Advances in Microbiology
Vol. 2 No. 2 (2012) , Article ID: 19664 , 17 pages DOI:10.4236/aim.2012.22007
Cholesterol Oxidase and Its Applications
Department of Biotechnology, Himachal Pradesh University, Shimla, India
Email: *kanwarss2000@yahoo.com
Received January 3, 2012; revised February 22, 2012; accepted March 2, 2012
Keywords: Microbial cholesterol oxidase; cholesterol; flavoprotein; diagnostic enzyme; steroids
ABSTRACT
Cholesterol oxidase (COX), a bi-functional FAD-containing microbial enzyme belongs to the family oxidoreductases. COX catalyses the oxidation of cholesterol into 4-cholesten-3-one. In recent time, cholesterol oxidase has received great attention due to its wider use in clinical (determination of serum cholesterol) laboratories practice and in the biocatalysis for the production of a number of steroids. COX has been shown to possess potent insecticidal activity, besides its use to track cell cholesterol. Moreover, COX is also implicated in the manifestation of some of the diseases of bacterial (tuberculosis), viral (HIV) and non-viral prion origin (Alzheimer’s). These applications and disease mechanisms have promoted the need of screening, isolation and characterization of newer microbes from diverse habitats as a source of COX to learn more about its structural and functional aspects. In this review, we discuss microbial sources of COX, its structure and important biochemical properties besides its broad range of biological functions and applications.
1. Introduction
Cholesterol oxidase (COX, EC 1.1.3.6) a monomeric bi-functional FAD-containing (flavoenzyme) enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with oxygen as acceptor. COX catalyzes the oxidation of 3β-hydrox-ysteroids and the isomerization of the intermediate, Δ5-6-ene- 3β-ketosteroid (cholest-5-en-3-one) to produce Δ3-4-ene- 3β-ketosteroid (cholest-4-en-3-one). The systematic name of this enzyme class is cholesterol: oxygen oxidoreductases and other common names in use are cholesterol-O2 oxidoreductases, 3β-hydroxy steroid oxidoreductases, and 3β-hydroxysteroid: oxygen oxidoreductases. COX was first isolated and characterized from Rhodococcus erythropolis [1]. Later, it was found to be produced by many bacteria both Gram-positive and Gram-negative, but the Actinomycetes come under most prolific COX-group.
Now the COX has become one of the most widely used enzymes in clinical laboratories as it participates in bile acid biosynthesis. COX enzyme is simple, specific, and highly sensitive; its use has become widespread in the determination of serum cholesterol that has direct implications in atherosclerosis, coronary heart disease and other lipid disorders, and for determining the risk of heart attack and thrombosis. However, this is only one of the applications of this type of oxidase. This enzyme that shows potent insecticidal activity has been used to track cell cholesterol and has also been found to be a potent parricide [2,3]. It has also been developed as a pest control in the agricultural industry especially in transgenic crops [2]. Cholesterol oxidase exists in two forms in Brevibacterium sterolicum, one in which the FAD cofactor was non-covalently bound to the enzyme (BCO1) and the other in which the cofactor was covalently linked (BCO2). Biochemical and structural studies of enzyme revealed the involvement of enzyme in interaction with lipid bilayer [4,5].
2. Production of Cholesterol Oxidase
The ability to decompose cholesterol is widespread among microorganisms that have been explored as free or immobilized cells [6,7] or as a source of cholesterol oxidase [8] in steroid biotransformations. COX is an intra-cellular or an extra-cellular enzyme produced by various microorganisms. Cholesterol oxidases may be intrinsic membrane bound enzymes located on the outside of the cell or can be isolated from broth filtrate as an extracellular enzyme.
3. Microbial Sources
Several microorganisms of diverse environments have ability to produce COX. In 1944, Turfitt was the first to isolate the enzyme from the microbe Rhodococcus erythropolis. He explored its effect as an oxidant of cholesterol. Different workers isolated COX from different of Mycobacterium species isolated from soil [9]. The cholesterol was degraded by incubation with a cell free extract of Mycobacterium to produce 4-cholesten-3-one [10]. Various species belonging to the genus Rhodococcus have also been found to have cholesterol-degrading ability [11]. COX producing bacteria have also been isolated from the foods of animal origin e.g. chicken fat, pork fat, butter and bacon, and most of them belong to Rhodococcus spp. [12]. COX from various microorganisms are characterized by broad substrate specificity within the cholestan family (Table 1). Rhodococcus spp. has similar cholesterol degrading ability, but the only difference is in the production of cholesterol oxidase. An extracellular COX isolated from Rhodococcus sp. [13] revealed a 99% sequence homology with other bacterial COX genes. Another extracellular COX produced by Micrococcus sp. was reported with high COX activity (3.68 U/mL). This was the first COX producing bacterium reported from Micrococcus spp. that possessed an optimum pH 7.0 and temperature 50˚C [14]. Another method reported by researcher to degrade the cholesterol level in egg yolk involved an ultrasonic-assisted enzyme process-using COX without affecting the quality of the yolk [15] Another method involved an ultrasonic-assisted enzyme process-using COX was found to degrade the cholesterol level in egg yolk affecting the quality of the yolk [15].
COX producing bacteria are mainly classified into two subclasses: 1) pathogenic and 2) non-pathogenic. Nonpathogenic bacteria use cholesterol as their carbon source (e.g. Streptomyces and fast-growing Mycobacteria), while pathogenic bacteria require COX for infection of the host macrophages (e.g. Rhodococcus equi and slow-growing Mycobacteria). The pathogenic bacteria require COX for infection of the host macrophages because of its ability to alter the physical structure of the lipid membrane by converting cholesterol into cholest-4-en-3-one. Both pathogenic and nonpathogenic bacteria up-regulate the expression of COX in the presence of cholesterol. COX from Gram-negative bacteria such as Burkholderia and Chromobacterium spp. have also been reported [16]. These enzymes possess thermal, detergent and organic solvent tolerance. There are two forms of COX, one contains a FAD-cofactor non-covalently bound to the enzyme (class I) and the other contains the cofactor covalently linked to the enzyme (class II). These two enzymes have no significant homology. The phylogenetic analyses show that both class I and class II enzymes can be further divided into at least two groups.

Table 1. Cholesterol oxidase producing microorganisms.
4. Production of COX
Due to low production of COX by various microorganisms, many workers have attempted to improve the yield of COX have introduced different steps in the methodology. Enhancing the cell-mass production via alteration of growth medium can increase the production. The effect of oleic acid adsorption onto the cell surface of Schizophyllum commune on yields of cholesterol oxidase was examined [41]. It was found that cholesterol oxidase production was affected by the amount of oleic acid in the culture broth, and that it was the level of insoluble oleic acid, rather than the soluble component adsorbed onto the cell, that was directly influencing the formation of COX. In addition, an increase in the rate of agitation correlated with the extent of COX production.
Many workers revealed the gene structures of several sources of COX and also attempted to clone and express COX from various sources to produce a bacterial strain that could be efficient for commercial enzyme production [36,42-54]. COX produced by native soil Rhodococcus strain was isolated and showed maximum activity at 35˚C with optimum pH of 7.0. The enzyme had a molecular mass of 55 kDa and was cloned in STV28 vector [55]. The COX was modified with polyethylene-glycol to allow the enzyme to function in organic solvents such as benzene, which would normally denature, and thus inactivate the COX protein [56,57].
5. Properties of Cholesterol Oxidase
5.1. Action of COX
Previous work has reported the bi-functional properties of the COX. COX catalyses not only the oxidation of D5-ene-3-hydroxysteroids with a trans A-B ring junction to the corresponding D5-3-ketosteroid, but also to the D4-3β-ketosteroid. In the case of Brevibacterium sterolicum, the occurrence of isomerization via a cis-diaxial intra-molecular transfer of protons from the 4β to 6β position has been reported [58]. Although the enzymes exhibited a broad range of steroid specificities dependent on bacterial source and/or chemical modification, the presence of a 3β-hydroxyl group was an essential requirement for substrate activity in all cases [59-61]. The first isolation of the oxidation product was reported using an NAD+ and NADP+ independent soil mycobacterium [62]. Enzymes produced by Nocardia and other species are generally referred as oxidases because of the requirement of oxygen for their mode of action while others are known as dehydrogenases as they are true NADdependent [63], e.g. the coupled enzyme 3β-hydroxysteroid: NAD(P) oxidoreductase-3-ketosteroid D4, D5- isomerase (3β-hydroxysteroid dehydrogenase: D5-isomerase) from the mitochondrial fraction of human placenta [64]. In bacteria, COX is the first enzyme in the catalytic pathway that yields propionate and acetate as final products. Importantly, there is no mammalian homolog of COX.
5.2. Biochemical Properties of COX
COX has great commercial value as it is routinely used in laboratories for serum cholesterol assay [65,66]. Since the monitoring of high-density lipoprotein (HDL) cholesterol and low-density lipoprotein (LDL) cholesterol in serum is important for the diagnosis of hyperlipemia or atherosclerotic diseases, several methods for the separation of HDL or LDL cholesterol with various detergents have been devised [67-69]. A detergent-tolerant COX from γ-Proteobacterium Y-134 was employed for the assay of HDL and LDL cholesterol in serum [70]. The COX with highest stability in the presence of a wide range of detergents helps in the improvement of a differential assay method for HDL and LDL. COX was also used for the optical resolution of non-steroidal compounds and allylic alcohols [71,72] and the bioconversion of a number of 3β hydroxysteroids in the presence of organic solvents [73-76]. Organic solvents as well as detergents are also employed to solublize the steroids. COX with organic solvent-tolerance would be useful for these applications.
5.3. Structure of COX
To perform different biological functions, proteins generally use tightly-bound cofactors and now up to 30 flavo-enzymes have been reported which are covalently linked to histidine, cysteine or tyrosine side chain [76]. Mainly crystal structures of COX belonging to two distinct structural classes’ type-I and type-II [77] have been studied [78-80]. Type-I, as those from Streptomyces sp. SA-COO and R. equi [77] have their cofactor tightly bound to the enzyme via non-covalent bonding. Type-II COX from Brevibacterium sterolicum was completely different from Type-I. In Type-II the cofactor was covalently linked to the protein via a bond between the 8- methyl group of the isoalloxazine ring and the ND1 atom of His69. COX of both the class-I and II had same catalytic activity, but differed in their sequence homology and flavin binding. Both had significant differences in their redox and kinetic activities [81-83]. A C406A mutant of monoamine oxidase that showed covalent flavinbinding markedly improved the structural stability of the enzyme [84]. Another COX belonged to the family of vanillyl-alcohol oxidases contained a fold proposed to favor covalent flavinylation [56]. The covalent flavin linkage of COX was studied in a mutant in which His69 →Ala exchange prevented formation of the histidyl-FAD bond without the loss of catalytic activity and also carried the isomerization step of the intermediate cholest- 5-en-3-one to the final product [85]; while non-covalent flavin binding resulted in a 35-fold decrease in turnoverrate and had much lower flavin midpoint redox potential (204 mV, compared with 101 mV for wild-type). Due to this fact, it was concluded that the flavin 8 linkage to a (N1) histidine is a central factor in the modulation of redox properties of COX to increase its oxidative power. The 3-D structure of the COX (Figure 1) with solvent accessible surface has been deciphered [86].
