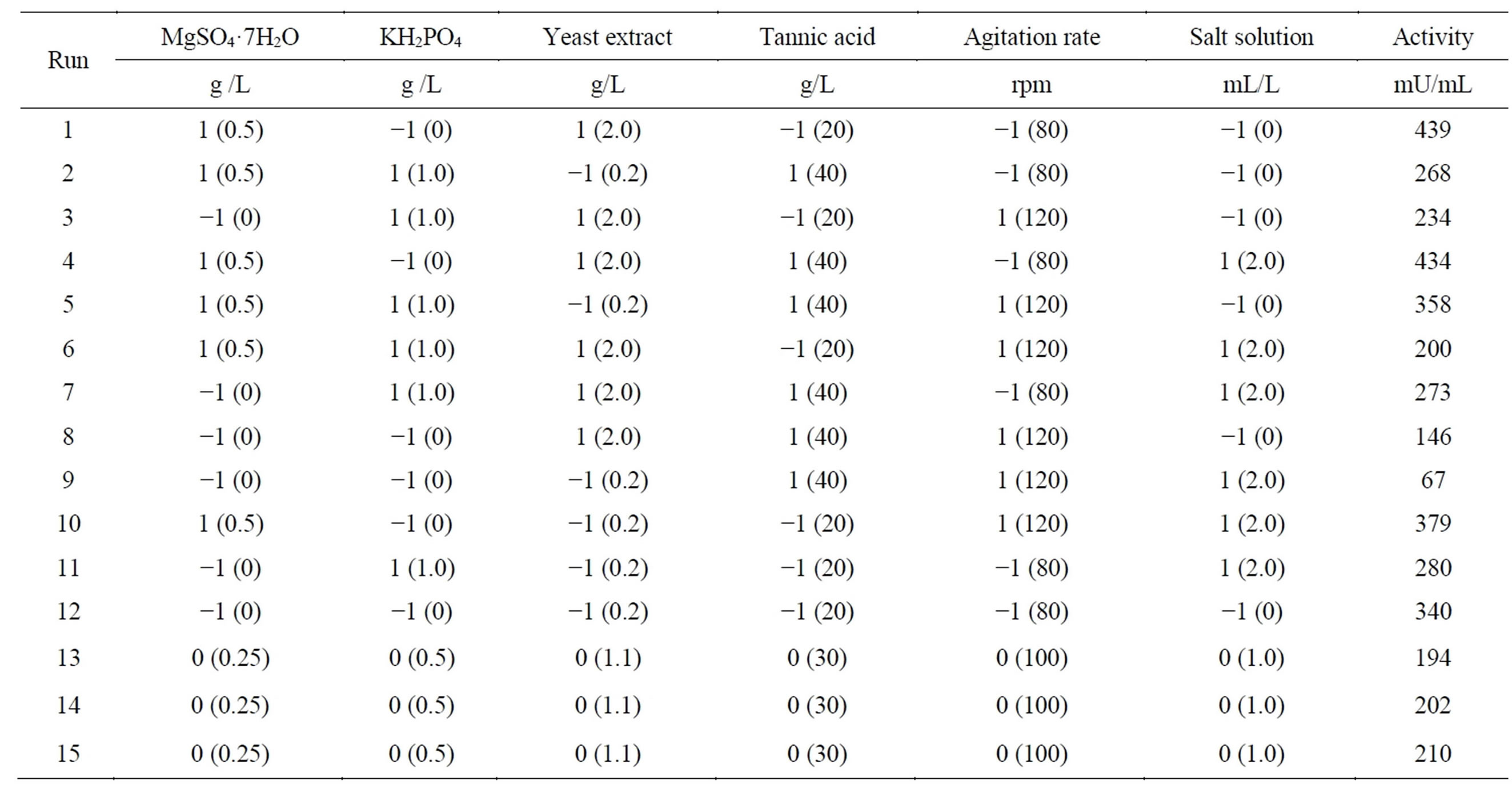
Table 1. Plackett-Burman design matrix with six independent variables containing real and coded values and tannase activity as the response.
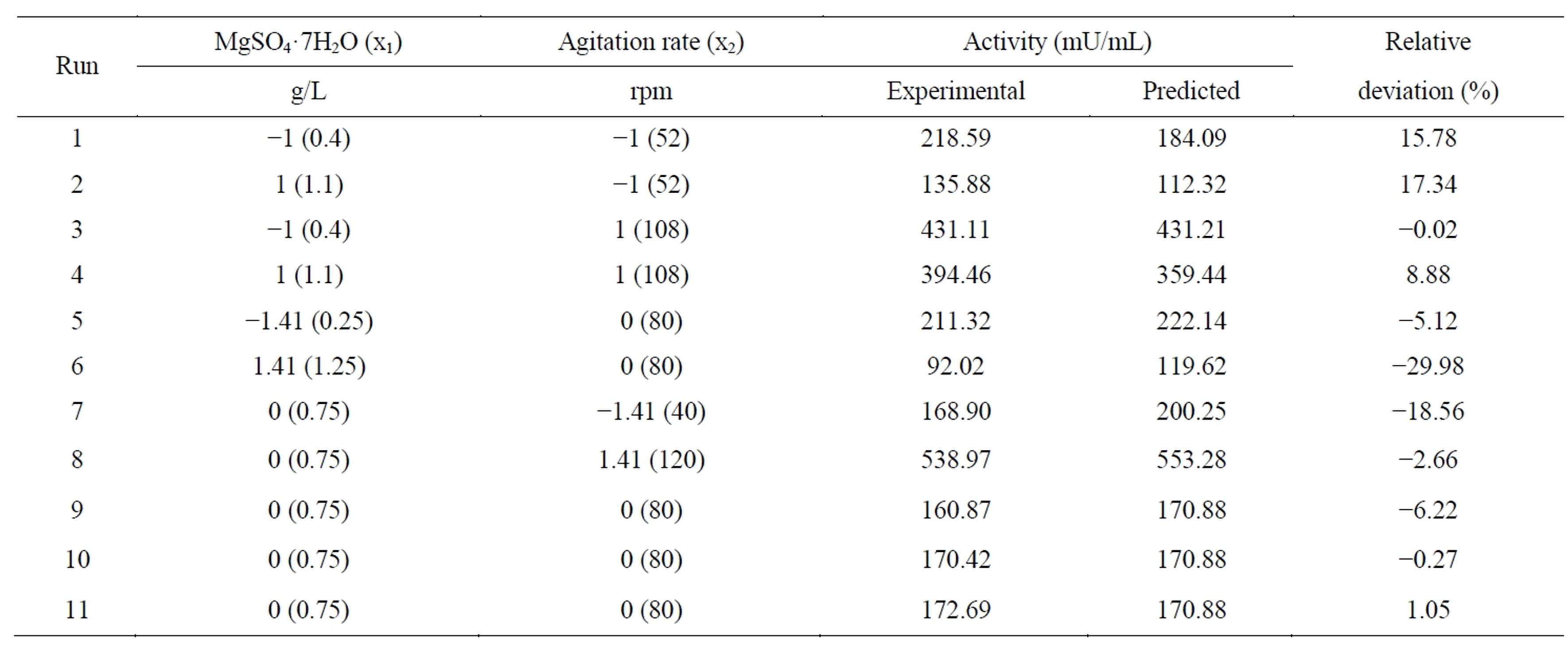
Table 2. The levels of the two independent variables and the central composite rotatable design for the optimization of tannase activity.
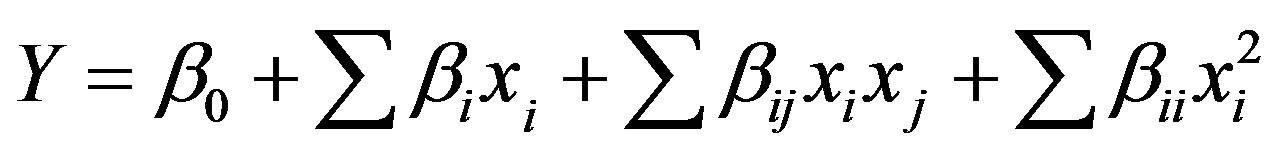 (1)
(1)
where Y is the predicted value, represented by the enzymatic activity; β0 is the constant term; βi is the linear coefficient; βij is the interaction coefficient; βii is the quadratic coefficient; and xi and xj are the independent variables in the coded values. The significance of all terms in the polynomial function was assessed statistically using the F-value at a probability (p) of 0.05. The model was then used to generate response surfaces and contour curves to determine the optimal operating ranges of the independent variables.
3. Results
Three species of Aspergillus genera were cultured in Adams, Czapeck, Khanna, M5 and Vogel culture media. Adams medium was more favorable for enzyme production for Aspergillus japonicus 246A (19.8 U/mg) and Aspergillus sp. GM4 (12.0 U/mg) (Table 3), followed by Khanna and Czapeck. M5 medium was not favorable for tannase production, which could have been due to the composition of the medium, with yeast extract and peptone as additional carbon sources. A. tamarii 3 presented a lower tannase activity compared to the other species, with a maximum production of 3.0 U/mg in Czapeck medium. Protein content of and tannase production by Aspergillus japonicus 246A and Aspergillus sp. GM4 were very similar. Many studies have been conducted with A. japonicus for tannase production. Therefore, we selected Aspergillus sp. GM4 for optimization of tannase production culture conditions.
The potential of some substrates to induce tannase activity was measured (Table 4). The Aspergillus sp. GM4 was initially cultured in modified Adams medium containing 2% (w/v) glucose, and a basal tannase activity (1.29 U/mg) was observed. The mycelia mass was transferred to modified Adams medium containing gallic acid, green tea, methyl gallate, tannic acid or glucose to examine induction or repression by these carbon sources.