5.4. Purification of COX
The procedures applied for the extraction and purification of COX are similar to those applied to enzymes, in general. However, subtle differences in the extraction and purification procedures are generally required in order to obtain a sufficient yield of enzyme from COX producing bacteria to improve yield and/or clarification. The techniques adopted for extraction are largely dependent on the nature of the secretion of the enzyme, i.e. whether intracellular or extracellular. The COX oxidase from Nocardia rhodochrous is believed to be an intrinsic membrane-bound enzyme located on the outside of the cell. The enzyme may be extracted from bacterial cells by treatment with Triton X-100 or trypsin, or to a lesser extent with phosphate buffer, but not by mechanical cell disruption or by changes in pH or ionic strength of the buffer [84]. The enzyme possesses closely associated 3β- hydroxy steroid oxidase and 3-oxo steroid A4-A5-isomerase activities [24]. The use of the Nocardia in the presence of high concentrations of organic solvents and the immobilization of cholesterol oxidase and its use in aqueous/organic solvent emulsions have been described previously [87,88]. COX from Streptomyces species for serum assay was reported to be superior to those from other micro-organisms [89] due to lower cost of production and longer shelf life. It is currently being used in organic synthesis [90,91] since it is stable in water-immiscible solvents at high concentrations and shows excellent stereoand enantio-selectivity in the oxidation of non-steroidal compounds. COX produced by Streptomyces sp. has been reported to have insecticidal activity against insects like boll weevil larvae [92]. The earlier researchers [93,94] have reported intracellular [89,95] and extracellular [96,97] production of COX by Streptomyces lavendulae NCIM 2421 that produced both intracellular as well as extracellular enzyme.
The purification of COX involved differential ammonium sulphate precipitation, ion-exchange and gel filtration chromatography. The COX produced by Pseudomonas sp. strain CO629 was purified 2400-fold to homogeneity in an overall yield of 60% from culture broth. The enzyme was a monomer with a molecular weight of 56 kDa, as estimated by SDS-PAGE and Sephadex G-150 gel column chromatography. The properties of the enzyme purified from strain CO629 were largely different from those reported for several other microorganisms. On the other hand, the molecular weight of the enzyme from S. violascens [98], B. sterolicum [99] and Schizophyllum commune [100] were reported to be 61, 31 and 53 kDa, respectively. All these enzymes consisted of a single polypeptide chain, whilst the molecular weights were apparently distinct from the origin of the enzymes. It was demonstrated using site-directed mutagenesis that the two domains of Streptomyces cholesterol oxidase could be separated from each other [101].
Extra-cellular COX from the culture supernatants of Rhodococcus sp. GK1 was purified to 21-fold, in specific activity terms, by affinity-like chromatography on the Kieselguhr-cholesterol column [31]. Impurities in the applied supernatant were eluted from the column throughout the sample application by washing with the phosphate
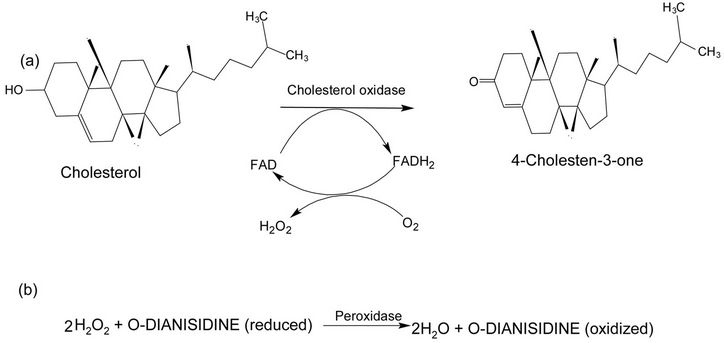
Figure 1. Mechanism of reaction catalyzed by cholesterol oxidase. (a) The cholesterol oxidase oxidizes cholesterol into 4-cholestene-3-one and H2O2; (b) H2O2 and o-dianisidine (reduced) with peroxidase enzyme oxidized o-dianisidine and releases water molecule. Part (b) shows the principle of an enzymatic assay.
buffer. The enzyme was retained on the Kieselguhrcholesterol and was subsequently eluted with buffer containing Triton X-100. Protein monitoring during enzyme elution was not possible because of the high absorbance of the detergent at 280 nm. The purified COX had a specific activity of 5.5 U/mg, and was revealed to be apparently homogeneous on SDS-PAGE. The recovery of the purified COX in the collected samples was around 30% relative to the crude samples. The sequence of ten amino acids from the N-terminal segment of COX was determined by the Edman degradation method and was found to be: H2N-Ala-Pro-Pro-Val-Ala-Ser-X-Arg-Tyr-X-(Phe). The residues “X” may be two cystiene molecules according to a preliminary determination. The first six amino acids of the sequence could be sufficient for constructing a nucleotide probe to isolate the COX gene.
To achieve purification of cholesterol oxidase from the Rhodococcus equi, the culture supernatant was initially ultra-filtered to give a concentrated retentate enriched with COX. The progressive purification of COX, as determined by synergistic hemolysis showed a representative elution profile of COX fractions following desalting of acetone-precipitated material on Sephadex G-25 column. Synergistic hemolysis was present only within the small peak. Desalting on Sephadex G-25 removed residual acetone and markedly concentrated the COX. A cation-exchanger isolated COX to near homogeneity on Superose 12-HR gel filtration chromatography, which purified the exo-enzyme. This purified COX induced total lyses of sheep red blood cells sensitized with Staphylococcus aureus β-toxin (Sphingomyelinase C) or with Corynebacterium pseudotuberculosis phospholipase D. It also oxidized aqueous suspensions of cholesterol phospholipase C [102]. A recovery of 0.3% and a purification of 32.8-fold was recorded by the SH hemolytic activity of the COX material. COX of Pseudomonas sp. strain ST-200 purified from the culture supernatant was found to be extracellular and was bound to thin enzyme of 60 kDa flavin and was categorized as a 3β-hydroxysteroid oxidase, converting 3β hydroxyl groups to keto groups. This strain exhibited a 3 to 3.5 fold increase in COX activity in the presence of an organic solvent than that found in the absence of organic solvent, with log Pow values (partition coefficients of the organic solvent between n-octanol and water), in the range of 2.1 to 4.2, compared with that in the absence of organic solvents [103]. An extracellular COX degrading strain B4 reported form Bordetella genus was first of its kind to produce a cellular form of COX (~450 U/L) which reached 1700 U/L in 24 h at 37˚C. At 7.0 pH, the yield was higher than that of Rhodococcus equi No. 23 (~240 U/L after 60 - 72 h cultivation [104].
Brevibacterium COX expressed in E. coli could be purified using one-step of Ni-affinity chromatography [105]. For expression of recombinant Brevibacterium COX in E. coli BL21 (DE3), an affinity protocol was developed using riboflavin and lumiflavin as the affinity ligand. The affinity separation provided 98% yield with riboflavin affinity column and 99.5% with HPLC Vydac C4 column via single step protein purification process [106].
6. Stability and Activity of COX
A characteristic of the cholesterol oxidase is the marked stability under extreme conditions such as high temperatures and rather acidic and basic pHs. The heat stability of this enzyme is of a great advantage for clinical use. The temperature profile of the COX from Brevibacterium sp. had an optimum temperature for enzyme activity at around 53˚C [107] which was higher than the earlier reported optimum temperature for COX enzyme from Rhodoccocus equi (47˚C) and Corynebacterium cholesterolicum (40˚C) [108]. It was closer to that for the purified enzyme from Streptomyces violascens, with an optimum temperature at around 50˚C and plateau between 40˚C and 60˚C [109]. The optimum temperature for COX sourced from Bacillus sp., Arthrobacter simplex and Streptomyces violascens appeared to range between 50˚C - 60˚C and the enzymes appeared to be thermo-tolerant (Table 2). COX produced by Brevibacterium sp. had similar optimal pH to that reported for the same enzyme from other microorganism.
The action of COX is strongly influenced by the environment of its substrate even in simple detergent solutions [110]. COX from Brevibacterium sp. showed maximum activity at pH 7.5 [107], that moderately decreased to 80% at pH range of 6.0 - 8.7. Studies revealed that cholesterol is the best carbon source for COX production and can be enhanced by emulsification of cholesterol with Tween-80. To further improve the thermal stability of COX, site direct mutagenesis was applied to the Brevibacterium sp. grown at 50˚C for 2 h. It was observed that three amino acid residues (Q153E, F128L and S143H) located near the FAD-binding site of the enzyme were responsible for an increase in specific activity by 11.6% and 47% of two mutated sites Q153E and F128L, respectively [111]. Further studies on this bacterium showed that 0.15% (w/v) cholesterol emulsified with 0.05% Tween-80 induced the highest amount of COX production in comparison to use of other detergents [112].
Among various nitrogen sources, the yeast extract (0.3% w/v) and di-ammonium hydrogen phosphate were the most favorable nitrogen sources for COX production by Rhodococcus sp. 2C, while ammonium chloride and ammonium acetate had inhibitory effects on the COX production by R. equi No. 23 [113] and A. simplex [114], respectively. The results reported from Nocardia rhodochrous showed that the amount of the enzyme extracted
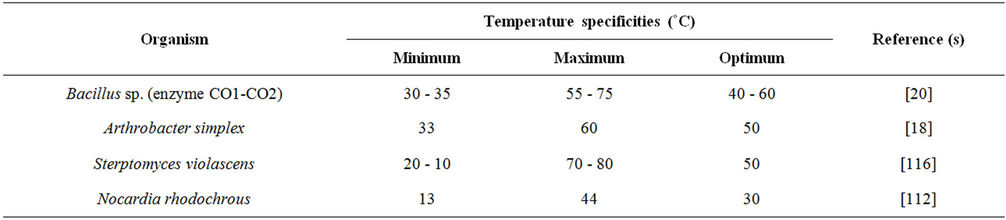
Table 2. Temperature specificities of cholesterol oxidase enzyme.
increased with detergent concentration up to 10% (v/v) and that more enzymes was solubilized using detergent concentrations higher than 0.5% (v/v). Additionally, low levels of surfactant in combination with a metal salt could elute cholesterol oxidase, effectively. Many reagents including Ca2+-ions were found to inhibit the COX activity strongly. Except methanol, all other alcohols and mineral salts addition depressed the system’s activity [115]. The inhibitors of pyruvate oxidation and of the TCA cycle also depressed cholesterol oxidation.
7. Reactions Catalyzed by Cholesterol Oxidase
The metabolism of cholesterol in mammals is extremely complex. Cholesterol is also metabolized by microorganisms as a carbon or an energy source [32,117]. COX attacks sterols at the 3β-hydroxyl positions to form 4-cholestenone and H2O2. This is the first step of microbial degradation of cholesterol in the oxidation of the 3β-hydroxyl group by COX [117], followed by the degradation of the side-chain [118,119] by enzyme-complex possibly involving cytochrome P-450 and lyases leading to 4-androstene-3, 17-dione, or 1, 4-androstadiene-3 or 17-dione, and eventual breakdown of the steroid moiety with carbon dioxide and water being the final products. The direct mode of action of COX was reported which involved the enzymatic oxidation mid-gut epithelial membrane cholesterol by demonstration that the COX oxidized cholesterol in isolated boll weevil mid-gut brush border membrane vesicles, with some concomitant changes in activity of brush border membrane vesicles marker enzymes [120].
8. Broader Applications of COX
The flavoenzymes including COX of microbial origin (Table 3) are the enzymes of great interest in the present era. COX is widely used in clinical diagnosis and determining lipid disorders [121]. COX is used as an insecticide [122] and also plays a role in lysis of macrophages and leukocytes [123]. Arthrobacter spp. produced a large variety of medically important metabolites and extracellular enzymes, in particular cholesterol oxidase.
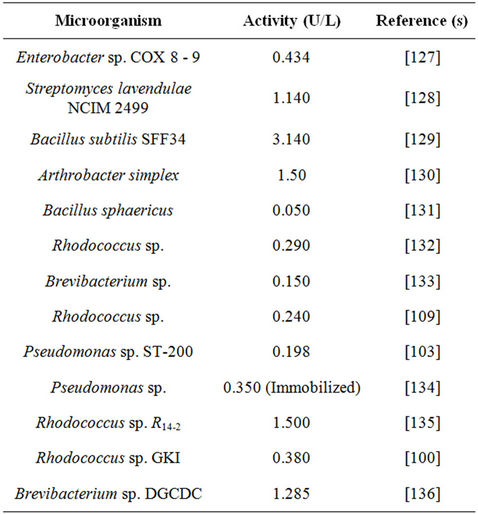
Table 3. Cholesterol oxidase activity produced by different microorganisms.
8.1. Clinical Applications
Bacterial COX exhibits a wide range of clinical and industrial applications, such as the determination of cholesterol concentration for arteriosclerosis diagnosis, steroids analysis in food samples, determination of stearic configuration of 3β-hydroxysteroids and 3-ketosteroids preparation from corresponding steroids [124]. An increased plasma cholesterol concentration leads to an increased risk of developing coronary heart disease [41]. Keeping in view the importance of COX in various biological functions, disease manifestation or viral pathogenesis (Table 4), much attention has been given by researchers to clone COX of diverse microbial origin in E. coli. The enzyme production may be improved through genetic engineering, in order to reduce the cost of diagnostic kits, which directly benefits the mankind. COX gene (choA) of Streptomyces sp. SA-COO has been cloned into S. lividans with the vector pIJ702 [125], and choA complete nucleotide sequence was determined in
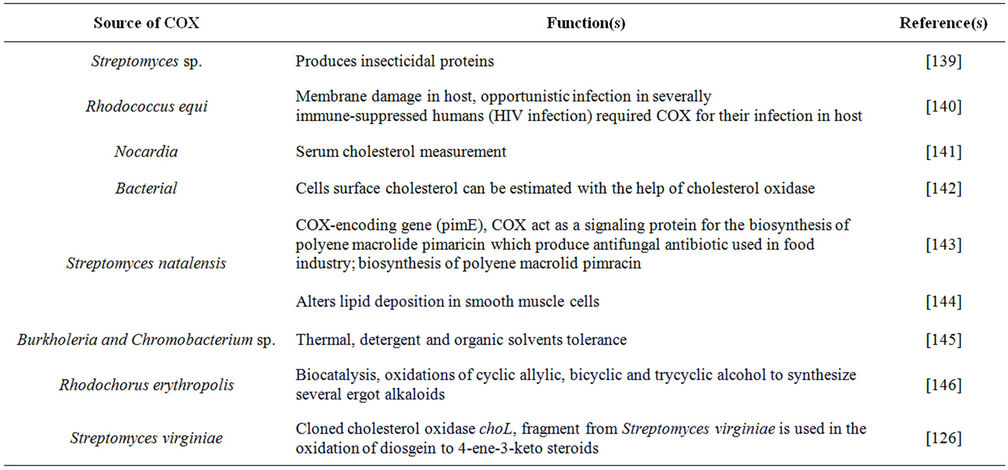
Table 4. Broader functions of microbial cholesterol oxidase(s).
1989 (Enzyme Nomenclature Elsevier 1972). Afterwards, the COX gene (choB) of Bacillus sterolicum has also been cloned and sequenced [126].
A 3178-bp DNA fragment containing full-length choL was cloned, sequenced and aligned with a few known genes of similar type [125]. Sequence analysis revealed the complete sequence of the choL ORF with GTG as the start codon and 70% G + C content. The choL gene consisted of 1629 nucleotides and encodes a deduced protein of 542 amino acids. As a rule, the deduced ORF of Cho often contains a signal peptide in its N-terminal, which contains a central hydrophobic region and a more polar C-terminal region. In comparison with the signal peptide of ChoA, the predicted signal sequence also contains the typical positively charged amino acids such as His and Arg-Arg-Arg. For the mature protein of choL, the consensus sequence for FAD binding, i.e., GxGxGxxxxA xxxxxxG is located near the N-terminal end at amino acids 16 - 32. Interestingly, the third invariant Gly in the FAD-binding motif is substituted with Ala in ChoL and in previously published cholesterol oxidases from Streptomyces spp. The molecular weight of ChoL was estimated to be 58.9 kDa, and the pI value was calculated to be 8.52 by the ExPASy computer pI/Mw program algorithm. BLAST analysis showed that ChoL had the highest sequence identity (85%) with ChoA, a commercial Cho from Streptomyces sp. SA-COO. COX gene (choM) that had an insecticidal product has been cloned, sequenced and expressed in Escherichia coli and in tobacco plant [51]. The importance of COX in the metabolism of diosgenin in S. virginiae IBL-14 was demonstrated by cloning and expression of choL gene in E. coli [91].
8.2. Insecticidal Activity
Bacterial COX has potent insecticidal activity against the cotton boll weevil (Anthonomus grandis). Generally, cotton boll weevil is difficult to control because it does not come in direct contact with chemical sprays, larvae feed and sheltered within immature flower buds or fruit (bolls) of cotton (Gossypium hirsutum). Upon ingestion, this protein causes developmental arrest and death of boll weevil larvae, and decrease in fecundity of female adult boll weevils [137]. COX also exhibits insecticidal effect against several species (Table 4) of lepidopteron cotton insects which occurs during membrane fusion, since the curvature [138] including tobacco budworm (Heliothis virescens), corn earworm (Helicoverpa zea), and pink bollworm (Pectinophora gossypiella).
8.3. Organization of Biological Membranes
Cholesterol is the main constituent of eukaryotic cell membrane. Cholesterol-enriched membrane micro-domain is used by some viruses (e.g. influenza virus, some retroviruses and filoviruses) for assembly and cell entry [147-152]. Cholesterol is expected to promote and stabilize the local bi-layer bending, which was suppose to take place during membrane fusion, since the curvature stress is towards the negative side [77]. Many workers have studied the role of cholesterol in membrane organization that has used COX as probe [153]. The inaccessibility of COX was also demonstrated by [154] for the outer-membrane surface of human erythrocytes [155] and virus [127]. Susceptibility of cholesterol oxidation by COX depends upon the removal of polar head group of phospholipids by phospholipase C from the exterior surface of viral, erythrocyte, or vesicle membrane [154]. Cholesterol promotes the formation and growth of the contact sites between membranes that allow lipid mixing which increase the efficiency of fusion via expanding the fusion pore [77].
8.4. Pathogenic Action
COX is used in the treatment of bacterial infections because of its ability to alter the physical structure of the cell membrane due to the conversion of cholesterol into cholest-4-en-3-one. COX is produced by a wide variety of microorganism including some life threatening pathogens such as Rhodococcus equi, Mycobacterium tuberculosis and M. leprae [155]. Rhodococcus equi is a common soil-dwelling Gram-positive bacterium that frequently infects young horses [127]. Some pathogenic bacteria require cholesterol oxidase to infect their host macrophage, probably because of the ability of cholesterol oxidase to alter the physical structure of the membrane by converting cholesterol to cholesten-4-en-3-one [156]. As these enzymes are unique to bacteria, they represent a potential target for a new class of antibiotics. Recently, it has been demonstrated that Alzheimer’s disease β-amyloid selectively oxidizes cholesterol at the C-3 hydroxyl group and catalytically produces 4-cholesten-3- one. Therefore, it mimics the activity of cholesterol oxidase [128]. Cholesterol is also known to play a key role in the entry of some viruses. Removal of cellular cholesterol rendered primary cells and cell lines highly resistant to HIV-1-mediated syncytium formation and to infection by both CXCR4- and CCR5-specific viruses. Thus, it appears that cholesterol may be critical to the HIV-1 coreceptor function of chemokine receptors and is required for infection of cells by HIV-1 [129].
8.5. COX Biosensor
Cholesterol detection is important in clinical and food analysis. Different electrochemical biosensors have been proposed for cholesterol detection [130,131]. In recent years the importance of cholesterol determination has increased as reflected in the number of articles dealing with the development of electrochemical devices for cholesterol analysis. Different methods such as screen printed electrode [132], hydrogel membranes [133], polymeric membrane [134], self-assembled monolayers [135], composite sol-gel membrane [136], liquid crystal cubic phase matrices [139] and films prepared by the layer-by-layer technique [140] have been reported.
In electrochemical biosensor the detection was generally monitored on the basis of consumption of oxygen and H2O2 production. Previously, no reference of carbon nanotubes electrode (CNT) was reported for cholesterol detection. Novel amperometric biosensors, by immobilizing COX in sol-gel layer on CNT-Pt modified electrodes have been fabricated [140,141]. The biosensor detected cholesterol based on the reduction of H2O2. The COX/CNT-Pt biosensor was rapid in current response and was also selective against electro-active interference. This biosensor was successfully used for serum cholesterol determination. In 2003, biosensors were compared on the entrapment [141] basis of COX and cholesterol esterase in electro-polymerized films of polypyrole and diaminophthalene (DAN) derivatives for amperometric determination of cholesterol [142]. The poly (diaminonaphthalene) [P(DAN)] layers did not destroy the catalytic activity of the enzyme COX and cholesterol esterase if adequate preparation condition were used.
In 2010, a new electrochemical biosensor was introduced [142] for cholesterol determination that combined a Fourier transformation continuous cycle voltmeter [FFTCCV] technique in a flow injection analysis [142, 157]. This was the first application of FFTCCV method for cholesterol biosensor based on COX. The COX was immobilized onto MnO2 nano-particles and multi-walled carbon-nanotubes (MWCNTs) and placed on a glassy carbon electrode surface using nafion. This was first time when a very high-sensitivity and low detection limit detector was used for cholesterol biosensor on MWCNT/ MnO2 NPs modified electrode. Biosensor showed longterm storage stability up to 30 days, 92.3% sensitivity was retained after 54 days; afterwards there was a decrease in sensitivity due to the loss of catalytic activity.
A surface plasma resonance based biosensor for simple, label-free, highly selective and sensitive detection of cholesterol employing the flavo-enzyme COX as a sensing element has been proposed [158]. The COX along with bovine serum albumin was immobilized onto the gold-plated sensor chip covalently attached with 11-mercaptoundecanoic acid. The analyte brought changes in the sensor surface that could be monitored in real time. The biosensor was specific for cholesterol and showed no significant interference from potent molecules such as ascorbic acid and uric acid.
A novel amperometric cholesterol biosensor immobilize with COX on electrochemically polymerized polypyrole-polyvinlyulphonate (PPy-PVS) film entrapped on platinum electrode was fabricated [159]. Untill now, different conducting polymers have been used for immobilization of bio-molecules such as polyacetylene, polythiohpene (PPy), polyindole and polyaniline [160-164]. A PPy-PVs composite membrane was used because of its unique featured “charge controlled membrane” in which the fixed charge were electrochemically checked by an internal electronic state [160]. Mostly, cholesterol biosensors have been used in biochemical analysis owing to their good selectivity, low cost, small size, fast response and long term stability. The cited literature based on cholesterol biosensors appeared to be mainly focused on diagnosing disorders [133,141,166,167].