Tannase activity increased 3.25-fold when Aspergillus sp. GM4 was grown with 2% (w/v) tannic acid compared to 1% (w/v). Methyl gallate presented a higher induction ratio than all of the other additives except 2% (w/v) tan nic acid. In the presence of green tea, Aspergillus sp. GM4 presented low tannase activity. No activity was detected after 48 h incubation in the medium containing glucose, suggesting that tannase production by Aspergillus sp. GM4 is substrate induced and is repressed in the presence of glucose. Aspergillus sp. GM4 produced tannase with gallic acid as the sole carbon source in the medium. This result was interesting because it suggests that the tannase production by Aspergillus sp. GM4 can continue even with an increase in product content in the culture medium. Adams medium containing 2% (w/v)
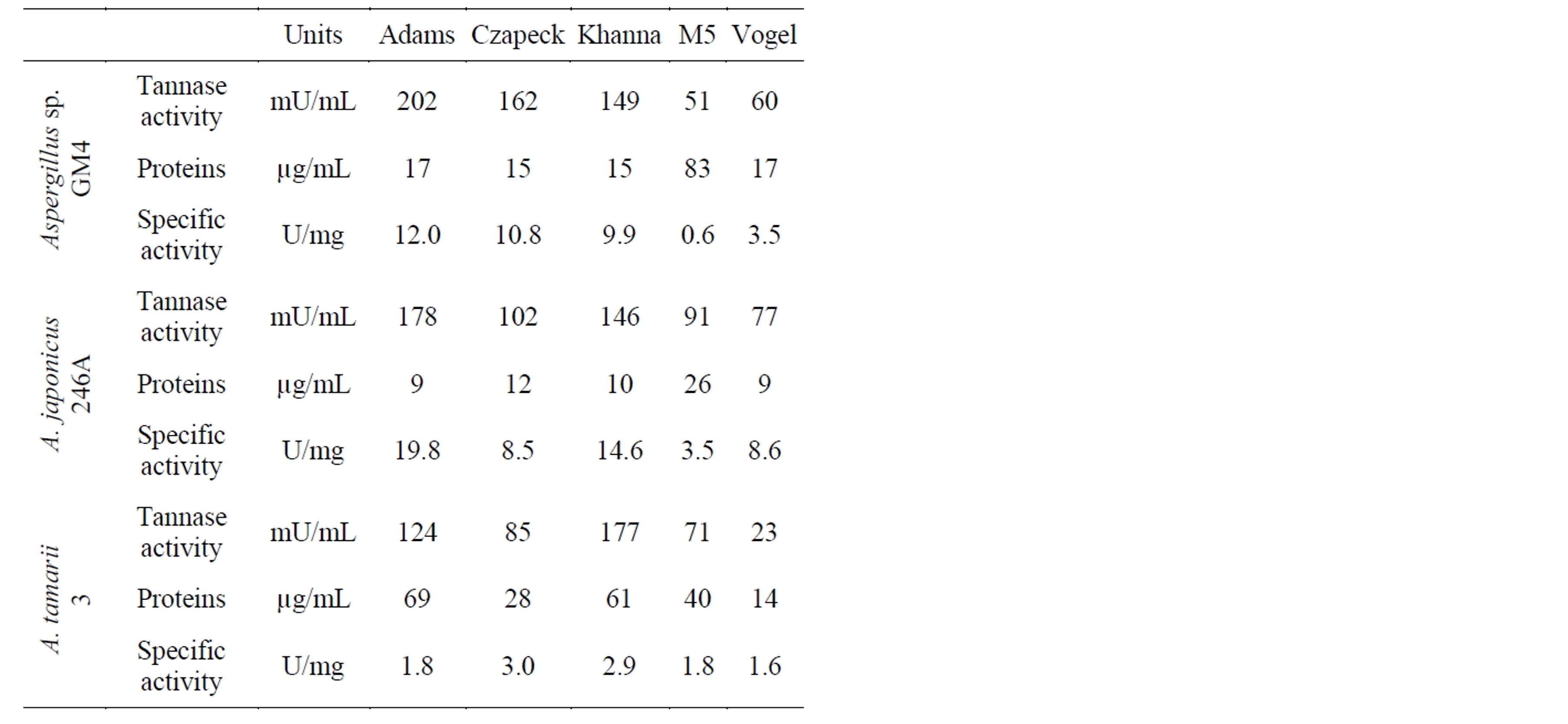
Table 3. Production of tannase by Aspergillus sp. GM4, A. japonicus 246A and A. tamarii 3 in different culture media.
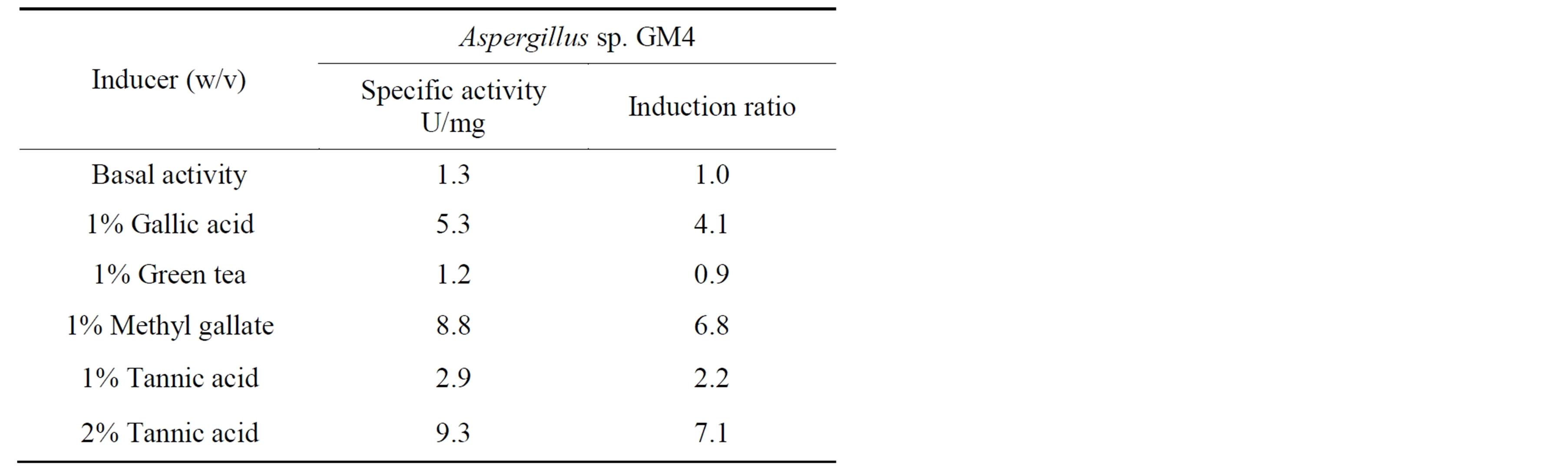
Table 4. The specific enzyme activity and the induction ratio in the presence of different inducers.
tannic acid was used to cultivate Aspergillus sp. GM4 for 24, 48, 72, 96 and 120 h. The extracellular tannase activity was the highest after 72 h of incubation, and the activity decreased after this period. Therefore, 72 h was fixed as the best incubation time for tannase production from Aspergillus sp. GM4.
The results of the PB design are shown in Table 1. The tannase activity varied from 67 mU/mL (run 9) to 439 mU/mL (run 1). The positive or negative effects of the independent variables MgSO4·7H2O, KH2PO4, yeast extract, tannic acid, agitation rate and salt solution were valued and are shown in Figure 1.
Yeast extract, salt solution, KH2PO4 and tannic acid have no statistically significant effect on tannase production (Figure 1). It was found that low concentrations of KH2PO4, tannic acid and salt solution increase tannase production, but the effects are not significant. Furthermore, the results indicate that the enzyme production by Aspergillus sp. GM4 is not affected by yeast extract concentration in the tested range (0.2 to 2.0 g/L).