A new and alternative approach was proposed [133] that utilized a bi-enzyme (oxides and peroxidase) system to circumvent the interference problem to improve the sensitivity of the biosensor [168]. A silica-polyaniline membrane that included single step fabrication process involving electrochemical polymerization of N [3-(trimethoxysilyl) propyl] aniline to form poly N [3-trimethoxysilyl) propyl] aniline (PTMSPA) and simultaneous immobilization of two enzymes; namely horseradish peroxidase (HRP) and COX into PTMSPA matrix was created [133]. The matrix reported PTMSPA possessed features suited for the fabrication of third generation biosensors [133]. Recently, a novel COX biosensor was fabricated by co-immobilizing three enzymes; namely COX, cholesterol esterase and HRP on nanoporous gold network directly grown on titanium substrate [169]. This biosensor possessed a wide linear range up to 300 mg/dL in a physical condition (pH 7.4), which made it very effective for clinical determination of cholesterol. Another work reported multi-walled carbon nanotubes/COX film modified glassy carbon electrode (COX/MWCNT/GCE) that was prepared and its electro-catalytic behavior towards cholesterol was investigated using cyclic voltametry. The modified electrode showed electrocatalytic activity only towards cholesterol and an attempt was also made to utilize (COX/MWCNT/GCE) electrode to measure the cholesterol that made it more promising for cholesterol determination [170].
The microchip capillary electrophoresis (MCE) was also used to demonstrate the rapid detection of cholesterol in serum, using (MCE) fabricated from poly (dimethylsiloxane) (PDMS) microchip channel successfully applied to quantify cholesterol levels. The method provided a very sensitivity (1 nm) and rapidness for cholesterol detection (less than 100 seconds). Also, the developed method was used to measure cholesterol in a bovine serum standard solution. The developed polymer micro-fluid biochip showed more advantages like, compact size, high sensitivity, and high selectivity, low cost and fast response that appeared to be beneficial to perform routine analysis in clinical laboratory [172]. To improve COX biosensor techniques, a disposable amperometric biosensor was fabricated that comprised, a sensing electrode and a reference electrode in simultaneous contact with an integrated reagent layer, formed by working ink containing cholesterol esterase, cholesterol dehydrogenase, coenzyme, redox mediator, surfactant, stabilizer, filler and an aqueous thickening agent to analyze the total cholesterol content in a minute or so [172]. Another new approach made to create a COX biosensor used a crystalline bacterial cell surface (S-layer protein extracted from B. sphaericus using chaotropic agent i.e. guanidine hydrochloride to produce monomolecular film with no further purification) monomolecular film of COX and mixed films of COX/S-Layer proteins and DPPC/S-Layer (Dipalmitoyl phosphatidyl choline) that were produce using Langmuir Blodgett technique [173].
9. Conclusion
Cholesterol oxidase of diverse microbial sources is rapidly becoming an enzyme of great interest today. Problems such as low production of COX, however, have caused researchers to improve the yield of COX by introducing new and novel isolation steps. A broad-range of clinical and industrial applications of COX have generated a renewed interest in exploring various natural habitat/environments for discovering newer microbial sources as potential producers of stable COX.
REFERENCES
- G. E. Turfitt, “The Microbiological Degradation of Steroids. Oxidation of Cholesterol by Proactinomyces spp.,” Journal of Biochemistry, Vol. 38, No. 5, 1944, pp. 49-62.
- D. R. Corbin, J. T. Greenplate, E. Y. Wong and J. P. Purcell, “Cloning of an Insecticidal Cholesterol Oxidase Gene and Its Expression in Bacteria and Plant Protoplasts,” Applied and Environmental Microbiology, Vol. 60, No. 12, 1994, pp. 4239-4244.
- J. P. Purcell, J. T. Greenplate, M. G. Jennings, J. S. Ryerse, J. C. Pershing, S. R. Sims, M. J. Prinsen, D. R. Corbin, M. Tran and R. D. Sammons, “Cholesterol Oxidase: A Potent Insecticidal Protein Active against Boll Weevil Larvae,” Biochemical and Biophysical Research Communications, Vol. 196, No. 3, 1993, pp. 1406-1413. doi:10.1006/bbrc.1993.2409
- J. Li, A. Vrielink, P. Brick and D. M. R. Blow, “Crystal Structure of Cholesterol Oxidase Complexed with a Steroid Substrate: Implications for Flavin Adenine Dinucleotide Dependent Alcohol Oxidases,” Biochemistry, Vol. 32, No. 43, 1993, pp. 11507-11515. doi:10.1021/bi00094a006
- N. S. Sampson, I. J. Kass and K. B. Ghoshroy, “Assessment of the Role of V Loop of Cholesterol Oxidase: A Truncated Loop Mutant Has Altered Substrate Specificity,” Biochemistry, Vol. 37, No. 16, 1998, pp. 5770-5778. doi:10.1021/bi973067g
- T. Yamane, H. Nakatani and E. Sada, “Steroid Bioconversion in Water-Soluble Organic Solvents: ∆1-Dehydrogenation by Free Microbial Cells and by Cells Entrapped in Hydrophilic or Lipophilic Gel,” Biotechnology and Bioengineering, Vol. 21, No. 11, 1979, pp. 2133-2145.
- C. Lee and W. Liu, “Production of Androsta-1, 4-Diene-3, 17-Dione from Cholesterol Using Immobilized Growing Cells of Mycobacterium sp. NRRL B-3683 Adsorbed on Solid Carriers,” Applied Microbiology and Biotechnology, Vol. 36, No. 5, 1992, pp. 598-603. doi:10.1007/BF00183235
- A. Constantinidis, “Steroid Transformation at High Substrate Concentrations Using Immobilized Corynebacterium simplex Cells,” Biotechnology and Bioengineering, Vol. 22, No. 1, 1980, pp. 119-136. doi:10.1002/bit.260220110
- A. Schatz, K. Savard and I. J. Pinter, “The Ability of Soil Microorganisms to Decompose Steroids,” Journal of Bacteriology, Vol. 58, No. 2, 1949, pp. 117-125.
- T. C. Stadtman, A. Cherkes and C. B. Anfinsen, “Studies on the Microbial Degradation of Cholesterol,” Journal of Biological Chemistry, Vol. 206, 1954, pp. 522-523.
- N. P. Ferreira and R. P. Tracey, “Numerical Taxonomy of Cholesterol-Degrading Soil Bacteria,” Journal of Applied Bacteriology, Vol. 57, No. 3, 1984, pp. 429-446. doi:10.1111/j.1365-2672.1984.tb01409.x
- K. H. Watanabe, S. H. Aihara, R. Nakamura, K. I. Suzuki and K. Komagata, “Isolation and Identification of Cholesterol Degrading Rhodococuss Strain from Food of Animal Origin and Their Cholesterol Oxidase Activities,” Journal of General Applied Microbiology, Vol. 32, No. 2, 1986, pp. 137-147. doi:10.2323/jgam.32.137
- H. Lashkarian, J. Raheb, K. Shahzamani, H. Shahbani and M. Shamsara, “Extracellular Cholesterol Oxidase from Rhodococcus sp.: Isolation and Molecular Characterization,” Iranian Biomedical Journal, Vol. 14, No. 1-2, 2010, pp. 49-57.
- R. Kanchana, D. Correia, S. Sarkar, P. Gawde and A. Rodrigues, “Production and Partial Characterization of Cholesterol Oxidase from Micrococcus sp. Isolated from Goa, India,” International Journal of Applied Biology and Pharmaceutical Technology, Vol. 2, No. 2, 2011, pp. 393-398.
- Y. Sun, H. Yang, X. Zhong, L. Zhang and W. Wang, “Ultrasonic-Assisted Enzymatic Degradation of Cholesterol in Egg Yolk,” Innovative Food Science and Emerging Technologies, accepted manuscript, 2011.
- N. Doukyu and R. Aono, “Purification of extracellular Cholesterol Oxidase with High Activity in the Presence of Organic Solvent from Pseudomonas sp. Strain ST-200,” Applied Environmental Microbiology, Vol. 64, No. 5, 1998, pp. 1929-1932.
- N. Doukyu, “Characteristics and Biotechnological Applications of Microbial Cholesterol Oxidase,” Applied Microbiology and Biotechnology, Vol. 83, No. 5, 2009, pp. 825-837. doi:10.1007/s00253-009-2059-8
- P. S. J. Cheetham, P. Dunnill and M. D. Lilly, “The Characterization and Inter-Conversion of Three Forms of Cholesterol Oxidase Extracted from Nocardia rhodochrous,” Journal of Biochemistry, Vol. 201, No. 3, 1982, pp. 515- 521.
- Y. A. Petrova, G. M. Podsukhina, T. A. Dikun and A. A. Selezneva, “Conditions of Isolation of Cholesterol Oxidase from Actinomyces lavendulae mycelium,” Applied Biochemistry and Microbiology, Vol. 15, No. 2, 1979, pp. 125-128.
- W. H. Liu, M. H. Meng and K. S. Chen, “Purification and Some Properties of Cholesterol Oxidases Produced by an Inducible and a Constitutive Mutant of Arthrobacter simplex,” Agricultural and Biological Chemistry, Vol. 52, No. 2, 1988, pp. 4130-4138. doi:10.1271/bbb1961.52.413
- T. Uwajima, H. Yabi and O. Terada, “Properties of Crystalline 3β-Hydroxysteroid Oxidase of Brevibacterium sterolicum,” Agricultural and Biological Chemistry, Vol. 38, No. 6, 1974, pp. 1149-1156. doi:10.1271/bbb1961.38.1149
- T. Uwajima and O. Terada, “On the Kinetics of Cholesterol Oxidase from Brevibacterium sterolicum in the Presence of Detergent,” Agricultural and Biological Chemistry, Vol. 38, No. 6, 1978, pp. 1149-1156. doi:10.1271/bbb1961.38.1149
- N. Croteau and A. Vrielink, “Crystallization and Preliminary X-Ray Analysis of Cholesterol Oxidase from Brevibacterium sterolicum Containing Covalently Bound FAD,” Journal of Structural Biology, Vol. 116, No. 2, 1996, pp. 317-319. doi:10.1006/jsbi.1996.0047
- L. Motteran, M. S. Pilone, G. Molla, S. Ghisla and L. Pollegioni, “Cholesterol Oxidase from Brevibacterium sterolicum the Relationship between Covalent Flavinylation and Redox Properties,” The Journal of Biological Chemistry, Vol. 276, 2001, pp. 18024-18030. doi:10.1074/jbc.M010953200
- C. H. Rhee, K. P. Kim and H. D. Park, “Two Novel Extracellular Cholesterol Oxidases of Bacillus sp. Isolated from Fermented Flatfish,” Biotechnology Letters, Vol. 24, No. 17, 2002, pp. 1385-1389. doi:10.1023/A:1019888428005
- Y. Shirokane, K. Nakamura and K. Mizusawa, “Purification and Some Properties of an Extracellular 3b-Hydroxysteroid Oxidase Produced by Corynebacterium cholesterolicum,” Journal of Fermentation Technology, Vol. 55, 1977, pp. 337-346.
- A. G. Smith and C. J. W. Brooks, “Application of Cholesterol Oxidase in the Analysis of Steroids,” Journal of Chromatography, Vol. 101, No. 2, 1974, pp. 373-378. doi:10.1016/S0021-9673(00)82854-X
- B. C. Buckland, W. Richmond, P. Dunnill and M. D. Lilly, “The Large-Scale Isolation of Intracellular Microbial Enzymes: Cholesterol Oxidase from Nocardia,” Industrial Aspects of Biochemistry, Federation of European Biochemical Societies, Amsterdam, 1974, pp. 65-79.
- S. Lee, H. Rhee, W. Tae, J. Shin and B. Parlk, “Purification and Characterization of Cholesterol Oxidase from Pseudomonas sp. and Taxonomic Study of the Strain,” Applied Microbiology and Biotechnology, Vol. 31, No. 5-6, 1989, pp. 542-546. doi:10.1007/BF00270791
- F. Cheillan, H. Lafont, E. Termine, F. Fernandez, P. Sauve and G. Lesgards, “Molecular Characteristics of the Cholesterol Oxidase and Factors Influencing Its Activity,” Biochimica et Biophysica Acta, Vol. 999, No. 3, 1989, pp. 233-238. doi:10.1016/0167-4838(89)90002-2
- N. Doukyu and R. Aono, “Purification of Extracellular Cholesterol Oxidase with High Activity in the Presence of Organic Solvents from Pseudomonas sp. Strain ST-200,” Applied and Environmental Microbiology, Vol. 64, No. 5, 1998, pp. 1929-1932.
- M. P. Bokoch, A. Devadaoo, M. S. Palencsar and J. D. Burgess, “Steady-State Oxidation of Cholesterol Catalysed by Cholesterol Oxidase in Liquid Bilayer Membranes on Platinum Electrodes,” Analytica Chimica Acta, Vol. 519, No. 1, 2004, pp. 47-55. doi:10.1016/j.aca.2004.03.030
- T. L. Johnson and G. A. Somkuti, “Isolation of Cholesterol Oxidase from Rhodococcus equi ATCC 33706,” Biotechnology and Applied Biochemistry, Vol. 13, No. 2, 1991, pp. 196-204.
- H. Fukuda, Y. Kawakami and S. Nakamura, “A Method to screen Anti-Cholesterol Substances Produced by Microbes and a New Cholesterol Oxidase Produced by Streptomyces violescens,” Chemical and Pharmaceutical Bulletin, Vol. 21, No. 9, 1973, pp. 2057-2060. doi:10.1248/cpb.21.2057
- M. Fukuyama and Y. Miyake, “Purification and Properties of Cholesterol Oxidase from Schizophyllum with Covalently Bound Flavin,” Journal of Biochemistry, Vol. 85, No. 5, 1979, pp. 1183-1193.
- Y. Nishiya, N. Harada, S. Teshima, M. Yamahita, I. Fuji, N. Hirayama and Y. Murooka, “Improvement of Thermal Stability of Sterptomyces Choletsterol Oxidase by Random Mutagenesis and a Structure Interpretation,” Protein Engineering, Vol. 10, No. 3, 1997, pp. 231-235. doi:10.1093/protein/10.3.231
- Y. Kimberley, J. K. Ignatius, N. S. Sampson and A. Vrielink, “Crystal Structure Determination of Cholesterol Oxidase from Streptomyces and Structural Characterization of Key Active Site Mutants,” Biochemistry, Vol. 38, No. 14, 1999, pp. 4277-4286. doi:10.1021/bi982497j
- K. B. Ghoshroy, W. Zhu and N. S. Sampson, “Investigation of Membrane Disruption in the Reaction Catalyzed by Cholesterol Oxidase,” Biochemistry, Vol. 36, No. 20, 1997, pp. 6133-6140. doi:10.1021/bi962190p
- L. N. Sampson and A. Vrielink, “Sub-Atomic Resolution Crystal Structure of Cholesterol Oxidase, what Atomic Resolution Crystallography Revels about Enzyme Mechanism and the Role of the FAD Cofactor in Redox Activity,” Journal of Molecular Biology, Vol. 326, No. 5, 2003, pp. 1635-1650. doi:10.1016/S0022-2836(03)00054-8
- K. Isobe, N. Mori and N. Wakao, “The Second Cholesterol Oxidase Produced by γ-Proteobacterium Y-134,” Journal of Bioscience and Bioengineering, Vol. 96, No. 3, 2003, pp. 257-261.
- M. Takagi, T. Yoshida and H. Taguchi, “Effect of Oleic Acid Adsorption onto Cell Surface on Cholesterol Oxidase Production by Schizophyllum commune,” Journal of Fermentation Technology, Vol. 60, No. 1, 1982, pp. 31-35.
- T. Ohta, K. Fujishoro, K. Yamaguchi, Y. Tamura, K. Aisaka, T. Uwajima and M. Hasegawa, “Sequence of Gene choB Encoding Cholesterol Oxidase of Brevibacterium sterolicum: Comparison with choA of Streptomyces sp. SA-COO,” Gene, Vol. 103, No. 1, 1991, pp. 93-96. doi:10.1016/0378-1119(91)90397-T
- T. Ohta, K. Fujishiro, K. Yamaguchi, T. Uwajima, K. Aisaka and M. Hasegawa, “Hyperexpression and Analysis of choB Encoding Cholesterol Oxidase of Brevibacterium sterolicum in Escherichia coli and Streptomyces lividans,” Bioscience, Biotechnology, and Biochemistry, Vol. 56, No. 11, 1992, pp. 1786-1791. doi:10.1271/bbb.56.1786
- T. Ishizaki, N. Hirayama, H. Shinkawa, O. Nimi and Y. Murooka, “Nucleotide Sequence of the Gene for Cholesterol Oxidase from a Streptomyces sp.,” Journal of Bacteriology, Vol. 171, No. 2, 1989, pp. 596-601.
- K. Fujishoro, T. Ohta, M. Hasegawa, K. Yamaguchi, T. Mizukami and T. Uwajima, “Isolation and Identification of the Gene of Cholesterol Oxidase from Brevibacterium Sterolicum ATCC 21387 a Widely Used Enzyme in Clinical Analysis,” Biochemical and Biophysical Research Communications, Vol. 172, 1990, pp. 721-727. doi:10.1016/0006-291X(90)90734-5
- M. Horii, T. Ishizaki, S. Y. Paik, T. Manome and Y. Murooka, “An Operon Containing the Genes for Cholesterol Oxidase and Acytochrome P-450 Like Protein from a Streptomyces,” Journal of Bacteriology, Vol. 172, No. 7, 1990, pp. 3644-3653.
- I. Molnar, K. P. Choi, N. Hayashi and Y. Murooka, “Bacterial Cholesterol Oxidases Are Able to Act as Flavoprotein-Linked Ketosteroid Monooxygenases that Catalyse the Hydroxylation of Cholesterol to 4-Cholesten- 6-ol-3-one,” Journal of Fermentation and Bioengineering, Vol. 72, 1991, pp. 368-372.
- D. K. Solaiman and G. A. Somkuti, “Expression of Streptomyce Cholesterol Oxidase in Escherichia coli,” Journal of Industrial Microbiology, Vol. 8, No. 4, 1991, pp. 253-258. doi:10.1007/BF01576063
- D. K. Solaiman, G. A. Somkuti and D. H. Steinberg, “Construction and Characterization of Shuttle Plasmids for Lactic Acid Bacteria and Escherichia coli,” Plasmid, Vol. 28, No. 1, 1992, pp. 25-36. doi:10.1016/0147-619X(92)90033-7
- D. K. Solaiman and G. A. Somkuti, “Expression of Streptomyces melC and choA Genes by a Cloned Streptococcus thermophilus Promoter STP2201,” Journal of Industrial Microbiology, Vol. 15, No. 1, 1995, pp. 39-44. doi:10.1007/BF01570011
- D. K. Solaiman and G. A. Somkuti, “Isolation and Characterization of Transcription Signal Sequences from Streptococcus thermophilus,” Current Microbiology, Vol. 34, No. 4, 1997, pp. 216-219. doi:10.1007/s002849900171
- P. Brigidi, F. Bolognani, M. Rossi, C. Cerre and D. Matteuzzi, “Cloning of the Gene for Cholesterol Oxidase in Bacillus sp., Lactobacillus reuteri and Its Expression in Escherichia coli,” Letters in Applied Microbiology, Vol. 17, No. 2, 1993, pp. 61-64. doi:10.1111/j.1472-765X.1993.tb00371.x
- Y. Murooka, T. Ishizaki, O. Nimi and N. Maekawa, “Cloning and Expression of a Streptomyces Cholesterol Oxidase Gene in Streptomyces lividans with Plasmid pIJ702,” Applied Environmental Microbiology, Vol. 52, No. 6, 1996, pp. 1382-1385.
- G. A. Somkuti, D. K. Solaiman, T. L. Johnson and D. H. Steinberg, “Transfer and Expression of a Streptomyces cholesterol Oxidase Gene in Streptococcus thermophilus,” Biotechnology and Applied Biochemistry, Vol. 13, No. 2, 1991, pp. 238-245.
- H. E. Lashkarian, J. Raheb, K. Shahzamani, H. Shahbani, M. Shamsara and M. Hajipour, “Isolation and Identification of a Native Rhodococcus Strain Producing Cholesterol Oxidase from Soil,” International Journal of Biotechnology and Biochemistry, Vol. 6, 2010, pp. 205- 217.
- T. Yoshimoto, A. Ritani, K. Ohwada, K. Takahashi, Y. Kodera, A. Matsushima, Y. Saito and Y. Inada, “Polyethylene Glycol Derivative-Modified Cholesterol Oxidase Soluble and Active in Benzene,” Biochemical and Biophysical Research Communications, Vol. 148, No. 2, 1987, pp. 876-882. doi:10.1016/0006-291X(87)90957-0
- A. Gupte, R. Nagarajan and A. Kilara, “Block Copolymer Micro Domains: A Novel Medium for Enzymatic Reactions,” Biotechnology Progress, Vol. 7, No. 4, 1991, pp. 348-354. doi:10.1021/bp00010a009
- I. J. Kass and N. S. Sampson, “The Isomerization Catalyzed by Brevibacterium sterolicum Cholesterol Oxidase Proceeds Stereo Specifically with One Base,” Biochemical and Biophysical Research Communications, Vol. 206, No. 2, 1995, pp. 688-693. doi:10.1006/bbrc.1995.1097
- A. G. Smith and C. J. W. Brooks, “Studies on the Substrate Specificity of Cholesterol Oxidase from Nocardia erythropolis in the Oxidation of 3-Hydroxy Steroids,” Biochemical Society Transactions, Vol. 3, No. 5, 1975, pp. 675-677.
- T. Kamei, Y. Takiguchi, H. Suzuki, M. Matsuzaki and S. Nakamura, “Purification and Properties of Streptomyces violascens Origin by Affinity Chromatography on Cholesterol,” Chemical and Pharmaceutical Bulletin, Vol. 26, No. 9, 1978, pp. 2799-2804. doi:10.1248/cpb.26.2799
- Y. Inouye, K. Taguchi, A. Fuji, K. Ishimaru, S. Nakamura and R. Nomi, “Purification and Characterization of Extracellular 3b-Hydroxysteroid Oxidase Produced by Streptoverticillium cholesterolicum,” Chemical and Pharmaceutical Bulletin, Vol. 30, No. 3, 1982, pp. 951-958. doi:10.1248/cpb.30.951
- Enzyme Nomenclature Elsevier, Amsterdam, Netherlands, 1972.
- D. P. Edwards, J. L. Conner, E. D. Bransome and W. E. Braselton, “Human Placental 3β-Hydroxysteroid Dehydrogenase: D5-Isomerase,” The Journal of Biological Chemistry, Vol. 251, 1976, pp. 1632-1638.
- A. Noma and K. Nakayama, “Comparative Studies on Cholesterol Oxidases from Different Sources,” Clinica Chimica Acta, Vol. 73, No. 3, 1976, pp. 487-496. doi:10.1016/0009-8981(76)90152-2
- W. Richmond, “Preparation and Properties of Bacterial Cholesterol Oxidase from Nocardia sp. and Its Application to the Enzymatic Assay of Total Cholesterol in Serum,” Clinical Chemistry, Vol. 19, No. 12, 1973, pp. 1350- 1356.
- K. Hino, M. Nakamura, K. Nakanishi and M. Manabe, “A New Method for the Homogeneous Assay of Serum HDL,” Clinical Chemistry, Vol. 42, 1996, pp. 296-299.
- K. Shirai, T. Nema, Y. Hiroh, Y. Itoh, Y. Miyashita and H. Watanabe, “Clinical Efficacy of the Direct Assay Method Using Polymers for Serum High Density Lipoprotein Cholesterol,” Journal of Laboratory and Clinical Medicine, Vol. 11, No. 2, 1997, pp. 82-86.