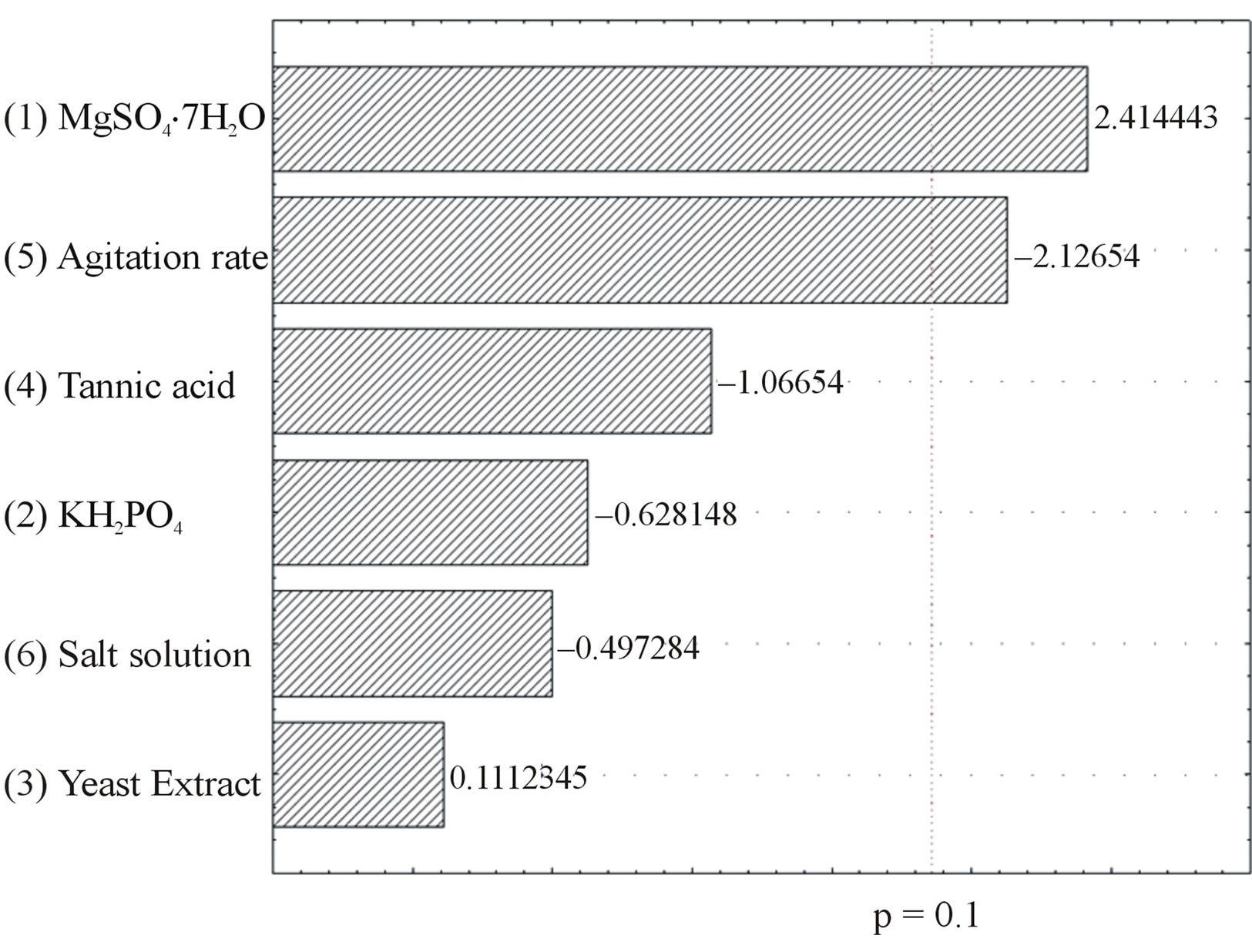
Figure 1. Pareto chart of the effects of the independent variables on tannase production by Aspergillus sp. GM4.
The variables MgSO4 and agitation rate have a significant effect on tannase production (p < 0.1), and these variables were selected for the CCRD. MgSO4 has shown a positive effect on enzyme production, indicating that an increase in this compound in the culture medium can favor enzyme production. The agitation rate showed a significant negative effect, which indicates that lower agitation levels yield higher levels of tannase production. The salt solution was not part of the initial composition of the culture medium and was removed from the medium for the next step to minimize the enzyme production costs.
The tannase characterization of Aspergillus sp. GM4 (data not shown) showed that the tannase activity is slightly inhibited in the presence of some metal ions (1 mM), such as K+, Zn2+, Mn2+, and Fe3+, with a greater effect for Mn2+. Although supplementing the medium with trace elements can be important for fungal growth, the presence of these metal ions may have negative effects on the secreted enzyme activity. Yeast extract, KH2PO4 and tannic acid were fixed. From this point, the medium was modified to the following composition (g/L): tannic acid, 20.0; KH2PO4, 0.5 and yeast extract, 1.0. The pH was adjusted to 6.0, and the medium was incubated at 30˚C. The MgSO4 concentration and the agitation rate were studied. The CCRD matrix containing the assays and the predicted and observed results for each assay are presented in Table 2.
The estimated effect for each variable and the interactions between the variables were determined and are reported in Table 5. Both variables are relevant to tannase production at a 95% confidence level. The correlation analysis has shown that the MgSO4 interaction versus the agitation rate and the MgSO4 quadratic term were not statistically significant. The agitation rate quadratic term and both linear terms are significant, which affects the curvature in the response surface plot. The analyses of the results show that the deviations were low in the desirable range where the activity value was high.
Table 6 shows the analysis of variance (ANOVA) for tannase production. The terms with no significance were removed from the ANOVA. The results of the ANOVA show good reproducibility of the data observed with a very low error value. The determination coefficient (R2) explained 97.6% of the variability of the data, and the F-test results demonstrated that the fit of the model was very good (F-regression >3.71).