- M. Okada, H. Matsui, Y. Ito, A. Fujiwara and K. Inano, “Low-Density Lipoprotein Cholesterol Can Be Chemically Measured: A New Superior Method,” Journal of Laboratory and Clinical Medicine, Vol. 132, No. 3, 1998, pp. 195-201. doi:10.1016/S0022-2143(98)90168-8
- J. Biellmann, “Resolution of Alcohols by Cholesterol Oxidase from Rhodococcus erythropolis: Lack of Enantiospecificity for the Steroids,” Chirality, Vol. 13, No. 1, 2001, pp. 34-39. doi:10.1002/1520-636X(2001)13:1<34::AID-CHIR7>3.0.CO;2-O
- K. Isobe, K. Shoji, Y. Nakanishi, M. Yokoe and N. Wakao, “Purification and Some Properties of Cholesterol Oxidase Stable in Detergents from Gamma-Proteobacterium Y-134,” The Journal of Bioscience and Bioengineering, Vol. 95, No. 3, 2003, pp. 257-263.
- S. Dieth, D. Tritsch and J. F. Biellmann, “Resolution of Allylic Alcohols by Cholesterol Oxidase Isolated from Rhodococcus erythropolis,” Tetrahedron Letters, Vol. 36, No. 13, 1995, pp. 2243-2246. doi:10.1016/0040-4039(95)00235-5
- R. Aono and N. Doukyu, “Stereo Specific Oxidation of 3b-Hydroxysteroids by per Solvent Fermentation with Pseudomonas sp. ST-200,” Bioscience, Biotechnology, and Biochemistry, Vol. 60, No. 7, 1996, pp. 1146-1151. doi:10.1271/bbb.60.1146
- R. Aono, N. Doukyu, H. Kobayashi, H. Nakajima and K. Horikoshi, “Oxidative Bioconversion of Cholesterol by Pseudomonas sp. Strain ST-200 in a Water-Organic Solvent Two-Phase System,” Applied Environmental Microbiology, Vol. 60, No. 7, 1994, pp. 2518-2523.
- R. Z. Kazandjian, J. S. Durdich and A. M. Kilbanov, “Enzymatic Analysis in Organic Solvents,” Biotechnology and Bioengineering, Vol. 28, No. 3, 1986, pp. 417-421. doi:10.1002/bit.260280315
- M. Mewies, W. S. McIntire and N. S. Scrutton, “Covalent Attachment of Flavin Adenine Dinucleotide (FAD) and Flavin Mononucleotide (FMN) to Enzymes: The Current State of Affairs,” Protein Science, Vol. 7, No. 1, 1989, pp. 7-21.
- N. Lario, M. Fernandez, J. Aguero, B. G. Zorn, J. A. Vazquez-Boland and J. Navas, “Rapid Identification of Rhodococcus equi by a PCR Assay Targeting the choE Gene,” Journal of Clinical Microbiology, Vol. 41, No. 7, 2003, pp. 3241-3245. doi:10.1128/JCM.41.7.3241-3245.2003
- A. Vrielink, L. F. Lloyd and D. M. Blow, “Crystal Structure of Cholesterol Oxidase from Brevibacterium sterolicum Refined at 1.8 A Resolution,” Journal of Molecular Biology, Vol. 219, No. 3, 1991, pp. 533-554. doi:10.1016/0022-2836(91)90192-9
- Q. K. Yue, I. J. Kass, N. S. Sampson and A. Vrielink, “Crystal Structure Determination of Cholesterol Oxidase from Streptomyces and Structural Characterization of Key Active Site Mutants,” Biochemistry, Vol. 38, No. 14, 1999, pp. 4277-4286. doi:10.1021/bi982497j
- R. Coulombe, K. Q. Yue, S. Ghisla and A. Vrielink, “Oxygen Access to the Active Site of Cholesterol Oxidase through a Narrow Channel Is Gated by an Arg-Glu Pair,” The Journal of Biological Chemistry, Vol. 276, 2001, pp. 30435-30441. doi:10.1074/jbc.M104103200
- G. Gadda, G. Wels, L. Pollegioni, S. Zucchelli, D. Ambrosius, M. S. Pilone and S. Ghisla, “Characterization of Cholesterol Oxidase from Streptomyces hygroscopicus and Brevibacterium sterolicum,” European Journal of Biochemistry, Vol. 250, No. 2, 1997, pp. 369-376. doi:10.1111/j.1432-1033.1997.0369a.x
- L. Pollegioni, G. Wels, M. S. Pilone and S. Ghisla, “Kinetic Mechanism of Cholesterol Oxidase from Streptomyces hygroscopicus and Brevibacterium sterolicum,” European Journal of Biochemistry, Vol. 264, No. 1, 1999, pp. 140-151. doi:10.1046/j.1432-1327.1999.00586.x
- D. E. Edmondson and V. P. Newton, “The Covalent FAD of Monoamine Oxidase: Structural and Functional Role and Mechanism of the Flavinylation Reaction,” Antioxidants and Redox Signaling, Vol. 3, No. 5, 2010, pp. 789- 806. doi:10.1089/15230860152664984
- P. S. J. Cheetham, P. Dunnill and M. D. Lilly, “Extraction of Cholesterol Oxidase from Nocardia rhodochrous,” Enzyme and Microbial Technology, Vol. 2, No. 3, 1980, pp. 201-205. doi:10.1016/0141-0229(80)90047-2
- P. S. J. Cheetham, “The Identification, Properties, and Immobilization of the 3P-Hydroxysteroid Oxidase and 3-Oxosteroid A"-A'-Isomerase Components of cholesterol Oxidase from Nocardia rhodochrous,” Journal of Applied Biochemistry, Vol. 1, 1979, pp. 51-59.
- L. Motteran, M. S. Pilone, G. Molla, S. Ghisla and L. Pollegioni, “Cholesterol Oxidase from Brevibacterium sterolicum the Relationship between Covalent Flavinylation and Redox Properties,” The Journal of Biological Chemistry, Vol. 276, 2001, pp. 18024-18030. doi:10.1074/jbc.M010953200
- M. D. Lilly, P. S. J. Cheetham, D. J. Lewis, J. Yates and P. Dunnill, “Abstract in International Fermentation Symposium,” International Fermentation Symposium, Vol. 5, Berlin, 1976, p. 327.
- C. C. Allain, L. S. Poon, C. S. G. Chan, W. Richmond and P. C. Fu, “Enzymatic Determination of Total Serum Cholesterol,” Clinical Chemistry, Vol. 20, No. 4, 1974, pp. 470-475.
- P. Lolekha and Y. Jantaveesirirat, “Streptomyces, a Superior Sources of Cholesterol Oxidase Used in Serum Cholesterol Assay,” Journal of Clinical Laboratory Analysis, Vol. 6, No. 6, 1992, pp. 705-709. doi:10.1002/jcla.1860060612
- Y. Mitsuo, T. Mitsutoshi, O. Hisaya, F. Isao, H. Noriaki and M. Yoshikatsu, “Separation of Two Reactions, Oxidation and Isomerization Catalyzed by Streptomyces Cholesterol Oxidase,” Protein Engineering, Vol. 11, No. 11, 1998, pp. 1075-1081. doi:10.1093/protein/11.11.1075
- K. Dai and Y. Nihon, “Cholesterol Oxidase Properties and Application,” Nihon Yukagaka Gakkai, Vol. 48, 1999, pp. 3-14.
- J. T. Greenplate, D. R. Corbin and J. P. Purcell, “Cholesterol Oxidase Potent Boll Weevil Larvicidal & Ossalic Suitable for Transgenic Cotton Development,” Proceedings of Beltwide Cotton Conference, New Orleans, 7-10 January 1997, pp. 877-880.
- L. Y. A. Petrova, O. I. Glubkovskaya, G. M. Possukhina and A. A. Selezneva, “Study of the Conditions and Properties of Cholesterol Oxidase from Actinomyces lavendulae,” Biokhimiya, Vol. 46, 1981, pp. 1570-1575.
- G. A. Shirshova, L. N. Muntayen, T. S. Nazarova and L. E. Nikitin, “Comparative Study of Cholesterol Oxidase Biosynthesis of Two Strains of Streptomyces lavendulae,” Prikladnaya Biokhimiya i Microbiologiy, Vol. 28, 1992, pp. 711-715.
- P. Lorendano, G. Giovanni, A. Dorothea, G. Sandro and P. Mirella, “Cholesterol Oxidase from Streptomyces hygrocopicus and Brevibacterium Sterol Effect of Surfactants and Organic Solvents on Activity,” Biotechnology and Applied Biochemistry, Vol. 30, No. 1, 1999, pp. 27-33.
- M. Tabatabaei Yazdi, N. Kamranpour, M. Zahraei and K. Agaiepour, “Streptomyces fradiae, a Potent Producer of Cholesterol Oxidase,” Iranian Journal of Science and Technology Transaction, Vol. 10, No. 3, 1999, pp. 151-157.
- S. Lartillot and P. Kedziora, “Extracellular Cholesterol Oxidase from Streptomyces Strain,” Preperative Biochemistry and Biotechnology, Vol. 20, 1990, pp. 51-62.
- T. Kamei, Y. Takiguchi, H. Suzuki, M. Matsuzaki and S. Nakamura, “Purification and Properties of Streptomyces violascens Origin by Affinity Chromatography on Cholesterol,” Chemical and Pharmaceautical Bulletin, Vol. 26, No. 9, 1978, pp. 2799-2804. doi:10.1248/cpb.26.2799
- T. Uwajima, H. Yagi, S. Nakamura and O. Terada, “Isolation and Crystallization of Extracellular 3β-Hydroxysteroid Oxidase of Brevibacterium sterolicum Novel sp.,” Agricultural and Biological Chemistry, Vol. 37, No. 10, 1973, pp. 2345-2350. doi:10.1271/bbb1961.37.2345
- M. Yamashita, M. Toyama, H. Ono, I. Fujii, N. Hirayama and Y. Murooka, “Separation of the Two Reactions, Oxidation and Isomerization, Catalyzed by Streptomyces Cholesterol Oxidase,” Protein Engineering, Vol. 11, No. 11, 1998, pp. 1075-1081. doi:10.1093/protein/11.11.1075
- A. Elalami, J. Kerit, A. Filali-Maltouf, J. Boudrant and P. Germain, “Characterization of a Secreted Cholesterol Oxidase from Rhodococcus sp. GKI (CIP105 335),” World Journal of Microbiology and Biotechnology, Vol. 15, No. 5, 1999, pp. 579-585. doi:10.1023/A:1008958008333
- R. Linder and A. W. Bernheimer, “Oxidation of Macrophage Membrane Cholesterol by Intracellular Rhodococcus equi,” Veterinary Microbiology, Vol. 56, No. 3-4, 1997, pp. 269-276. doi:10.1016/S0378-1135(97)00095-3
- J. G. Terezinha, M. Salva1 Alcina Liserre1, L. Aloísia Moretto1, A. T. Marco Zullo, G. Ventrucci and J. B. Tobias Menezes, “Some Enzymatic Properties of Cholesterol Oxidase Produced by Brevibacterium sp.,” Revista de Microbiologia, Vol. 30, No. 4, 1999, pp. 315-323.
- N. Doukyu, K. Shibata, H. Ogino and M. Sagermann, “Cloning, Sequence Analysis, and Expression of a Gene Encoding Chromobacterium sp. DS-1 Cholesterol Oxidase,” Applied Microbiology and Biotechnology, Vol. 82, No. 3, 1989, pp. 479-490. doi:10.1007/s00253-008-1775-9
- Y. Lin, J. Fu and X. Song, “Purification and Characterization of an Extracellular Cholesterol Oxidase from a Bordetella Species,” Process Biochemistry, Vol. 45, No. 9, 2010, pp. 1563-1569. doi:10.1016/j.procbio.2010.06.005
- F. Volontè, L. Pollegioni, G. Molla, L. Frattini, F. Marinelli and L. Piubelli, “Production of Recombinant Cholesterol Oxidase Containing Covalently Bound FAD in Escherichia coli,” BMC Biotechnology, Vol. 10, 2010, p. 33. doi:10.1186/1472-6750-10-33
- Y. Xin, H. Yang, X. Xia, L. Zhang, C. Cheng, G. Mou, J. Shi, Y. Han and W. Wang, “Affinity Purification of a Cholesterol Oxidase Expressed in Escherichia coli,” Journal of Chromatography , Vol. 879, No. 13-14, 2011, pp. 853-858.
- H. Tomioka, M. Kagawa and S. Nakamura, “Some Enzymatic Properties of 3β-Hydroxysteroid Oxidase Produced by Streptomyces violascens,” Journal of Biochemistry, Vol. 79, No. 5, 1976, pp. 903-915.
- S. De Martinez and C. Green, “The Action of Cholesterol Oxidases on Cholesterol in Vesicles and Micelles,” Biochemical Society Transaction, Vol. 7, No. 5, 1979, pp. 978-979.
- M. T. Yazdi, M. Zahraei, K. Aghaepour and N. Kumaranpour, “Purification and Partial Characterization of Cholesterol Oxidase from Streptomyces fradiae,” Enzyme and Microbial Technology, Vol. 18, No. 4-5, 2001, pp. 410- 414. doi:10.1016/S0141-0229(00)00337-9
- M. T. Lee, W. C. Chen and C. C. Chou, “Medium Improvement by orthogonal designs for Cholesterol Oxidase Production by Rhodococcus equi No. 23,” Process Biochemistry, Vol. 32, No. 8, 1997, pp. 697-703. doi:10.1016/S0032-9592(97)00031-9
- Y. Sun, H. Yang and W. Wang, “Improvement of the Thermostability and Enzymatic Activity of Cholesterol Oxidase by Site-Directed Mutagenesis,” Biotechnology Letters, Vol. 33, No. 10, 2011, pp. 2049-2055. doi:10.1007/s10529-011-0669-6
- W. H. Liu, M. H. Meng and C. F. Huang, “Purification and Some Properties of Cholesterol Oxidase Produced by an Inducible and a Constitutive Mutant of Arthrobacter simplex,” Agricultural and Biological Chemistry, Vol. 52, No. 2, 1987, pp. 413-418. doi:10.1271/bbb1961.52.413
- S. J. Cheetham Peter, P. Dunnill and M. D. Lilly, “The Characterization and Interconversion of Three Forms of Cholesterol Oxidase Extracted from Nocardia rhodochrous,” The Journal of Biochemistry, Vol. 201, No. 3, 1982, pp. 515-521.
- K. Arima, M. Nagasawa, M. Bae and G. Tamura, “Microbial Transformation of Sterois Decomposition of Cholesterol by Microorganisms,” Agricutural and Biological Chemistry, Vol. 33, No. 11, 1969, pp. 1636-1643. doi:10.1271/bbb1961.33.1636
- M. Nagasawa, M. Bae, G. Tamura and K. Arima, “Microbial Transformation of Sterols. Part 11. Cleavage of Sterol Side Chains by Microorganisms,” Agricutural and Biological Chemistry, Vol. 33, No. 11, 1969, pp. 1644- 1650. doi:10.1271/bbb1961.33.1644
- K. Watanabe, H. Aihara, Y. Nakagawa, R. Nakamura and T. Sasaki, “Properties of the Purified Extracellular Cholesterol Oxidase from Rhodoccocus equi No 23,” Journal of Agricultural and Food Chemistry, Vol. 37, No. 4, 1989, pp. 1178-1182. doi:10.1021/jf00088a079
- C. J. Sih, H. H. Tai and. Y. Tsong, “The Mechanism of Microbial Conversion of Cholesterol into 17-Keto Steroids,” Journal of American Chemical Society, Vol. 89, No. 8, 1967, pp. 1957-1958. doi:10.1021/ja00984a039
- Z. Shen, D. R. Corbin, J. T. Greenplate, R. J. Grebenok, D. W. Galbraith and J. P. Purcell, “Studies on the Mode of Action of Cholesterol Oxidase on Insect Midgut Membranes,” Archives of Insect Biochemistry and Physiology, Vol. 34, No. 4, 1997, pp. 429-442. doi:10.1002/(SICI)1520-6327(1997)34:4<429::AID-ARCH3>3.0.CO;2-N
- J. MacLachlan, A. T. L. Wotherspoon, R. O. Ansell and C. J. W. Brooks, “Cholesterol Oxidase: Sources, Physical Properties and Analytical Applications,” The Journal of Steroid Biochemistry and Molecular Biology, Vol. 72, No. 5, 2000, pp. 169-195. doi:10.1016/S0960-0760(00)00044-3
- D. R. Corbin, R. J. Grebenok, T. E. Ohnmeiss, J. T. Greenplate and J. P. Purcell, “Expression and Chloroplast Targeting of Cholesterol Oxidase in Transgenic Tobacco Plants,” Plant Physiology, Vol. 126, No. 3, 2001, pp. 1116-1128. doi:10.1104/pp.126.3.1116
- J. Navas, B. Gonzalez-Zorn, N. Ladron, P. Garrido and J. A. Vazquez-Boland, “Identification and Mutagenesis by Allelic Exchange of choE, Encoding a Cholesterol Oxidase from the Intracellular Pathogen Rhodococcus equi,” Journal of Bacteriology, Vol. 183, No. 16, 2001, pp. 4796- 4805. doi:10.1128/JB.183.16.4796-4805.2001
- G. Gadda, G. Wels, L. Pollegioni, S. Zucchelli, D. Ambrosius, M. S. Pilone and S. Ghisla, “Characterization of Cholesterol Oxidase from Streptomyces hygroscopicus and Brevibacterium sterolicum,” European Journal of Biochemistry, Vol. 250, No. 2, 1997, pp. 369-376. doi:10.1111/j.1432-1033.1997.0369a.x
- National Institutes of Health, “Third Report of the Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults,” NIH Publication No. 1, National Institutes of Health, Bethesda, 2001, p. 3670.
- Y. Murooka, T. Ishizaki, O. Nimi and N. Maekawa, “Cloning and Expression of a Streptomyces Cholesterol Oxidase Gene in Streptomyces lividans with Plasmid pIJ702,” Applied and Environmental Microbiology, Vol. 52, No. 6, 1986, pp. 1382-1385.
- B. Li, W. Wang, F.-Q. Wang and D.-Z. Wei, “Cholesterol Oxidase ChoL Is a Critical Enzyme that Catalyzes the Conversion of Diosgenin to 4-Ene-3-Keto Steroids in Streptomyces virginiae IBL-14,” Applied Microbiology and Biotechnology, Vol. 85, No. 6, 2010, pp. 1831-1838. doi:10.1007/s00253-009-2188-0
- J. T. Greenplate, N. B. Duck, J. C. Pershing and J. P. Purcell, “Cholesterol Oxidase: An Oostatic and Larvicidal Agent Active against the Cotton Boll Weevil, Anthonomus grandis,” Entomologia Experimentalis et Applicata, Vol. 74, No. 3, 1995, pp. 253-258. doi:10.1111/j.1570-7458.1995.tb01898.x
- A. Brzostek, B. Dziadek, A. Rumijowska-Galewicz, J. Pawelczyk and J. Dziadek, “Cholesterol Oxidase Is Required for Virulence of Mycobacterium tuberculosis,” Federation of European Microbiological Societies Microbiology Letters, Vol. 275, No. 1, 2007, pp. 106-112.
- Z, Liao, L. M. Cimakasky, R. Hampton, D. H. Nguyen and J. E. K. Hildreth, “Lipid Raftes and HIV Pathogenesis: Host Membrane Cholesterol Is Required for Infection by HIV Type-I,” AIDS Research and Human Retrovirusis, Vol. 17, No. 11, 2004, pp. 1009-1019.
- A. Kumar, R. Malhotra, B. D. Malhotra and S. K. Grover, “CO-Immoblization of Cholesterol Oxidase and Horseradish Peroxidase in a Sol-Gel Film,” Analytica Chimica Acta, Vol. 414, No. 1-2, 2000, pp. 43-50. doi:10.1016/S0003-2670(00)00792-3
- R. C. Srivastava, R. Sahney, S. Upadhyay and R. L. Gupta, “Membrane Permeability Based Cholesterol Sensor—A New Possibility,” Journal of Membrane Science, Vol. 164, No. 1-2, 2000, pp. 45-49. doi:10.1016/S0376-7388(99)00201-X
- R. Foster, J. Cassidy and E. P. O’Donoghue, “Electrochemical Diagnostic Strip Device for Total Cholesterol and Its Subfractions,” Electroanalysis, Vol. 12, No. 9, 2000, pp. 716-721. doi:10.1002/1521-4109(200005)12:9<716::AID-ELAN716>3.0.CO;2-S
- S. Brahim, D. A. Nariresingh and G. Elie, “Amperometric Determination of Cholesterol in Serum Using a Biosensor of Cholesterol Oxidase Contained within a PolypyrroleHydrogel Membrane,” Analytica Chimica Acta, Vol. 448, No. 1-2, 2001, pp. 27-36. doi:10.1016/S0003-2670(01)01321-6
- C. Bongiovanni, T. Ferri, A. Poscia, M. Varalli, R. Santucci and A. Desideri, “An Electrochemical Multienzymatic Biosensor for Determination of Cholesterol,” Bioelectrochemistry, Vol. 54, No. 1, 2001, pp. 17-22. doi:10.1016/S0302-4598(01)00105-2
- T. Nakaminami, S. Ito, S. Kuwabata and H. Yoneyama, “Amperometric Determination of Total Cholesterol at Gold Electrode Conalently Modified with Cholesterol Oxidase and Cholesterol Esterase with Use of Thionin as an Electron Mediator, Analytical Chemistry, Vol. 71, No. 5, 1999, pp. 1068-1076. doi:10.1021/ac9805450
- T. Yao and K. Takashima, “Amperometric Biosensor with a Composite Membrane of Sol-Gel Derived Enzyme Film and Electrochemically Generated Poly(1,2-Diaminobenzene) Film,” Biosensors and Bioelectronics, Vol. 13, No. 1, 1998, pp. 67-73. doi:10.1016/S0956-5663(97)00076-6
- M. H. Ropers, R. Bilewicz, M. J. Stebe, A. Hamidi, A. Miclo and E. Rogalska, “Fluorinated and Hydrogenated Cubic Phases as Matrices for Immobilization of Cholesterol Oxidase on Electrodes,” Physical Chemistry Chemical Physics, Vol. 3, No. 2, 2001, pp. 240-245. doi:10.1039/b007718f
- A. Ono and E. O. Freed, “Plasma Membrane Rafts Play a Critical Role in HIV-1 Assembly and Release,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 98, No. 24, 2001, pp. 13925-13930. doi:10.1073/pnas.241320298
- S. Bavari, C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn and M. J. Aman, “Lipid Raft Micro Domains: A Gateway for Compartmentalized Trafficking of Ebola and Marburg Viruses,” Journal of Experimental Medicine, Vol. 195, No. 5, 2002, pp. 593-602. doi:10.1084/jem.20011500
- M. K. Ram, P. Bertoncello, H. Ding, S. Paddeu and S. C. Nicolini, “Cholesterol Biosensor Prepared by Layer-byLayer Technique,” Biosensors and Bioelectronic, Vol. 16, No. 9-12, 2001, pp. 849-856. doi:10.1016/S0956-5663(01)00208-1
- Q. C. Shi and Peng Tu Zhi, “A Novel Cholesterol Oxidase Biosensor Based on pt-Nanoparticle/Carbon Nanotube Modified Electrode,” Chinese Chemical Letters, Vol. 16, No. 8, 2005, pp. 1081-1084.