The regression analysis of the experimental data obtained after the ANOVA resulted in the adjusted second order polynomial Equation (2) below:
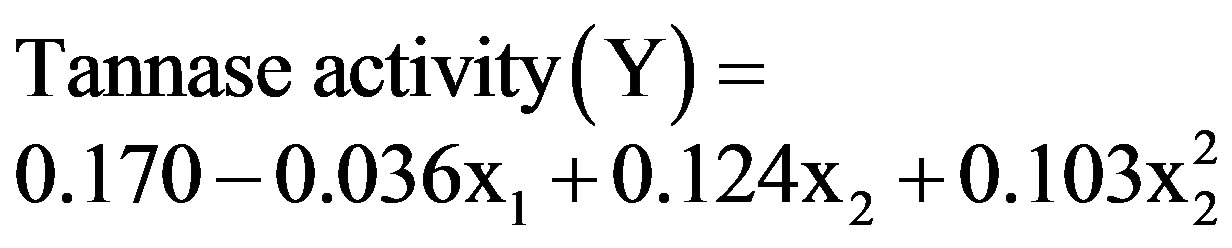 (2)
(2)
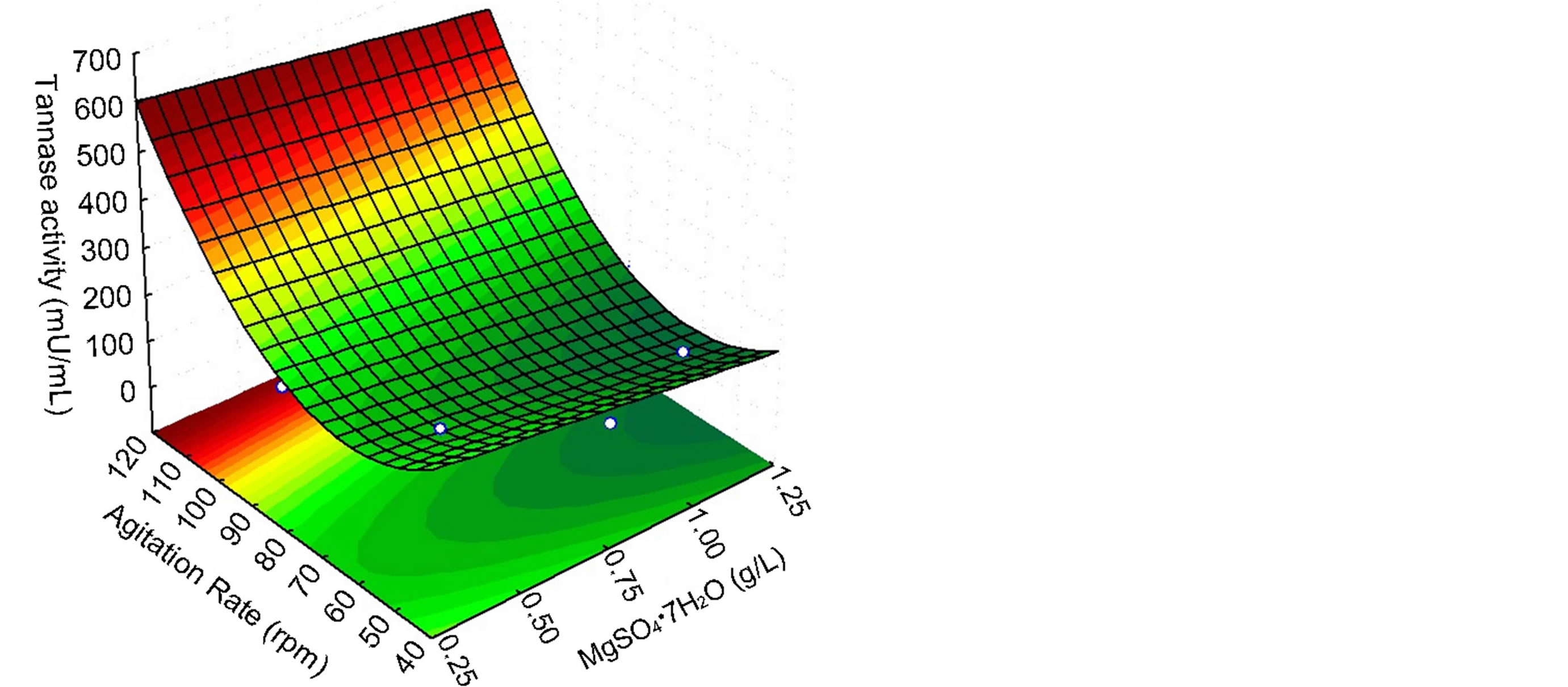
where Y is the tannase produced as a function of the coded levels of MgSO4·7H2O concentration (x1) and agitation rate (x2). The predicted values and the relative deviation between the real and predicted response for each assay are presented in Table 2, and the response surface and contour curve with the influence of the variables MgSO4 and agitation rate on enzyme activity are presented in Figure 2.
Figure 2 shows that the optimum conditions for tannase production are at the highest levels of agitation (between 110 and 120 rpm) and that the variations in the MgSO4 levels do not exhibit a greater influence on the tannase activity. This result indicates that the MgSO4 levels should be between 0.25 and 0.5 g/L. These results are different from the previous ones from the PlackettBurman design, most likely due to differences in the tested ranges.
In this simplified low cost medium, Aspergillus sp. GM4 tannase demonstrated an activity of 539 mU/mL, representing a 2.66-fold increase compared to the activity obtained before applying the experimental design. The experimental design reduced the number of medium components and slightly increased the tannase production.
4. Discussion
Tannase production by microorganisms such as bacteria and fungi has been conducted under different culture conditions to optimize the enzyme yield. The extracellular tannase production by Aspergillus sp. GM4 was 4-fold higher than that observed for A. tamarii 3 in the most optimal culture medium. Despite the higher enzymatic production of A. japonicus 246A compared to Aspergillus sp. GM4 in Adams medium, the latter was selected for the optimization of tannase production. The A. japonicus tannase is a well-characterized enzyme [19]. The influence of the medium composition on tannase production was also verified for A. flavus cultivation
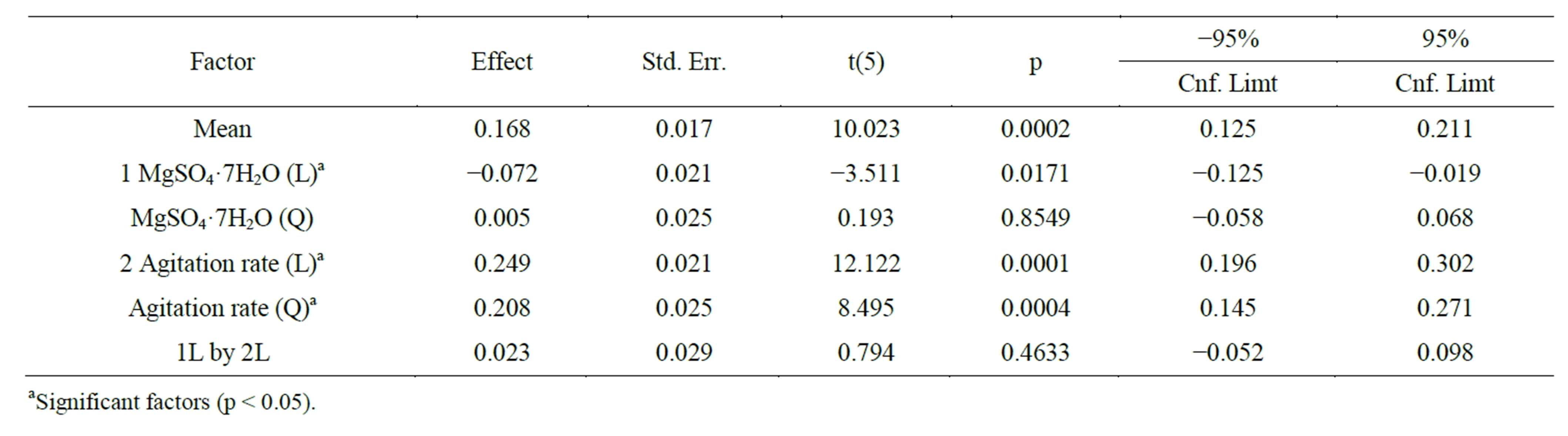
Table 5. Effect estimates for extracellular tannase production by Aspergillus sp. GM4.