- J. C. Vidal, R. E. Garcia, J. Espuelas, T. Aramendia and J. R. Castillo, “Comparison of Biosensors Based on Entrapment of Cholesterol Oxidase and Cholesterol Esterase in Electropolymerized Films of Polypyrrole and Diaminonaphthalene Derivatives for Amperometric Determination of Cholesterol,” Analytical and Bioanalytical Chemistry, Vol. 377, No. 2, 2003, pp. 273-280. doi:10.1007/s00216-003-2120-x
- P. Norouzi, F. Faridbod, E. Nasli-Esfahani, B. Larijani and M. R. Ganjali, “Cholesterol Biosensor Based on MWCNTs-MnO2 Nanoparticles Using FFT Continuous Cyclic Voltammetry,” International Journal of Electrochemical Science, Vol. 5, 2010, pp. 1008-1017.
- M. V. Mendes, E. Recio, N. Antón, S. M. Guerra, J. Santos-Aberturas, J. F. Martín and J. F. Aparicio, “Cholesterol Oxidases Act as signalling proteins for the Biosynthesis of the Polyene Macrolide Pimaricin,” Chemistry & Biology, Vol. 14, No. 3, 2007, pp. 279-290. doi:10.1016/j.chembiol.2007.01.010
- P. Daneshgar, P. Norouzi, F. Dousty, M. R. Ganjali and A. A. Moosavi-Movahedi, “Dysprosium Hydroxide Nanowires Modified Electrode for Determination of Rifampicin Drug in Human Urine and Capsules by Adsorptive Square Wave Voltammetry,” Current Pharmaceutical Analysis, Vol. 5, No. 3, 2009, pp. 246-255. doi:10.2174/157341209788922066
- N. Doukyu, K. Shibata, H. Ogino and M. Sagermann, “Purification and Characterization of Chromobacterium sp. DS-1 Cholesterol Oxidase with Thermal, Organic Solvent, and Detergent Tolerance,” Applied Microbiology and Biotechnology, Vol. 80, No. 1, 2008, pp. 59-70. doi:10.1007/s00253-008-1526-y
- M. R. Pourjavid, P. Norouzi and M. R. Ganjali, “Light lanthanides Determination by Fast Fourier Transformcontinuous Cyclic Voltammetry after Separation by Ion Exchange Chromatography,” International Journal of Electrochemical Science, Vol. 4, 2009, pp. 923-942.
- A. G. Smith and C. J. Brooks, “The Substrate Specificity and Stereochemistry, Reversibility and Inhibition of the 3-Oxo Steroid Delta 4-Delta 5-Isomerase Component of Cholesterol Oxidase,” Biochemical Journal, Vol. 167, No. 1, 1977, pp. 121-129.
- G. Del Real, S. Jimenez-Baranda, R. A. Lacalle, E. Mira, P. Lucas, C. Gomez-Mouton, A. C. Carrera, A. C. Martinez and S. Manes, “Blocking of HIV-1 Infection by Targeting CD4 to Nonraft Membrane Domains,” Journal of Experimental Medicine, Vol. 196, No. 3, 2002, pp. 293- 301. doi:10.1084/jem.20020308
- M. Guyader, E. Kiyokawa, L. Abrami, P. Turelli and D. Trono, “Role for Human Immunodeficiency Virus Type 1 Membrane Cholesterol in Viral Internalization,” Journal of Virology, Vol. 76, No. 20, 2002, pp. 10356-10364. doi:10.1128/JVI.76.20.10356-10364.2002
- X. Sun and G. R. Whittaker, “Role for Influenza Virus Envelope Cholesterol in Virus Entry and Infection,” Journal of Virology, Vol. 77, No. 23, 2003, pp. 12543-12551. doi:10.1128/JVI.77.23.12543-12551.2003
- M. Takeda, G. P. Leser, C. J. Russell and R. A. Lamb, “Influenza Virus Hemagglutinin Concentrates in Lipid Raft Micro Domains for Efficient Viral Fusion,” Proceedings of the National Academy of Sciences of the United States of America, Vol. 100, No. 25, 2003, pp. 14610-14617. doi:10.1073/pnas.2235620100
- B. Subrata, Y. Shu-Rong, P. S. Blank and J. Zimmerberg, “Cholesterol Promotes Hemifusion and Pore Widening in Membrane Fusion Induced by Influenza Hemagglutinin,” The Journal of General Physiology, Vol. 131, No. 5, 2008, pp. 503-513. doi:10.1085/jgp.200709932
- F. Jesus Aparicio and J. F. Martın, “Microbial Cholesterol Oxidases: Bioconversion Enzymes or Signal Proteins,” Journal of Royal Society of Chemistry, Vol. 4, No. 8, 2008, pp. 804-809.
- L. Pollegioni, L. Piubelli and G. Molla, “Cholesterol Oxidase,” Biotechnology Journal, Vol. 276, 2009, pp. 6857- 6870.
- D. Ye, J. Lei, W. Li, F. Ge, K. Wu, W. Xu and B. Yong, “Purification and Characterization of Extracellular Cholesterol Oxidase from Enterobactor sp.,” World Journal of Microbiology and Biotechnology, Vol. 24, No. 10, 2008, pp. 2227-2233. doi:10.1007/s11274-008-9734-2
- L. Puglielli, L. Friedlich Avi, D. R. Setchell Kenneth, S. Nagano, C. Opazo, A. R. Cherny, K. J. Barnham, J. D. Wade, S. Melov, D. M. Kovacs and A. I. Bush, “Alzheimer Disease β-Amyloid Activity Mimics Cholesterol Oxidase,” Journal of Clinical Investigation, Vol. 115, No. 9, 2005, pp. 2556-2563. doi:10.1172/JCI23610
- Q. C. Shi and T. P. Zhi, “A Novel Cholesterol Oxidase Biosensor Based on pt-Nanoparticle/Carbon Nanotube Modified Electrode,” Chinese Chemical Letters, Vol. 16, No. 8, 2005, pp. 1081-1084.
- R. Gehlot, K. Sharma, M. Mathew and S. Kumbhat, “Surface Plasmon Resonance Based Biosensor for Label Free Detection of Cholesterol,” Indian Journal of Chemistry, Vol. 47, No. 12, 2008, pp. 1804-1808.
- F. Yıldırımoglu, F. Arslan, S. Cete and A. Yasar, “Preparation of a Polypyrrole-Polyvinylsulphonate Composite Film Biosensor for Determination of Cholesterol Based on Entrapment of Cholesterol Oxidase,” Sensor, Vol. 9, No. 8, 2009, pp. 6435-6445. doi:10.3390/s90806435
- S. Çete, A. Yaşar, F. Arslan, “An Amperometric Biosensor for Uric Acid Determination Prepared from Uricase Immobilized in Polypyrrole Film,” Artificial Cells, Blood Substitutes and Biotechnology, Vol. 34, No. 3, 2006, pp. 367-380. doi:10.1080/10731190600684116
- S. Cete, A. Yasar and F. Arslan, “An Amperometric Biosensor for Uric Acid Determination Prepared from Uricase Immobilized in Polypyrrole Film,” Artificial Cells, Blood Substitutes and Biotechnology, Vol. 35, No. 3, 2007, pp. 607-620.
- F. Arslan, “An Amperometric Biosensor for Uric Acid Determination Prepared from Uricase Immobilized in Polyaniline-Polypyrrole Film,” Sensors, Vol. 8, No. 9, 2008, pp. 5492-5500. doi:10.3390/s8095492
- T. Shimidzu, A. Ohtani and K. Honda, “Charge-Controllable Polypyrrole/Polyelectrolyte Composite Membranes. III: Electrochemical Deionization System Constructed by Anion-Exchangeable and Cation-Exchangeable Polypyrrole Electrodes,” Journal of Electroanalytical Chemistry, Vol. 251, No. 2, 1988, pp. 323-337. doi:10.1016/0022-0728(88)85193-3
- G. Li, Y. Wang and H. Xu, “A Hydrogen Peroxide Sensor Prepared by Electropolymerization of Pyrrole Based on Screen-Printed Carbon Paste Electrodes,” Sensors, Vol. 7, No. 3, 2007, pp. 239-250. doi:10.3390/s7030239
- M. L. Moraes, N. C. de Souza, O. H. Caio, F. Marystela, P. Ubirajara, F. Rodrigues, R. Jr. Antonio, Z. Valtencir and N. O. Osvaldo Jr., “Immobilization of Cholesterol Oxidase in LbL Films and Detection of Cholesterol Using ac Measurements,” Materials Science and Engineering, Vol. 29, No. 2, 2009, pp. 442-447.
- M. H. Yang, Y. Yang, H. F. Yang, G. Shen and R. Q. Yu, “Layer-by-Layer Self-Assembled Multilayer Films of Carbon Nanotubes and Platinum Nanoparticles with Polyelectrolyte for the Fabrication of Biosensors,” Biomaterials, Vol. 27, No. 2, 2006, pp. 246-255. doi:10.1016/j.biomaterials.2005.05.077
- M. M. Kalayil, S. Padmanabhan, I. G. Anantha and L. Kwang-Pill, “Silica-Polyaniline Based Bienzyme Cholesterol Biosensor: Fabrication and Characterization,” Electroanalysis, Vol. 20, No. 20, 2010, pp. 2467-2474.
- A. K. Chauhan, A. S. Survase, J. Kishenkumar and S. U. Annapure, “Medium Optimization by Orthogonal Array and Response Surface Methodology for Cholesterol Oxidase Production by Sterptomyces lavendulae NCIM 2499,” Journal of General and Applied Microbiology, Vol. 55, No. 3, 2009, pp. 171-180. doi:10.2323/jgam.55.171
- A. Ahmadalinezhad and A. Chen, “High-Performance Electrochemical Biosensor for the Detection of Total Cholesterol,” Biosensors and Bioelectronics, Vol. 26, No. 11, 2011, pp. 4508-4513. doi:10.1016/j.bios.2011.05.011
- J. Y. Yang, Y. Li, S. M. Chen and K. C. Lin, “Fabrication of a Cholesterol Biosensor Based on Cholesterol Oxidase and Multiwall Carbon Nanotube Hybrid Composites,” International Journal of Electrochemical Science, Vol. 6, 2011, pp. 2223-2234.
- N. Ruechaa, W. Siangprohb and O. Chailapakul, “A Fast and Highly Sensitive Detection of Cholesterol Using Polymer Microfluidic Devices and Amperometric System,” Talanta, Vol. 84, No. 5, 2011, pp. 1323-1328. doi:10.1016/j.talanta.2011.02.040
- C. Fang, J. He and Z. Chen, “A Disposable Amperometric Biosensor for Determining Total Cholesterol in Whole Blood,” Sensors and Actuators, Vol. 155, No. 2, 2011, pp. 545-550. doi:10.1016/j.snb.2011.01.005
- H. C. Ferraza, J. A. Guimaraesa, T. L. M. Alvesa and C. J. L. Constantino, “Monomolecular Films of Cholesterol Oxidase and S-Layer Proteins,” Applied Surface Science, Vol. 257, No. 15, 2011, pp. 6535-6539. doi:10.1016/j.apsusc.2011.01.143
NOTES
*Corresponding author.