Table 6. Analysis of variance for tannase production.
using tannic acid as a carbon source [6]. Generally, the production of this enzyme occurs during the first steps of microbial growth because tannase is a primary metabolite [13,20]. According to Aguilar et al. [21], the maximal tannase production by Aspergillus sp. is attained between 1 to 3 days of cultivation, as observed for the Aspergillus sp. GM4 extracellular tannase. The decrease in the tannase production after three days of cultivation may be due to the reduction in substrate availability and the secretion of proteases that can act on secreted tannase.
Tannase can be produced either constitutively or under induction, depending on the strain and fermentation conditions. The enzyme production by Aspergillus sp. GM4 was induced in the presence of 2% (w/v) tannic acid, methyl gallate and gallic acid. However, a basal level of tannase production was observed when glucose was used as a carbon source. According to Bradoo et al. [19], the fungus A. japonicus produced tannase constitutively in culture medium containing simple or complex sugars, but the production is increased when tannic acid is the sole carbon source [19]. There is some controversy regarding the tannase regulation mechanism and the specific roles of some compounds in the induction and repression of tannase expression. Tannic acid cannot act directly as an inducer because tannic acid is a large molecule that cannot cross the cell membrane, and there is no known tannic acid transporter. Therefore, the basal levels of tannase synthesis are important for initiating tannic acid hydrolysis to release the intermediary compounds that can act as true inducers [3]. In addition, some studies have shown that up to 0.2% (w/v) glucose concentration may favor the production of tannase, but
 (a)
(a)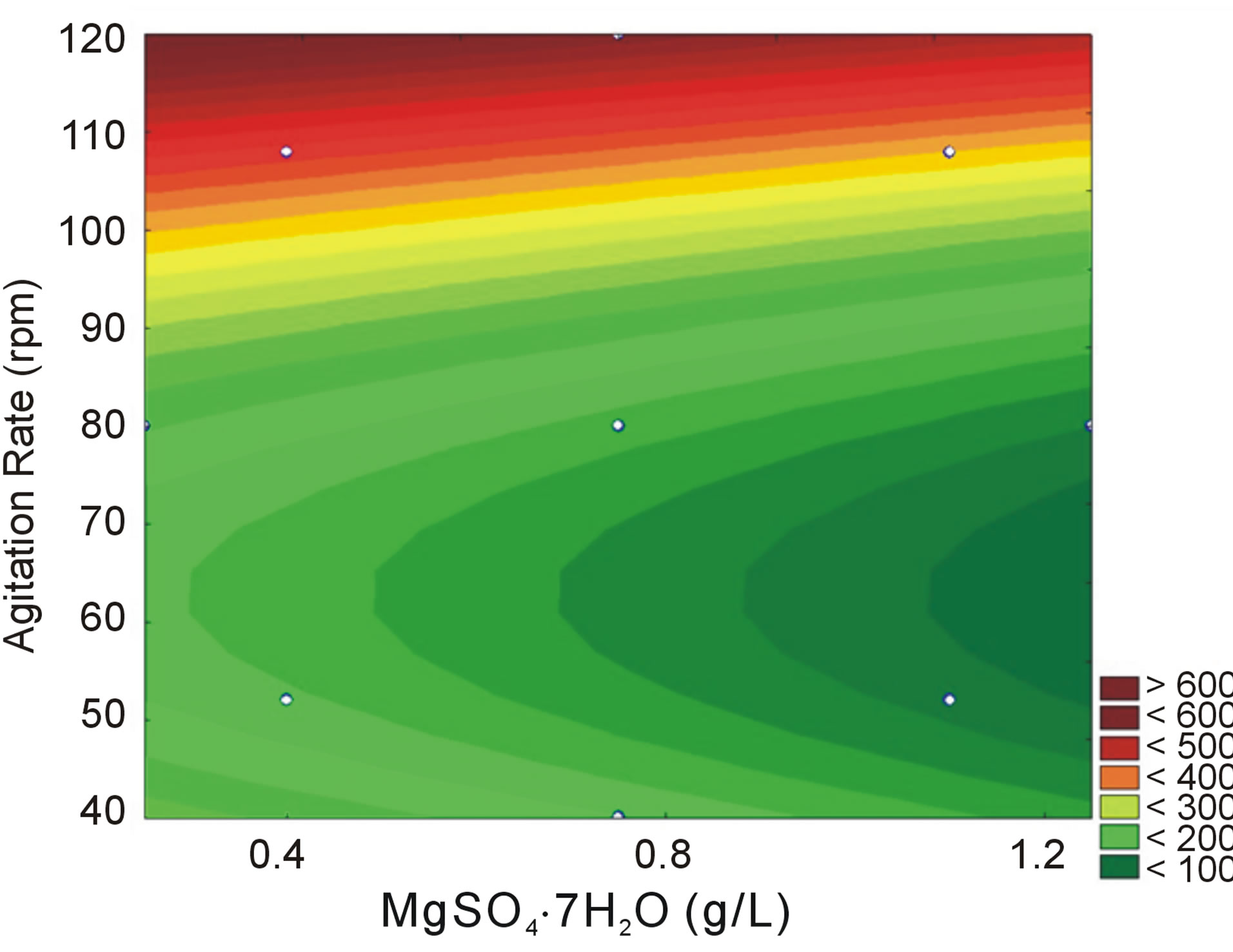 (b)
(b)
Figure 2. Three-dimensional response surface plot and contour curve for tannase activity showing the effects of agitation rate and MgSO4·7H2O.
above 2% (w/v), this carbon source exhibits a strong catabolic repression of enzyme synthesis [13].
Over cultivation time, the substrate availability decreases while there is an increase in the hydrolysis product. This condition may be unfavorable if the enzyme production is regulated by the end-product. The tannase production by A. niger [13] and A. japonicus [19] are repressed by gallic acid, suggesting that the enzyme production in these strains are regulated in this manner. In contrast, Aspergillus sp. GM4 tannase production is induced by gallic acid in cultivation medium, which demonstrates that the presence of the product in the medium (at the tested concentrations) does not repress the production process. This response to gallic acid was also observed for tannase produced by Emericella nidulans [22] and Bacillus massiliensis [7]. Gallic acid has been used to induce tannase production and has also been linked to the regulation of this process.
The use of statistical methodology to optimize the culture conditions and to improve enzyme production has been a useful tool for biotechnology and has recently received more attention. Some important parameters should be considered for microorganism growth and secretion of biotechnological products of industrial interest. The RSM can be used to analyze the influence of different parameters on enzyme production, to determine the statistical significance level, and to examine the statistical interactions among the factors involved in the process. The “one-at-a-time” approach showed that four factors (tannic acid, sodium nitrate, agitation rate and incubation period) have the most influence on tannase production in A. niger, and the use of the CCRD and RSM approaches increased the enzymatic production 2-fold (from 9.8 U/mL to 19.7 U/mL) [23]. However, the tannase production by Aspergillus sp. GM4 increased 2.66-fold using the RSM under optimized conditions in submerged fermentation. Tannase production in submerged fermentation by Aspergillus sp. improves at high aeration rates [20], as observed in studies with A. niger and Aspergillus sp. GM4. The agitation rate shows an important effect on the supply of nutrients, especially oxygen, for filamentous fungi cultivation. Furthermore, good mixing, mass and heat transfer require a threshold level of agitation. However, high agitation rates can lead to high energy dissipation rates and to high shear stress, which may result in fragmentation and cell and mycelial network damage [24]. Therefore, it is important to find the optimal agitation rate for enzyme production without inducing mycelium damage.
Others factors that are important to tannase production are the supplementation of the culture medium with an adequate concentration of carbon, nitrogen and mineral source (besides cultivation in an ideal pH), temperature and incubation time. Aspergillus sp. GM4 tannase production was increased with low MgSO4 supplementation. Many enzymes require metal ion activators to achieve full catalytic activity. At low concentrations, metal ions act as enzymatic cofactors, thereby increasing the catalytic activity of the enzyme, whereas at high concentrations, metal ions can reduce catalytic activity [25]. Mg2+ is a well-known enzyme cofactor. The supplementation of the culture medium with salts containing magnesium is important for adequate action of magnesium-dependent enzymes, which are present in the general metabolic pathways and in synthesis of nucleic acids.
5. Conclusion
The Adams medium favored the Aspergillus sp. GM4 tannase production and is a low cost medium compared to the other tested culture media. Aspergillus sp. GM4 produces tannase in the presence of tannic acid and methyl gallate inducers. Gallic acid as the sole carbon source induced tannase production. This result suggests that tannase production by Aspergillus sp. GM4 can continue in the presence of increasing product in the culture medium and that the end product does not regulate enzyme production. In addition, the Plackett-Burman and Central Composite Rotatable designs were shown to be effective in reducing the number of components in the medium composition and increased tannase production 2.66-fold.
Acknowledgements
The authors sincerely thank the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG, Brazil), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil) and Conselho de Desenvolvimento Científico e Tecnológico (CNPq, Brazil) for the financial aid that made the development of the present work possible.
REFERENCES
- P. K. Lekha and B. K. Lonsane, “Production and Application of Tannin Acyl Hydrolase: State of the Art,” In: S. Neidleman and A. Laskin, Eds., Advances in Applied Microbiology, Academic Press, Inc., San Diego, 1997, pp. 215-260.
- S. Sharma, L. Agarwal and R. K. Saxena, “Purification, Immobilization and Characterization of Tannase from Penicillium variable,” Bioresource Technology, Vol. 99, No. 7, 2008, pp. 2544-2551. http://dx.doi.org/10.1016/j.biortech.2007.04.035
- M. Chávez-González, L. V. Rodríguez-Durán, N. Balagurusamy, A. Prado-Barragán, R. Rodríguez, J. C. Contreras and C. N. Aguilar, “Biotechnological Advances and Challenges of Tannase: An Overview,” Food Bioprocess Technology, Vol. 5, No. 2, 2012, pp. 445-459. http://dx.doi.org/10.1007/s11947-011-0608-5
- B. Bajpai and S. Patil, “Induction of Tannin Acyl Hydrolase (EC 3.1.1.20) Activity in Some Members of Fungi imperfecti,” Enzyme and Microbial Technology Enzyme and Microbial Technology, Vol. 20, No. 8, 1997, pp. 612-614. http://dx.doi.org/10.1016/S0141-0229(96)00206-2
- B. Kar and R. Banerjee, “Biosynthesis of Tannin Acyl Hydrolase from Tannin-Rich Forest Residue under Different Fermentation Conditions,” Journal of Industrial Microbiology and Biotechnology, Vol. 25, No. 1, 2000, pp. 29-38. http://dx.doi.org/10.1038/sj.jim.7000011
- R. Paranthaman, R. Vidyalakshmi, S. Murugesh and K. Singaravadivel, “Optimization of Various Culture Media for Tannase Production in Submerged Fermentation by Aspergillus flavus,” Advances in Biological Research, Vol. 3, 2009, pp. 34-39.
- P. D. Belur, R. Goud and D. C. Goudar, “Optimization of Culture Medium for Novel Cell-Associated Tannase Production from Bacillus massiliensis Using Response Surface Methodology,” Journal of Microbiology and Biotechnology, Vol. 22, No. 2, 2012, pp. 199-206. http://dx.doi.org/10.4014/jmb.1106.06004
- P. Khanna, S. S. Sundari and N. J. Kumar, “Production, Isolation and Partial Purification of Xylanases from an Aspergillus sp.,” World Journal of Microbiology and Biotechnology, Vol. 11, No. 2, 1995, p. 42. http://dx.doi.org/10.1007/BF00704661
- A. Wiseman, “Handbook of Enzyme Biotechnology,” John Wiley and Sons, Halsted Press, Inc., New York, 1985, p. 148.
- R. M. Peralta, H. F. Terenzi and J. A. Jorge, “Β-D-Glycosidase Activities of Humicola Grisea: Biochemical and Kinetic Characterization of a Multifunctional Enzyme,” Biochimica et Biophysica Acta, Vol. 1033, No. 3, 1990, pp. 243-249.
- P. R. Adams, “Mycelial Amylase Activities of Thermophilic Species of Rhizomucor, Humicola and Papulaspora,” Mycopathologia, Vol. 112, No. 1, 1990, pp. 35-37. http://dx.doi.org/10.1007/BF01795178
- H. J. Vogel, “Distribution of Lysine Pathways among Fungi: Evolutionary Implications,” The American Naturalist, Vol. 98, No. 903, 1964, pp. 435-446. http://dx.doi.org/10.1086/282338
- C. N. Aguilar, C. Augur, E. Favela-Torres and G. Viniegra-González, “Induction and Repression Patterns of Fungal Tannase in Solid-State and Submerged Cultures,” Process Biochemistry, Vol. 36, No. 6, 2001, pp. 565-570. http://dx.doi.org/10.1016/S0032-9592(00)00251-X
- A. Deschamps, G. Otuk and J. Lebeault, “Production of Tannase and Degradation of Chestnut Tannin by Bacteria,” Journal of Fermentation Technology, Vol. 61, No. 1, 1983, pp. 55-59.
- M. M. Bradford, “A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding,” Analytical Biochemistry, Vol. 72, 1976, pp. 248-254. http://dx.doi.org/10.1016/0003-2697(76)90527-3
- R. L. Plackett and J. P. Burman, “The Design of Optimum Multifactorial Experiments,” Biometrika, Vol. 33, No. 4, 1946, pp. 305-325. http://dx.doi.org/10.1093/biomet/33.4.305
- C. F. Silva, S. L. Arcuri, C. R. Campos, D. M. Vilela, J. G. Alves and R. F. Schwan, “Using the Residue of Spirit Production and Bio-Ethanol for Protein Production by Yeasts,” Waste Management, Vol. 31, No. 1, 2011, pp. 108-114. http://dx.doi.org/10.1016/j.wasman.2010.08.015
- J. F. M. Burkert, R. R. Maldonado, F. Maugeri Filho and M. I. Rodrigues, “Comparison of Lipase Production by Geotrichum candidum in Stirring and Airlift Fermenters,” Journal of Chemical Technology and Biotechnology, Vol. 80, No. 1, 2005, pp. 61-67. http://dx.doi.org/10.1002/jctb.1157
- S. Bradoo, R. Gupta and R. K. Saxena, “Parametric Optimization and Biochemical Regulation of Extracellular Tannase from Aspergillus japonicus,” Process Biochemistry, Vol. 32, No. 2, 1996, pp. 135-140. http://dx.doi.org/10.1016/S0032-9592(96)00056-8
- R. Belmares, J. Contrera-Esquivel, R. Rodriguez-Herrera, A. Coronel and C. N. Aguilar, “Microbial Production of Tannase: An Enzyme with Potential Use in Food Industry,” Lebensmittel-Wissenschaft Und-Technologie, Vol. 37, No. 8, 2004, pp. 857-864.
- C. N. Aguilar, R. Rodriguez, G. Gutierrez-Sanchez, C. Augur, E. Favela-Torres, L. A. Prado-Barragan, A. Ramirez-Coronel and J. C. Contreras-Esquivel, "Microbial Tannases: Advances and Perspectives,” Applied Microbiology and Biotechnology, Vol. 76, No. 1, 2007, pp. 47-59. http://dx.doi.org/10.1007/s00253-007-1000-2
- H. B. Gonçalves, A. J. Riul, H. F. Terenzi, J. A. Jorge and L. H. S. Guimarães, “Extracellular Tannase from Emericella nidulans Showing Hypertolerance to Temperature and Organic Solvents,” Journal of Molecular Catalysis B: Enzymatic Journal of Molecular Catalysis B: Enzymatic, Vol. 71, No. 1-2, 2011, pp. 29-35. http://dx.doi.org/10.1016/j.molcatb.2011.03.005
- S. Sharma, L. Agarwal and R. K. Saxena, “Statistical Optimization for Tannase Production from Aspergillus niger under Submerged Fermentation,” Indian Journal of Microbiology, Vol. 47, No. 2, 2007, pp. 132-138. http://dx.doi.org/10.1007/s12088-007-0026-6
- S. Kelly, L. H. Grimm, J. Hengstler, E. Schultheis, R. Krull and D. C. Hempel, “Agitation Effects on Submerged Growth and Product Formation of Aspergillus niger,” Bioprocess and Biosystems Engineering, Vol. 26, No. 5, 2004, pp. 315-323. http://dx.doi.org/10.1007/s00449-004-0368-y
- B. Kar, R. Banerjee and B. C. Bhattacharyya, “Effect of Additives on the Behavioral Properties of Tannin Acyl Hydrolase,” Process Biochemistry, Vol. 38, No. 9, 2003, pp. 1285-1293. http://dx.doi.org/10.1016/S0032-9592(02)00329-1
NOTES
*Corresponding author.

